Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Processing
2.2. IHC Intensity Scoring
2.3. Digital Image Analysis
2.4. Comparison between Semi-Quantitative Scoring and Digital Image Analysis
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Capobianco, E.; Iacoviello, L.; de Gaetano, G.; Donati, M.B. Editorial: Trends in Digital Medicine. Front. Med. 2020, 7, 116. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Kayser, K.; GĂśrtler, J.; Bogovac, M.; Bogovac, A.; Goldmann, T.; Vollmer, E.; Kayser, G. AI (Artificial Intelligence) in Histopathology--from Image Analysis to Automated Diagnosis. Folia Histochem. Cytobiol. 2010, 47, 355–361. [Google Scholar] [CrossRef][Green Version]
- Csonka, T.; Murnyák, B.; Szepesi, R.; Bencze, J.; Bognár, L.; Klekner, Á.; Hortobágyi, T. Assessment of Candidate Immunohistochemical Prognostic Markers of Meningioma Recurrence. Folia Neuropathol. 2016, 54, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Hortobágyi, T.; Bencze, J.; Varkoly, G.; Kouhsari, M.C.; Klekner, Á. Meningioma Recurrence. Open Med. 2016, 11, 168–173. [Google Scholar] [CrossRef]
- Bencze, J.; Szarka, M.; Bencs, V.; Szabó, R.N.; Módis, L.V.; Aarsland, D.; Hortobágyi, T. Hortobágyi Lemur Tyrosine Kinase 2 (LMTK2) Level Inversely Correlates with Phospho-Tau in Neuropathological Stages of Alzheimer’s Disease. Brain Sci. 2020, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.; Yue, W. Semi-Quantitative Determination of Protein Expression Using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio-Protocol 2019, 9, e3465. [Google Scholar] [CrossRef]
- Hanna, W.; O’Malley, F.P.; Barnes, P.; Berendt, R.; Gaboury, L.; Magliocco, A.; Pettigrew, N.; Robertson, S.; Sengupta, S.; Têtu, B.; et al. Updated Recommendations from the Canadian National Consensus Meeting on HER2/Neu Testing in Breast Cancer. Curr. Oncol. 2007, 14, 149–153. [Google Scholar] [CrossRef]
- Attems, J.; Toledo, J.B.; Walker, L.; Gelpi, E.; Gentleman, S.; Halliday, G.; Hortobagyi, T.; Jellinger, K.; Kovacs, G.G.; Lee, E.B.; et al. Neuropathological Consensus Criteria for the Evaluation of Lewy Pathology in Post-Mortem Brains: A Multi-Centre Study. Acta Neuropathol. 2021, 141, 159–172. [Google Scholar] [CrossRef]
- Kovacs, G.G.; Xie, S.X.; Lee, E.B.; Robinson, J.L.; Caswell, C.; Irwin, D.J.; Toledo, J.B.; Johnson, V.E.; Smith, D.H.; Alafuzoff, I.; et al. Multisite Assessment of Aging-Related Tau Astrogliopathy (ARTAG). J. Neuropathol. Exp. Neurol. 2017, 76, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Alafuzoff, I.; Thal, D.R.; Arzberger, T.; Bogdanovic, N.; Al-Sarraj, S.; Bodi, I.; Boluda, S.; Bugiani, O.; Duyckaerts, C.; Gelpi, E.; et al. Assessment of β-Amyloid Deposits in Human Brain: A Study of the BrainNet Europe Consortium. Acta Neuropathol. 2009, 117, 309–320. [Google Scholar] [CrossRef]
- Módis, L.V.; Varkoly, G.; Bencze, J.; Hortobágyi, T.G.; Módis, L.; Hortobágyi, T. Extracellular Matrix Changes in Corneal Opacification Vary Depending on Etiology. Mol. Vis. 2021, 27, 26–36. [Google Scholar]
- Walker, R.A. Quantification of Immunohistochemistry—Issues Concerning Methods, Utility and Semiquantitative Assessment I. Histopathology 2006, 49, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Bencze, J.; Szarka, M.; Bencs, V.; Szabó, R.N.; Smajda, M.; Aarsland, D.; Hortobágyi, T. Neuropathological Characterization of Lemur Tyrosine Kinase 2 (LMTK2) in Alzheimer’s Disease and Neocortical Lewy Body Disease. Sci. Rep. 2019, 9, 17222. [Google Scholar] [CrossRef] [PubMed]
- Skogseth, R.; Hortobágyi, T.; Soennesyn, H.; Chwiszczuk, L.; Ffytche, D.; Rongve, A.; Ballard, C.; Aarsland, D. Accuracy of Clinical Diagnosis of Dementia with Lewy Bodies versus Neuropathology. J. Alzheimer’s Dis. 2017, 59, 1139–1152. [Google Scholar] [CrossRef]
- Vitrolink. An Online Digital Image Analysis Platform. Available online: https://vitrolink.com/#/products (accessed on 12 December 2021).
