Retrospective Analysis of INRG Clinical and Genomic Factors for 605 Neuroblastomas in Japan: A Report from the Japan Children’s Cancer Group Neuroblastoma Committee (JCCG-JNBSG) †
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Data Variables
2.2. Genomic Profile
2.3. Survival Analyses
3. Results
3.1. Patient Characteristics
3.2. Prognostic Significance of Markers in 605 Patients in All INSS Stages
3.2.1. Prognostic Significance of Clinical Factors
3.2.2. Assessment of the Risk Score by Morgenstern et al. as a High-Risk Marker
3.2.3. Prognostic Significance of Genomic Factors 1p, 11q, and 17q
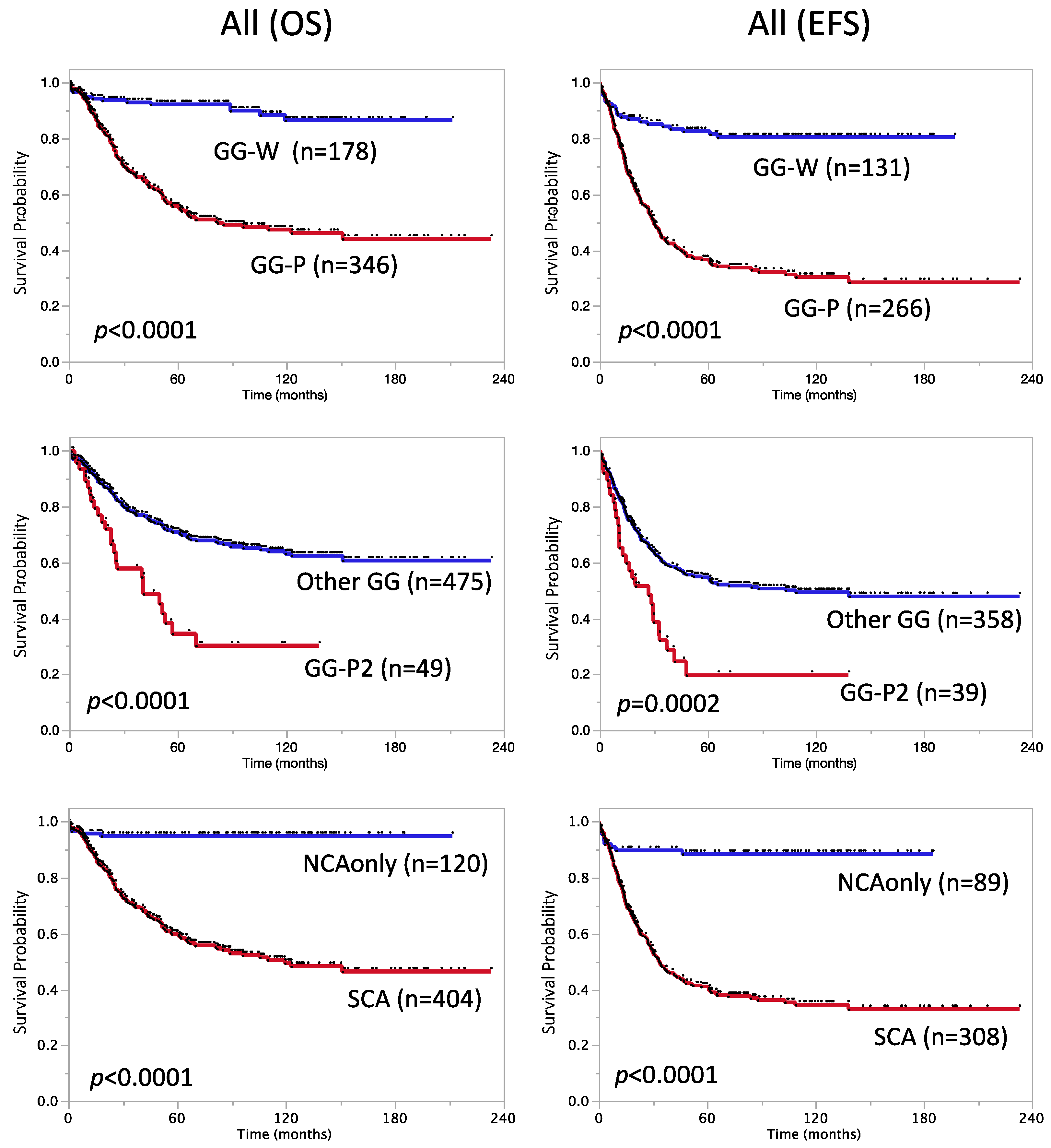
3.2.4. Prognostic Significance of SCA and Other Genome Subgroups
| Factor | N | 5-Year OS ± SE (%) | Log-Rank-p | N | 5-Year EFS ± SE (%) | Log-Rank-p |
|---|---|---|---|---|---|---|
| 1p loss | <0.0001 | <0.0001 | ||||
| Yes | 192 | 50 ± 3.9 | 151 | 36 ± 4.1 | ||
| No | 413 | 79 ± 2.2 | 299 | 62 ± 2.9 | ||
| 11q loss | <0.0001 | <0.0001 | ||||
| Yes | 191 | 57 ± 4.1 | 146 | 36 ± 4.3 | ||
| No | 414 | 75 ± 2.2 | 304 | 61 ± 2.9 | ||
| 17q gain | <0.0001 | <0.0001 | ||||
| Yes | 340 | 56 ± 3.0 | 262 | 37 ± 3.1 | ||
| No | 265 | 87 ± 2.2 | 188 | 76 ± 3.2 | ||
| Genome subgroup | <0.0001 | <0.0001 | ||||
| GG-P (Pa + Ps) | 346 | 56 ± 2.9 | 266 | 37 ± 3.1 | ||
| GG-W (Wa + Ws) | 178 | 92 ± 2.1 | 131 | 83 ± 3.4 | ||
| GG-P2 subgroup | <0.0001 | 0.0002 | ||||
| GG-P2 (P2a + P2s) | 49 | 34 ± 8.4 | 39 | 20 ± 7.6 | ||
| Other GG * | 475 | 71 ± 2.3 | 358 | 55 ± 2.7 | ||
| Genetic subtype | <0.0001 | <0.0001 | ||||
| NCA only | 120 | 95 ± 2.0 | 89 | 89 ± 3.4 | ||
| SCA (typSCA+atypSCA) | 404 | 60 ± 2.7 | 308 | 41 ± 2.9 | ||
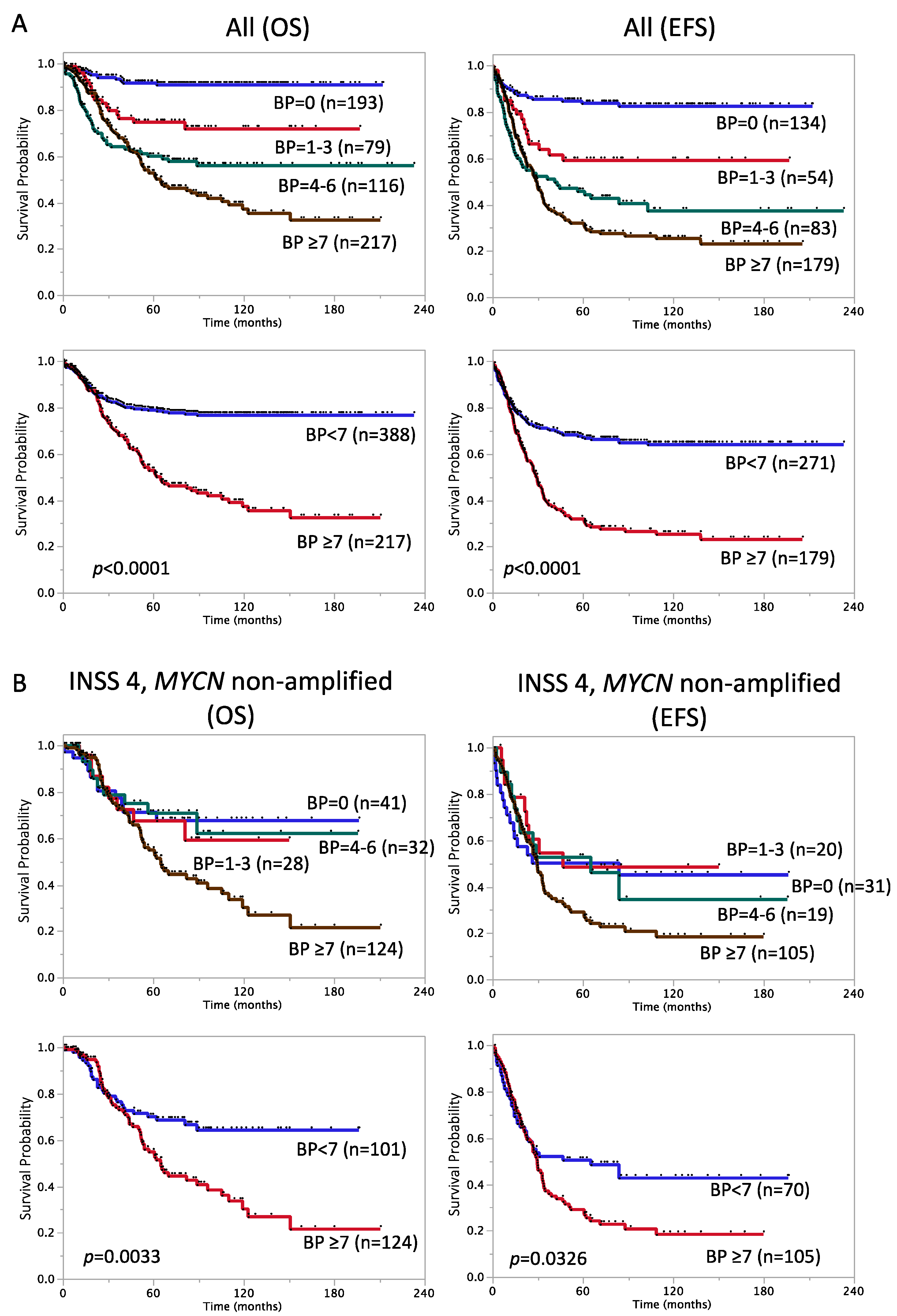
| Breakpoints | <0.0001 | <0.0001 | ||||
| <7 | 388 | 79 ± 2.2 | 271 | 67 ± 2.9 | ||
| ≥7 | 217 | 53 ± 3.8 | 179 | 32 ± 3.7 |
3.2.5. Prognostic Significance of the Number of BPs
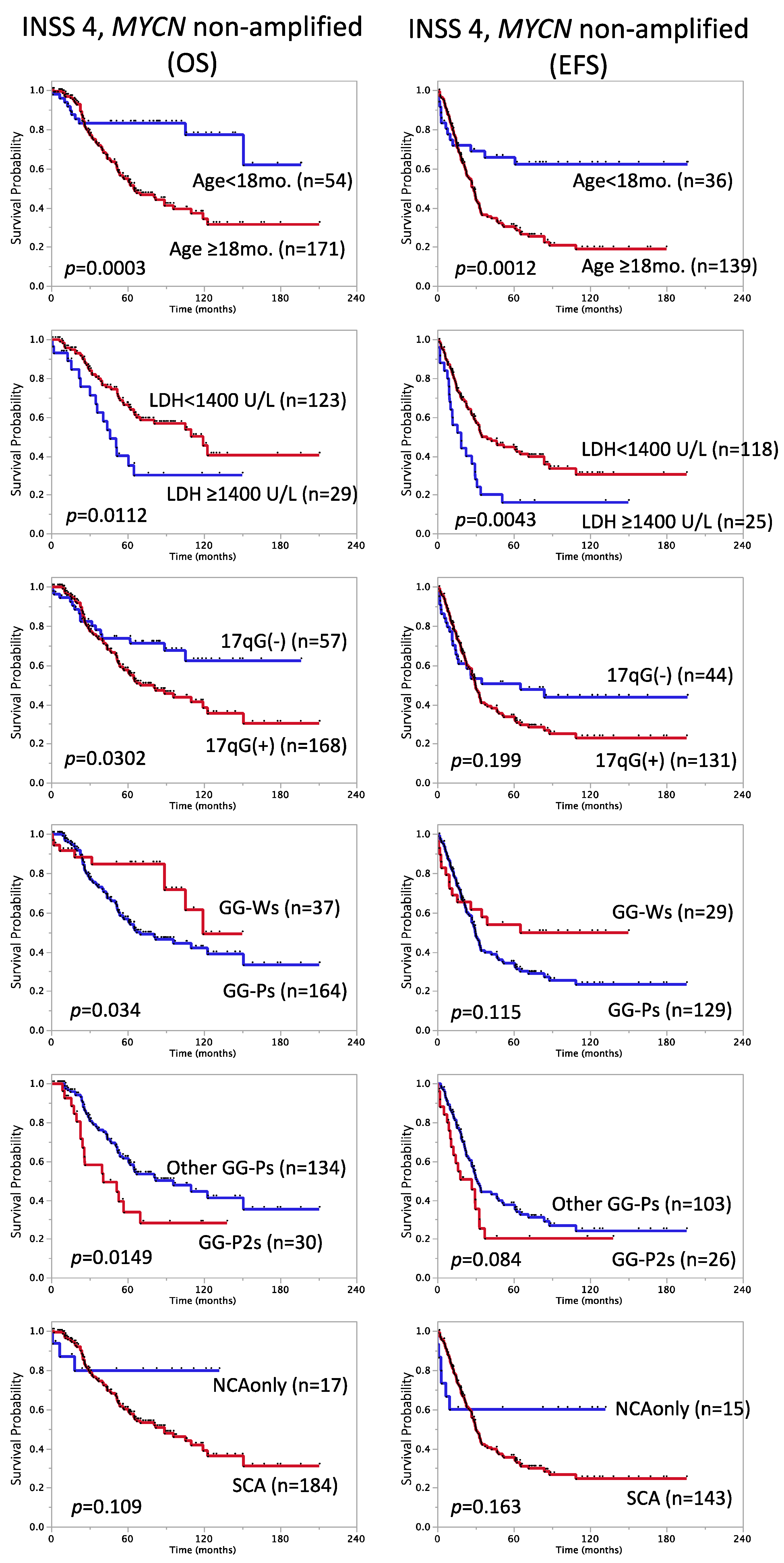
3.3. Factors with Prognostic Significance in Stage 4, MYCN Non-Amplified Cases
3.4. Multivariable Survival Analysis of Stage 4, MYCN Non-Amplified Cases
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brodeur, G.M. Neuroblastoma: Biological insights into a clinical enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef] [PubMed]
- Look, A.T.; Hayes, F.A.; Nitschke, R.; McWilliams, N.B.; Green, A.A. Cellular DNA content as a predictor of response to chemotherapy in infants with unresectable neuroblastoma. N. Engl. J. Med. 1984, 311, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Tomioka, N.; Kobayashi, H.; Kageyama, H.; Ohira, M.; Nakamura, Y.; Sasaki, F.; Todo, S.; Nakagawara, A.; Kaneko, Y. Chromosomes that show partial loss or gain in near-diploid tumors coincide with chromosomes that show whole loss or gain in near-triploid tumors: Evidence suggesting the involvement of the same genes in the tumorigenesis of high- and low-risk neuroblastomas. Genes Chromosomes Cancer 2003, 36, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Seeger, R.C.; Brodeur, G.M.; Sather, H.; Dalton, A.; Siegel, S.E.; Wong, K.Y.; Hammond, D. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 1985, 313, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Bown, N.; Cotterill, S.; Lastowska, M.; O’Neill, S.; Pearson, A.D.; Plantaz, D.; Meddeb, M.; Danglot, G.; Brinkschmidt, C.; Christiansen, H.; et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N. Engl. J. Med. 1999, 340, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Weiss, M.J.; Guo, C.; Gerbing, R.B.; Stram, D.O.; White, P.S.; Hogarty, M.D.; Sulman, E.P.; Thompson, P.M.; Lukens, J.N.; et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: A Children’s Cancer Group study. J. Clin. Oncol. 2000, 18, 1888–1899. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, E.F.; London, W.B.; Mosse, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Look, A.T.; Shimada, H.; et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005, 353, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Michon, J.; Huon, I.; d’Enghien, C.D.; Klijanienko, J.; Brisse, H.; Ribeiro, A.; Mosseri, V.; Rubie, H.; Munzer, C.; et al. Chromosomal CGH identifies patients with a higher risk of relapse in neuroblastoma without MYCN amplification. Br. J. Cancer 2007, 97, 238–246. [Google Scholar] [CrossRef]
- Mosse, Y.P.; Diskin, S.J.; Wasserman, N.; Rinaldi, K.; Attiyeh, E.F.; Cole, K.; Jagannathan, J.; Bhambhani, K.; Winter, C.; Maris, J.M. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer 2007, 46, 936–949. [Google Scholar] [CrossRef] [PubMed]
- Tomioka, N.; Oba, S.; Ohira, M.; Misra, A.; Fridlyand, J.; Ishii, S.; Nakamura, Y.; Isogai, E.; Hirata, T.; Yoshida, Y.; et al. Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene 2008, 27, 441–449. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Janoueix-Lerosey, I.; Schleiermacher, G.; Michels, E.; Mosseri, V.; Ribeiro, A.; Lequin, D.; Vermeulen, J.; Couturier, J.; Peuchmaur, M.; Valent, A.; et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J. Clin. Oncol. 2009, 27, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Ohira, M.; Nakagawara, A. Global genomic and RNA profiles for novel risk stratification of neuroblastoma. Cancer Sci. 2010, 101, 2295–2301. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Janoueix-Lerosey, I.; Ribeiro, A.; Klijanienko, J.; Couturier, J.; Pierron, G.; Mosseri, V.; Valent, A.; Auger, N.; Plantaz, D.; et al. Accumulation of segmental alterations determines progression in neuroblastoma. J. Clin. Oncol. 2010, 28, 3122–3130. [Google Scholar] [CrossRef]