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollar, P. Focal Loss for Dense Object Detection. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 42, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Beck, A.P. Fundamental Concepts for Semiquantitative Tissue Scoring in Translational Research. ILAR J. 2018, 59, 13–17. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Beck, A.P. Principles and Approaches for Reproducible Scoring of Tissue Stains in Research. Lab. Investig. 2018, 98, 844–855. [Google Scholar] [CrossRef]
- Gavrielides, M.A.; Gallas, B.D.; Lenz, P.; Badano, A.; Hewitt, S.M. Observer Variability in the Interpretation of HER2/Neu Immunohistochemical Expression With Unaided and Computer-Aided Digital Microscopy. Arch. Pathol. Lab. Med. 2011, 135, 233–242. [Google Scholar] [CrossRef]
- Cross, S.S.; Betmouni, S.; Burton, J.L.; Dubé, A.K.; Feeley, K.M.; Holbrook, M.R.; Landers, R.J.; Lumb, P.B.; Stephenson, T.J. What Levels of Agreement Can Be Expected Between Histopathologists Assigning Cases to Discrete Nominal Categories? A Study of the Diagnosis of Hyperplastic and Adenomatous Colorectal Polyps. Mod. Pathol. 2000, 13, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Aeffner, F.; Wilson, K.; Martin, N.T.; Black, J.C.; Hendriks, C.L.L.; Bolon, B.; Rudmann, D.G.; Gianani, R.; Koegler, S.R.; Krueger, J.; et al. The Gold Standard Paradox in Digital Image Analysis: Manual Versus Automated Scoring as Ground Truth. Arch. Pathol. Lab. Med. 2017, 141, 1267–1275. [Google Scholar] [CrossRef]
- Rizzardi, A.E.; Johnson, A.T.; Vogel, R.I.; Pambuccian, S.E.; Henriksen, J.; Skubitz, A.P.; Metzger, G.J.; Schmechel, S.C. Quantitative Comparison of Immunohistochemical Staining Measured by Digital Image Analysis versus Pathologist Visual Scoring. Diagn. Pathol. 2012, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Jasani, B.; Bänfer, G.; Fish, R.; Waelput, W.; Sucaet, Y.; Barker, C.; Whiteley, J.L.; Walker, J.; Hovelinck, R.; Diezko, R. Evaluation of an Online Training Tool for Scoring Programmed Cell Death Ligand-1 (PD-L1) Diagnostic Tests for Lung Cancer. Diagn. Pathol. 2020, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.M.B.; Castles, B.; Byrne, D.J.; Button, P.; Hendry, S.; Lakhani, S.R.; Sivasubramaniam, V.; Cooper, W.A.; Armes, J.; Millar, E.K.A.; et al. SP142 PD-L1 Scoring Shows High Interobserver and Intraobserver Agreement in Triple-Negative Breast Carcinoma but Overall Low Percentage Agreement with Other PD-L1 Clones SP263 and 22C3. Am. J. Surg. Pathol. 2021, 45, 1108–1117. [Google Scholar] [CrossRef]
- Chang, S.; Park, H.K.; Choi, Y.L.; Jang, S.J. Interobserver Reproducibility of PD-L1 Biomarker in Non-Small Cell Lung Cancer: A Multi-Institutional Study by 27 Pathologists. J. Pathol. Transl. Med. 2019, 53, 347–353. [Google Scholar] [CrossRef]
- Cooper, W.A.; Russell, P.A.; Cherian, M.; Duhig, E.E.; Godbolt, D.; Jessup, P.J.; Khoo, C.; Leslie, C.; Mahar, A.; Moffat, D.F.; et al. Intra- and Interobserver Reproducibility Assessment of PD-L1 Biomarker in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4569–4577. [Google Scholar] [CrossRef]