- Schleiermacher, G.; Michon, J.; Ribeiro, A.; Pierron, G.; Mosseri, V.; Rubie, H.; Munzer, C.; Benard, J.; Auger, N.; Combaret, V.; et al. Segmental chromosomal alterations lead to a higher risk of relapse in infants with MYCN-non-amplified localised unresectable/disseminated neuroblastoma (a SIOPEN collaborative study). Br. J. Cancer 2011, 105, 1940–1948. [Google Scholar] [CrossRef]
- Schleiermacher, G.; Mosseri, V.; London, W.B.; Maris, J.M.; Brodeur, G.M.; Attiyeh, E.; Haber, M.; Khan, J.; Nakagawara, A.; Speleman, F.; et al. Segmental chromosomal alterations have prognostic impact in neuroblastoma: A report from the INRG project. Br. J. Cancer 2012, 107, 1418–1422. [Google Scholar] [CrossRef]
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Liang, W.H.; Federico, S.M.; London, W.B.; Naranjo, A.; Irwin, M.S.; Volchenboum, S.L.; Cohn, S.L. Tailoring Therapy for Children With Neuroblastoma on the Basis of Risk Group Classification: Past, Present, and Future. JCO Clin. Cancer Inform. 2020, 4, 895–905. [Google Scholar] [CrossRef]
- Ambros, P.F.; Ambros, I.M.; Brodeur, G.M.; Haber, M.; Khan, J.; Nakagawara, A.; Schleiermacher, G.; Speleman, F.; Spitz, R.; London, W.B.; et al. International consensus for neuroblastoma molecular diagnostics: Report from the International Neuroblastoma Risk Group (INRG) Biology Committee. Br. J. Cancer 2009, 100, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Oberthuer, A.; Hero, B.; Berthold, F.; Juraeva, D.; Faldum, A.; Kahlert, Y.; Asgharzadeh, S.; Seeger, R.; Scaruffi, P.; Tonini, G.P.; et al. Prognostic impact of gene expression-based classification for neuroblastoma. J. Clin. Oncol. 2010, 28, 3506–3515. [Google Scholar] [CrossRef] [PubMed]
- Oberthuer, A.; Juraeva, D.; Hero, B.; Volland, R.; Sterz, C.; Schmidt, R.; Faldum, A.; Kahlert, Y.; Engesser, A.; Asgharzadeh, S.; et al. Revised risk estimation and treatment stratification of low- and intermediate-risk neuroblastoma patients by integrating clinical and molecular prognostic markers. Clin. Cancer Res. 2015, 21, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, S.; Fischer, M. Telomere Maintenance in Pediatric Cancer. Int. J. Mol. Sci. 2019, 20, 5836. [Google Scholar] [CrossRef]
- Morgenstern, D.A.; Bagatell, R.; Cohn, S.L.; Hogarty, M.D.; Maris, J.M.; Moreno, L.; Park, J.R.; Pearson, A.D.; Schleiermacher, G.; Valteau-Couanet, D.; et al. The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr. Blood Cancer 2019, 66, e27556. [Google Scholar] [CrossRef]