- Lidbury, J.A.; Rodrigues Hoffmann, A.; Ivanek, R.; Cullen, J.M.; Porter, B.F.; Oliveira, F.; van Winkle, T.J.; Grinwis, G.C.; Sucholdolski, J.S.; Steiner, J.M. Interobserver Agreement Using Histological Scoring of the Canine Liver. J. Vet. Intern. Med. 2017, 31, 778. [Google Scholar] [CrossRef]
- Rizzardi, A.E.; Zhang, X.; Vogel, R.I.; Kolb, S.; Geybels, M.S.; Leung, Y.-K.; Henriksen, J.C.; Ho, S.-M.; Kwak, J.; Stanford, J.L.; et al. Quantitative Comparison and Reproducibility of Pathologist Scoring and Digital Image Analysis of Estrogen Receptor Β2 Immunohistochemistry in Prostate Cancer. Diagn. Pathol. 2016, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.W.; Kim, L.G.; Kong, H.H.; Low, L.Y.; Wang, T.T.; Supriya, S.; Kathiresan, M.; Soong, R.; Salto-Tellez, M. Computer-Assisted Pathological Immunohistochemistry Scoring Is More Time-Effective than Conventional Scoring, but Provides No Analytical Advantage. Histopathology 2010, 56, 523–529. [Google Scholar] [CrossRef]
- Taylor, C.R.; Levenson, R.M. Quantification of Immunohistochemistry?Issues Concerning Methods, Utility and Semiquantitative Assessment II. Histopathology 2006, 49, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Cregger, M.; Berger, A.J.; Rimm, D.L. Immunohistochemistry and Quantitative Analysis of Protein Expression. Arch. Pathol. Lab. Med. 2006, 130, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- di Cataldo, S.; Ficarra, E.; Macii, E. Computer-Aided Techniques for Chromogenic Immunohistochemistry: Status and Directions. Comput. Biol. Med. 2012, 42, 1012–1025. [Google Scholar] [CrossRef]
- van der Loos, C.M. Multiple Immunoenzyme Staining: Methods and Visualizations for the Observation with Spectral Imaging. J. Histochem. Cytochem. 2008, 56, 313–328. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Arch. Pathol. Lab. Med. 2007, 131, 18–43. [Google Scholar] [CrossRef]
- Pell, R.; Oien, K.; Robinson, M.; Pitman, H.; Rajpoot, N.; Rittscher, J.; Snead, D.; Verrill, C. The Use of Digital Pathology and Image Analysis in Clinical Trials. J. Pathol. Clin. Res. 2019, 5, 81–90. [Google Scholar] [CrossRef]
- Aeffner, F.; Zarella, M.D.; Buchbinder, N.; Bui, M.M.; Goodman, M.R.; Hartman, D.J.; Lujan, G.M.; Molani, M.A.; Parwani, A.V.; Lillard, K.; et al. Introduction to Digital Image Analysis in Whole-Slide Imaging: A White Paper from the Digital Pathology Association. J. Pathol. Inform. 2019, 10, 9. [Google Scholar] [CrossRef]
- Farahani, N.; Parwani, A.V.; Pantanowitz, L. Whole Slide Imaging in Pathology: Advantages, Limitations, and Emerging Perspectives. Pathol. Lab. Med. Int. 2015, 7, 23–33. [Google Scholar] [CrossRef]
- Shrestha, P.; Kneepkens, R.; van Elswijk, G.; Vrijnsen, J.; Ion, R.; Verhagen, D.; Abels, E.; Vossen, D.; Hulsken, B.; Shrestha, P.; et al. Objective and Subjective Assessment of Digital Pathology Image Quality. AIMS Med. Sci. 2015, 2, 65–78. [Google Scholar] [CrossRef]