- Morgenstern, D.A.; London, W.B.; Stephens, D.; Volchenboum, S.L.; Simon, T.; Nakagawara, A.; Shimada, H.; Schleiermacher, G.; Matthay, K.K.; Cohn, S.L.; et al. Prognostic significance of pattern and burden of metastatic disease in patients with stage 4 neuroblastoma: A study from the International Neuroblastoma Risk Group database. Eur. J. Cancer 2016, 65, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Morgenstern, D.A.; Potschger, U.; Moreno, L.; Papadakis, V.; Owens, C.; Ash, S.; Pasqualini, C.; Luksch, R.; Garaventa, A.; Canete, A.; et al. Risk stratification of high-risk metastatic neuroblastoma: A report from the HR-NBL-1/SIOPEN study. Pediatr. Blood Cancer 2018, 65, e27363. [Google Scholar] [CrossRef]
- Moroz, V.; Machin, D.; Hero, B.; Ladenstein, R.; Berthold, F.; Kao, P.; Obeng, Y.; Pearson, A.D.J.; Cohn, S.L.; London, W.B. The prognostic strength of serum LDH and serum ferritin in children with neuroblastoma: A report from the International Neuroblastoma Risk Group (INRG) project. Pediatr. Blood Cancer 2020, 67, e28359. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.; Guo, D.; Irwin, M.S.; Berthold, F.; Hogarty, M.; Kamijo, T.; Morgenstern, D.; Pasqualini, C.; Ash, S.; Potschger, U.; et al. A nomogram of clinical and biologic factors to predict survival in children newly diagnosed with high-risk neuroblastoma: An International Neuroblastoma Risk Group project. Pediatr. Blood Cancer 2021, 68, e28794. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Tsuchida, Y.; Uchino, J.; Takeda, T.; Iwafuchi, M.; Ohnuma, N.; Mugishima, H.; Yokoyama, J.; Nishihira, H.; Nakada, K.; et al. Treatment results of advanced neuroblastoma with the first Japanese study group protocol. Study Group of Japan for Treatment of Advanced Neuroblastoma. J. Pediatr. Hematol. Oncol. 1999, 21, 190–197. [Google Scholar] [CrossRef]
- Kaneko, M.; Tsuchida, Y.; Mugishima, H.; Ohnuma, N.; Yamamoto, K.; Kawa, K.; Iwafuchi, M.; Sawada, T.; Suita, S. Intensified chemotherapy increases the survival rates in patients with stage 4 neuroblastoma with MYCN amplification. J. Pediatr. Hematol. Oncol. 2002, 24, 613–621. [Google Scholar] [CrossRef]
- Hishiki, T.; Matsumoto, K.; Ohira, M.; Kamijo, T.; Shichino, H.; Kuroda, T.; Yoneda, A.; Soejima, T.; Nakazawa, A.; Takimoto, T.; et al. Results of a phase II trial for high-risk neuroblastoma treatment protocol JN-H-07: A report from the Japan Childhood Cancer Group Neuroblastoma Committee (JNBSG). Int. J. Clin. Oncol. 2018, 23, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Iehara, T.; Hosoi, H.; Akazawa, K.; Matsumoto, Y.; Yamamoto, K.; Suita, S.; Tajiri, T.; Kusafuka, T.; Hiyama, E.; Kaneko, M.; et al. MYCN gene amplification is a powerful prognostic factor even in infantile neuroblastoma detected by mass screening. Br. J. Cancer 2006, 94, 1510–1515. [Google Scholar] [CrossRef]