- Tadrous, P.J. On the Concept of Objectivity in Digital Image Analysis in Pathology. Pathology 2010, 42, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Eggerschwiler, B.; Canepa, D.D.; Pape, H.C.; Casanova, E.A.; Cinelli, P. Automated Digital Image Quantification of Histological Staining for the Analysis of the Trilineage Differentiation Potential of Mesenchymal Stem Cells. Stem Cell Res. Ther. 2019, 10, 69. [Google Scholar] [CrossRef] [PubMed]
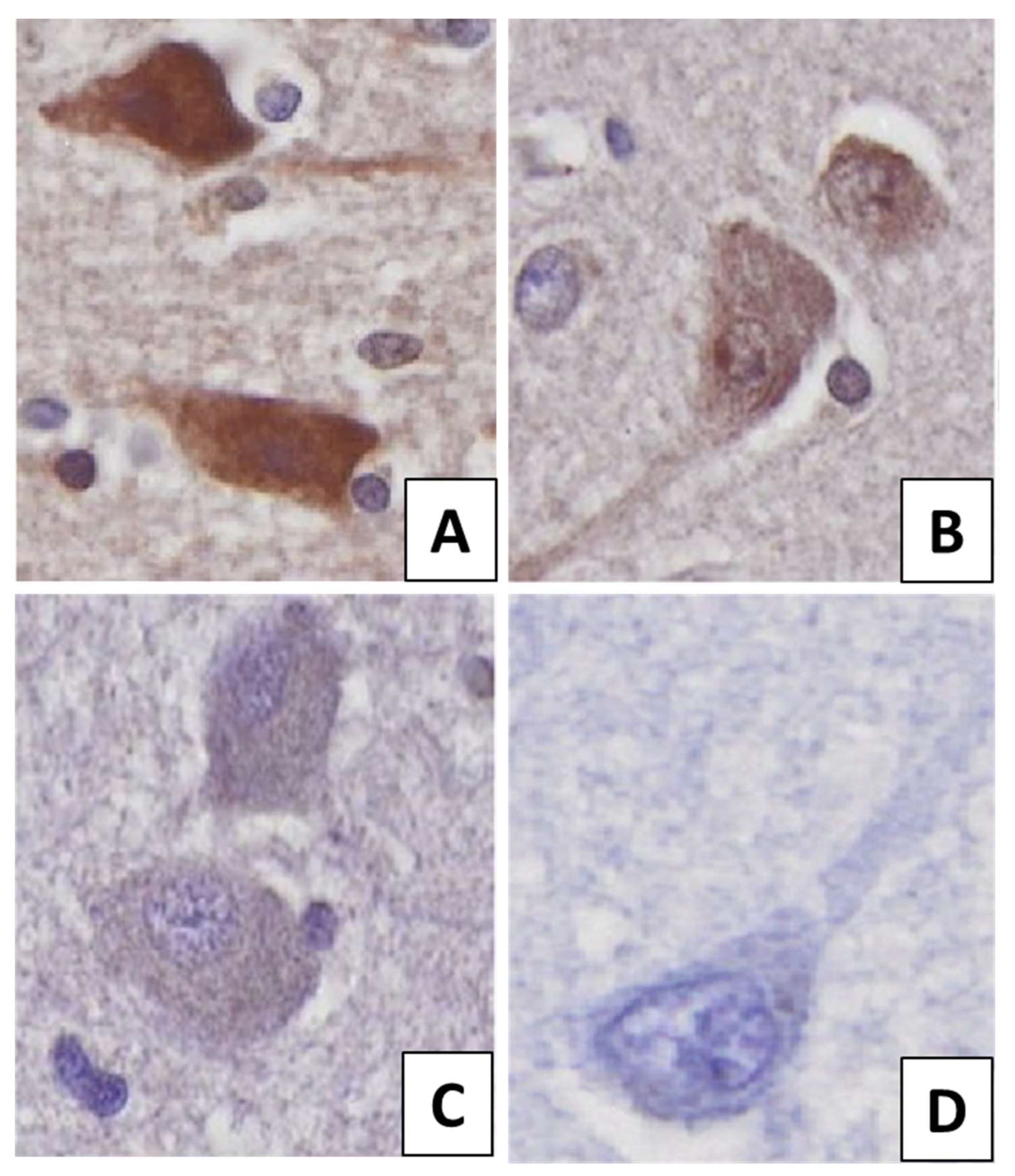
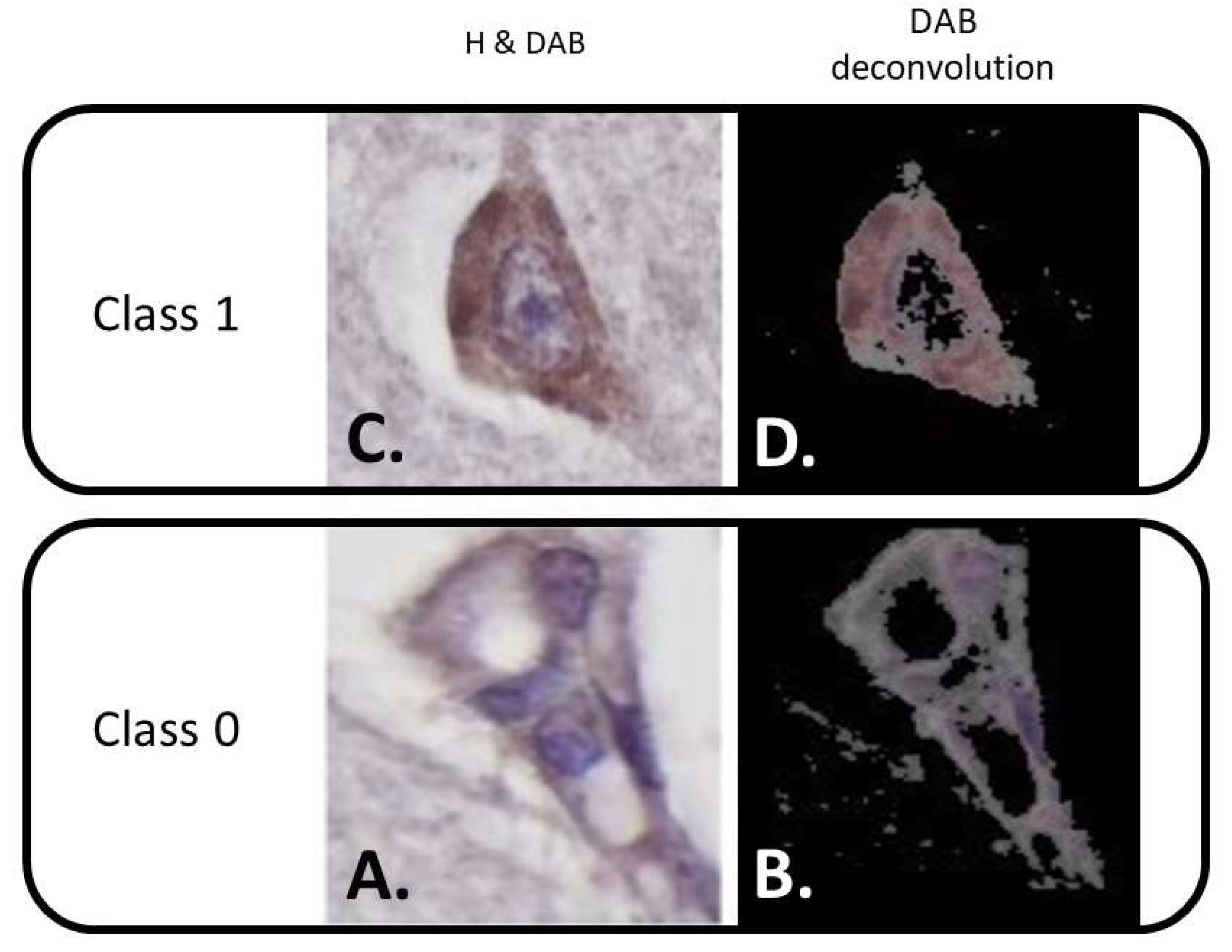

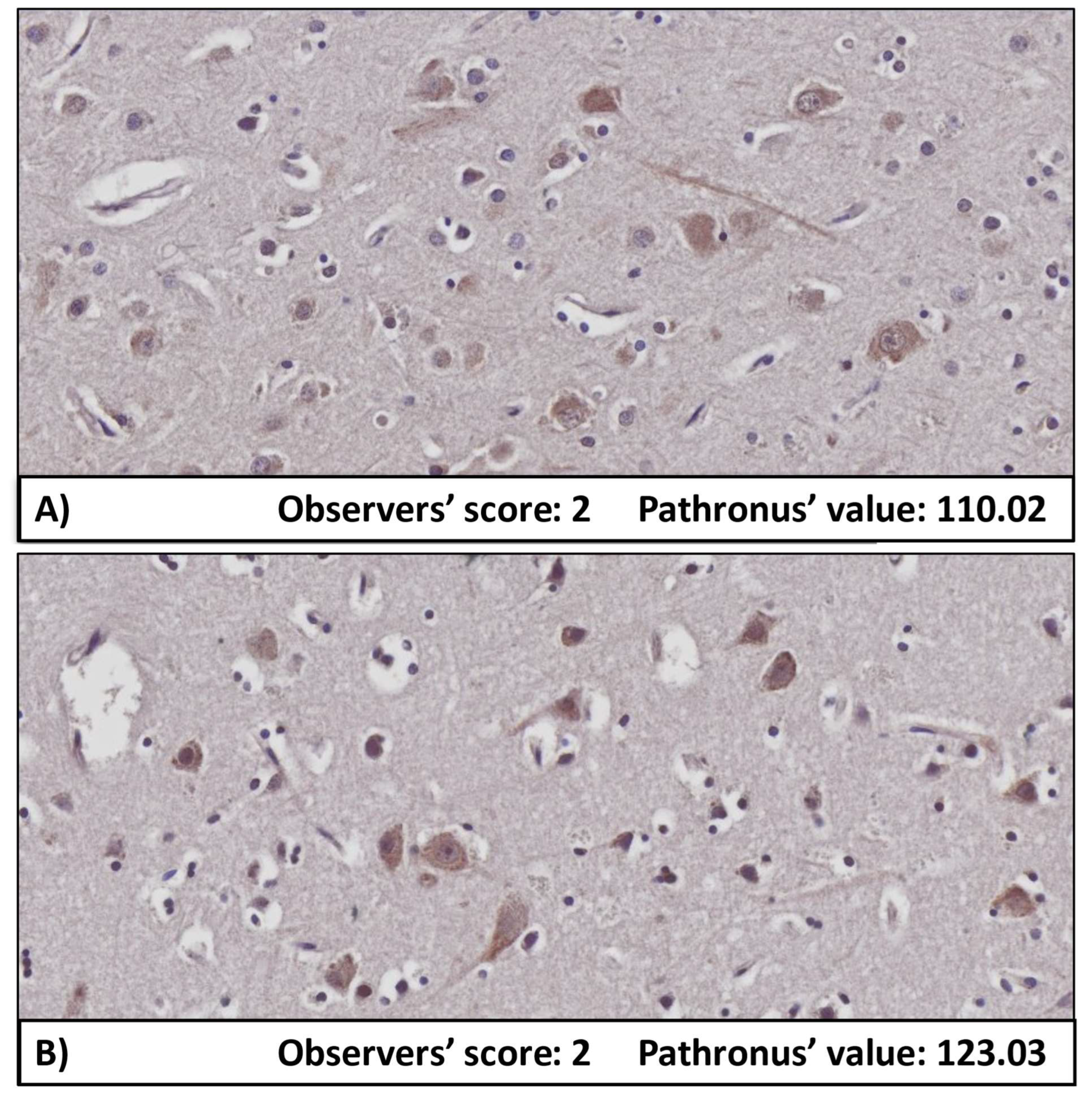
| Actual Class 1 | 21 (FN) | 441 (TP) |
| Actual Class 0 | 388 (TN) | 57 (FP) |
| Predicted Class 0 | Predicted Class 1 |
| (A) | |||||||
| CNT | Cohen’s kappa values | ||||||
| Strength of agreement | Observers | #1 | #2 | #3 | #4 | #5 | Pathronus |
| #1 | 0.091 | 0.103 | 0.6 | 0.048 | −0.01 | ||
| #2 | poor | 0.301 | 0.195 | 0.008 | −0.012 | ||
| #3 | poor | fair | 0.169 | −0.023 | −0.009 | ||
| #4 | moderate | poor | poor | −0.004 | −0.034 | ||
| #5 | poor | poor | poor | poor | 0.262 | ||
| Pathronus | poor | poor | poor | poor | fair | ||
| (B) | |||||||
| DLB | Cohen’s kappa values | ||||||
| Strength of agreement | Observers | #1 | #2 | #3 | #4 | #5 | Pathronus |
| #1 | 0.063 | 0.138 | 0.516 | 0.226 | 0.177 | ||
| #2 | poor | 0.316 | 0.141 | 0.087 | 0.048 | ||
| #3 | poor | fair | 0.114 | 0.143 | −0.022 | ||
| #4 | moderate | poor | poor | 0.286 | 0.196 | ||
| #5 | fair | poor | poor | fair | 0.270 | ||
| Pathronus | poor | poor | poor | poor | fair | ||
| (C) | |||||||
| AD | Cohen’s kappa values | ||||||
| Strength of agreement | Observers | #1 | #2 | #3 | #4 | #5 | Pathronus |