- Iehara, T.; Hiyama, E.; Tajiri, T.; Yoneda, A.; Hamazaki, M.; Fukuzawa, M.; Hosoi, H.; Sugimoto, T.; Sawada, T. Is the prognosis of stage 4s neuroblastoma in patients 12 months of age and older really excellent? Eur. J. Cancer 2012, 48, 1707–1712. [Google Scholar] [CrossRef] [PubMed]
- Iehara, T.; Hamazaki, M.; Tajiri, T.; Kawano, Y.; Kaneko, M.; Ikeda, H.; Hosoi, H.; Sugimoto, T.; Sawada, T.; Japanese Infantile Neuroblastoma Cooperative Study, G. Successful treatment of infants with localized neuroblastoma based on their MYCN status. Int. J. Clin. Oncol. 2013, 18, 389–395. [Google Scholar] [CrossRef]
- Chen, Y.; Takita, J.; Choi, Y.L.; Kato, M.; Ohira, M.; Sanada, M.; Wang, L.; Soda, M.; Kikuchi, A.; Igarashi, T.; et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 2008, 455, 971–974. [Google Scholar] [CrossRef]
- Depuydt, P.; Boeva, V.; Hocking, T.D.; Cannoodt, R.; Ambros, I.M.; Ambros, P.F.; Asgharzadeh, S.; Attiyeh, E.F.; Combaret, V.; Defferrari, R.; et al. Genomic Amplifications and Distal 6q Loss: Novel Markers for Poor Survival in High-risk Neuroblastoma Patients. J. Natl. Cancer Inst. 2018, 110, 1084–1093. [Google Scholar] [CrossRef]
- Hiyama, E.; Iehara, T.; Sugimoto, T.; Fukuzawa, M.; Hayashi, Y.; Sasaki, F.; Sugiyama, M.; Kondo, S.; Yoneda, A.; Yamaoka, H.; et al. Effectiveness of screening for neuroblastoma at 6 months of age: A retrospective population-based cohort study. Lancet 2008, 371, 1173–1180. [Google Scholar] [CrossRef]
- Oldridge, D.A.; Truong, B.; Russ, D.; DuBois, S.G.; Vaksman, Z.; Mosse, Y.P.; Diskin, S.J.; Maris, J.M.; Matthay, K.K. Differences in Genomic Profiles and Outcomes Between Thoracic and Adrenal Neuroblastoma. J. Natl. Cancer Inst. 2019, 111, 1192–1201. [Google Scholar] [CrossRef]
- Monclair, T.; Brodeur, G.M.; Ambros, P.F.; Brisse, H.J.; Cecchetto, G.; Holmes, K.; Kaneko, M.; London, W.B.; Matthay, K.K.; Nuchtern, J.G.; et al. The International Neuroblastoma Risk Group (INRG) staging system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 298–303. [Google Scholar] [CrossRef]
- Uryu, K.; Nishimura, R.; Kataoka, K.; Sato, Y.; Nakazawa, A.; Suzuki, H.; Yoshida, K.; Seki, M.; Hiwatari, M.; Isobe, T.; et al. Identification of the genetic and clinical characteristics of neuroblastomas using genome-wide analysis. Oncotarget 2017, 8, 107513–107529. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Zhang, J.; Lu, C.; Parker, M.; Bahrami, A.; Tickoo, S.K.; Heguy, A.; Pappo, A.S.; Federico, S.; Dalton, J.; et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 2012, 307, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Eleveld, T.F.; Oldridge, D.A.; Bernard, V.; Koster, J.; Colmet Daage, L.; Diskin, S.J.; Schild, L.; Bentahar, N.B.; Bellini, A.; Chicard, M.; et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 2015, 47, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ohira, M.; Zhou, Y.; Xiong, T.; Luo, W.; Yang, C.; Li, X.; Gao, Z.; Zhou, R.; Nakamura, Y.; et al. Genomic analysis-integrated whole-exome sequencing of neuroblastomas identifies genetic mutations in axon guidance pathway. Oncotarget 2017, 8, 56684–56697. [Google Scholar] [CrossRef] [PubMed]
| Factor | N | 5-Year OS ± SE (%) | Log-Rank-p | N | 5-Year EFS ± SE (%) | Log-Rank-p |
|---|---|---|---|---|---|---|
| Overall cohort | 605 | 70 ± 2.0 | 450 | 53 ± 2.5 | ||
| INSS stage | <0.0001 | <0.0001 | ||||
| stage 1,2,3,4s | 264 | 89 ± 2.0 | 185 | 79 ± 3.1 | ||
| stage 4 | 341 | 54 ± 3.0 | 265 | 35 ± 3.1 | ||
| Age at diagnosis | <0.0001 | <0.0001 | ||||
| <18 months | 254 | 81 ± 2.5 | 172 | 69 ± 3.6 | ||
| ≥18 months | 351 | 62 ± 2.8 | 278 | 43 ± 3.1 | ||
| MYCN amplification | <0.0001 | <0.0001 | ||||
| Not amplified (<10 copies) | 459 | 77 ± 2.2 | 337 | 60 ± 2.8 | ||
| Amplified (≥10 copies) | 146 | 47 ± 4.4 | 113 | 34 ± 4.6 | ||
| DNA ploidy | <0.0001 | <0.0001 | ||||
| Hyperdiploidy | 187 | 87 ± 2.7 | 138 | 75 ± 3.9 | ||
| Diploidy | 276 | 62 ± 3.2 | 196 | 43 ± 3.7 | ||
| Ferritin | 0.0003 | <0.0001 | ||||
| <250 ng/mL | 215 | 75 ± 3.2 | 195 | 62 ± 3.6 | ||
| ≥250 ng/mL | 94 | 56 ± 5.8 | 87 | 40 ± 5.7 | ||
| LDH | <0.0001 | <0.0001 | ||||
| <1400 U/L | 288 | 79 ± 2.6 | 274 | 65 ± 3.0 | ||
| ≥1400 U/L | 109 | 43 ± 5.2 | 96 | 29 ± 4.8 | ||
| Primary site of tumor | ||||||
| Adrenal | 0.0003 | 0.0032 | ||||
| No | 237 | 78 ± 2.9 | 174 | 63 ± 3.8 | ||
| Yes | 363 | 65 ± 2.7 | 273 | 47 ± 3.2 | ||
| Thorax | 0.0002 | 0.0017 | ||||
| No | 531 | 67 ± 2.2 | 398 | 50 ± 2.6 | ||
| Yes | 69 | 90 ± 3.8 | 49 | 78 ± 6.1 | ||
| Metastatic site (MET) | ||||||
| MET_Bone marrow | <0.0001 | <0.0001 | ||||
| No | 231 | 81 ± 2.7 | 218 | 72 ± 3.2 | ||
| Yes | 203 | 53 ± 3.9 | 188 | 34 ± 3.7 | ||
| MET_Bone | <0.0001 | <0.0001 | ||||
| No | 233 | 82 ± 2.7 | 221 | 71 ± 3.2 | ||
| Yes | 196 | 53 ± 3.9 | 181 | 35 ± 3.7 | ||
| MET_DLN | 0.0002 | <0.0001 | ||||
| No | 274 | 75± 2.8 | 262 | 63 ± 3.1 | ||
| Yes | 148 | 58 ± 4.4 | 134 | 39 ± 4.4 | ||
| MET_Liver | 0.0151 | 0.0653 | ||||
| No | 343 | 70 ± 2.7 | 320 | 55 ± 2.9 | ||
| Yes | 90 | 60 ± 5.4 | 83 | 50 ± 5.6 | ||
| MET_Lung | 0.0368 | 0.0314 | ||||
| No | 416 | 69 ± 2.4 | 388 | 55 ± 2.6 | ||
| Yes | 16 | 44 ± 13.3 | 15 | 33 ± 12.2 | ||
| Histological classification (INPC) | <0.0001 | <0.0001 | ||||
| Favorable | 131 | 84 ± 3.3 | 122 | 78 ± 3.9 | ||
| Unfavorable | 189 | 59 ± 4.0 | 177 | 40 ± 3.9 | ||
| Diagnosis (INPC) | 0.0188 | 0.0872 | ||||
| NB, GNB nodular | 316 | 68 ± 2.9 | 296 | 53 ± 3.1 | ||
| Others * | 28 | 92 ± 5.3 | 24 | 79 ± 8.6 | ||
| MKI (INPC) | 0.0015 | 0.0034 | ||||
| Low or Intermediate | 173 | 77 ± 3.6 | 164 | 63 ± 4.0 | ||
| High | 54 | 57 ± 7.4 | 51 | 38 ± 7.5 |
| Factor | N | 5-Year OS ± SE (%) | Log-Rank-p | HR | (95% CI) | N | 5-Year EFS ± SE (%) | Log-Rank-p | HR | (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|
| Stage 4, MYCN non-amplified | 225 | 62 ± 3.6 | 175 | 38 ± 3.9 | ||||||
| Age at diagnosis | 0.0003 | 3.2 | 1.7–6.5 | 0.0012 | 2.5 | 1.5–4.8 | ||||
| <18 months * | 54 | 83 ± 5.4 | 36 | 66 ± 8.1 | ||||||
| ≥18 months | 171 | 55 ± 4.3 | 139 | 30 ± 4.2 | ||||||
| DNA ploidy | 0.545 | 1.2 | 0.7–2.2 | 0.188 | 1.4 | 0.9–2.4 | ||||
| Hyperdiploidy * | 47 | 71 ± 7.4 | 40 | 45 ± 8.7 | ||||||
| Diploidy | 117 | 61 ± 5.1 | 87 | 39 ± 5.5 | ||||||
| Ferritin | 0.128 | 1.6 | 0.9–2.9 | 0.0471 | 1.6 | 1.0–2.6 | ||||
| <250 ng/mL * | 70 | 68 ± 6.2 | 64 | 47 ± 6.5 | ||||||
| ≥250 ng/mL | 55 | 57 ± 8.1 | 51 | 33 ± 7.5 | ||||||
| LDH | 0.0112 | 2.1 | 1.1–3.6 | 0.0043 | 2.0 | 1.2–3.2 | ||||
| <1400 U/L * | 123 | 66 ± 4.8 | 118 | 45 ± 4.9 | ||||||
| ≥1400 U/L | 29 | 40 ± 10.3 | 25 | 16 ± 7.3 | ||||||
| 1p loss | 0.07 | 1.5 | 0.9–2.4 | 0.203 | 1.3 | 0.8–2.0 | ||||
| Yes | 49 | 51 ± 8.0 | 43 | 29 ± 7.4 | ||||||
| No * | 176 | 65 ± 4.0 | 132 | 41 ± 4.5 | ||||||
| 11q loss | 0.0617 | 1.5 | 1.0–2.5 | 0.531 | 1.1 | 0.8–1.7 | ||||
| Yes | 141 | 57 ± 4.7 | 110 | 35 ± 4.9 | ||||||
| No * | 84 | 70 ± 5.5 | 65 | 42 ± 6.3 | ||||||
| 17q gain | 0.0302 | 1.8 | 1.1–3.2 | 0.199 | 1.4 | 0.9–2.2 | ||||
| Yes | 168 | 58 ± 4.3 | 131 | 34 ± 4.4 | ||||||
| No * | 57 | 74 ± 6.3 | 44 | 51 ± 7.8 | ||||||
| Genome Group | 0.034 | 2.1 | 1.1–4.5 | 0.115 | 1.6 | 0.9–2.9 | ||||
| GG-Ps | 164 | 57 ± 4.4 | 129 | 34 ± 4.4 | ||||||
| GG-Ws * | 37 | 85 ± 6.3 | 29 | 54 ± 9.5 | ||||||
| GG-P2s subgroup | 0.0149 | 2.0 | 1.1–3.4 | 0.084 | 1.6 | 0.9–2.6 | ||||
| GG-P2s | 30 | 34 ± 10.3 | 26 | 20 ± 8.8 | ||||||
| Other GG * | 134 | 62 ± 4.7 | 103 | 38 ± 5.0 | ||||||
| Genetic subtype | 0.109 | 2.5 | 0.9–10.2 | 0.163 | 1.8 | 0.9–4.6 | ||||
| NCA only * | 17 | 80 ± 10.5 | 15 | 60 ± 12.7 | ||||||
| SCA (typSCA+atypSCA) | 184 | 60 ± 4.1 | 143 | 36 ± 4.2 | ||||||
| Breakpoints | 0.0033 | 1.9 | 1.2–3.1 | 0.0326 | 1.5 | 1.0–2.3 | ||||
| <7 * | 101 | 70 ± 5.0 | 70 | 50 ± 6.2 | ||||||
| ≥7 | 124 | 55 ± 5.1 | 105 | 29 ± 4.8 |
| Combination of Variables | OS | EFS | ||||||
|---|---|---|---|---|---|---|---|---|
| N | HR | (95% CI) | p-Value | N | HR | (95% CI) | p-Value | |
| Age ≥ 18 months | 152 | 1.9 | 0.9–4.3 | 0.0789 | 143 | 2.4 | 1.3–5.0 | 0.004 |
| LDH ≥ 1400 U/L | 1.9 | 1.1–3.4 | 0.0323 | 1.9 | 1.1–3.0 | 0.0194 | ||
| Age ≥ 18 months | 164 | 2.5 | 1.1–7.1 | 0.0291 | 129 | 2.0 | 0.9–5.1 | 0.0799 |
| GG-P2s subgroup | 1.9 | 1.1–3.3 | 0.0325 | 1.6 | 0.9–2.7 | 0.0838 | ||
| Age ≥ 18 months | 225 | 2.7 | 1.4–5.6 | 0.0023 | 175 | 2.3 | 1.3–4.5 | 0.0035 |
| Breakpoints ≥ 7 | 1.5 | 0.9–2.4 | 0.0919 | 1.2 | 0.8–1.9 | 0.3361 | ||
| LDH ≥ 1400 U/L | 152 | 2.2 | 1.2–3.8 | 0.0138 | 143 | 2.1 | 1.3–3.4 | 0.006 |
| Breakpoints ≥ 7 | 2.0 | 1.2–3.6 | 0.0098 | 1.7 | 1.1–2.7 | 0.0181 | ||
| Age ≥ 18 months | 152 | 1.4 | 0.6–3.3 | 0.4604 | 143 | 2.0 | 1.0–4.4 | 0.0396 |
| LDH ≥ 1400 U/L | 2.1 | 1.1–3.7 | 0.0184 | 2.0 | 1.2–3.2 | 0.0131 | ||
| Breakpoints ≥ 7 | 1.8 | 1.0–3.5 | 0.0422 | 1.4 | 0.8–2.3 | 0.2123 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohira, M.; Nakamura, Y.; Takimoto, T.; Nakazawa, A.; Hishiki, T.; Matsumoto, K.; Shichino, H.; Iehara, T.; Nagase, H.; Fukushima, T.; et al. Retrospective Analysis of INRG Clinical and Genomic Factors for 605 Neuroblastomas in Japan: A Report from the Japan Children’s Cancer Group Neuroblastoma Committee (JCCG-JNBSG). Biomolecules 2022, 12, 18. https://doi.org/10.3390/biom12010018
Ohira M, Nakamura Y, Takimoto T, Nakazawa A, Hishiki T, Matsumoto K, Shichino H, Iehara T, Nagase H, Fukushima T, et al. Retrospective Analysis of INRG Clinical and Genomic Factors for 605 Neuroblastomas in Japan: A Report from the Japan Children’s Cancer Group Neuroblastoma Committee (JCCG-JNBSG). Biomolecules. 2022; 12(1):18. https://doi.org/10.3390/biom12010018
Chicago/Turabian StyleOhira, Miki, Yohko Nakamura, Tetsuya Takimoto, Atsuko Nakazawa, Tomoro Hishiki, Kimikazu Matsumoto, Hiroyuki Shichino, Tomoko Iehara, Hiroki Nagase, Takashi Fukushima, and et al. 2022. "Retrospective Analysis of INRG Clinical and Genomic Factors for 605 Neuroblastomas in Japan: A Report from the Japan Children’s Cancer Group Neuroblastoma Committee (JCCG-JNBSG)" Biomolecules 12, no. 1: 18. https://doi.org/10.3390/biom12010018
APA StyleOhira, M., Nakamura, Y., Takimoto, T., Nakazawa, A., Hishiki, T., Matsumoto, K., Shichino, H., Iehara, T., Nagase, H., Fukushima, T., Yoneda, A., Tajiri, T., Nakagawara, A., & Kamijo, T. (2022). Retrospective Analysis of INRG Clinical and Genomic Factors for 605 Neuroblastomas in Japan: A Report from the Japan Children’s Cancer Group Neuroblastoma Committee (JCCG-JNBSG). Biomolecules, 12(1), 18. https://doi.org/10.3390/biom12010018