| #1 | 0.204 | 0.179 | 0.457 | 0.034 | 0.195 | ||
| #2 | poor | 0.297 | 0.285 | 0.118 | 0.232 | ||
| #3 | poor | fair | 0.178 | 0.180 | 0.062 | ||
| #4 | moderate | fair | poor | 0.260 | 0.214 | ||
| #5 | poor | poor | poor | fair | 0.116 | ||
| Pathronus | poor | fair | poor | fair | poor | ||
| (D) | |||
| CNT | DLB | AD | |
| Fleiss’ kappa | 0.091 | 0.176 | 0.183 |
| p-value | <0.005 | <0.005 | <0.005 |
| Agreement | poor | poor | poor |
| Observers | #1 | #2 | #3 | #4 | #5 | Pathronus Converted | Pathronus Original | Reference Data |
|---|---|---|---|---|---|---|---|---|
| Strength of immunopositivity among groups | CNT > DLB > AD | CNT > DLB > AD | CNT > DLB > AD | CNT > DLB > AD | DLB > CNT > AD | CNT > DLB > AD | CNT > DLB > AD | CNT > DLB > AD |
| Statistical significance (p < 0.05) | CNT vs. DLB CNT vs. AD DLB vs. AD | CNT vs. AD | CNT vs. AD | CNT vs. DLB CNT vs. AD DLB vs. AD | - | CNT vs. AD DLB vs. AD | CNT vs. AD DLB vs. AD | CNT vs. AD DLB vs. AD |
| Digital Image Analysis | Semi-Quantitative Scoring | |
|---|---|---|
| Expensive | Cost | Cheap |
| Fast | Speed | Slow |
| Not required (except training period) | Histological experiment | Required |
| Objective (with standard settings) | Objectivity | Subjective |
| Based on software and settings | Inter-rater variability | Considerable |
| Not applicable | Intra-rater variability | Notable |
| Yes (except DAB labelling) | Quantification | Not applicable |
| Automatic (after training period) | Operation | Manual |
| Large | Data volume | Limited |
| IT background, slide scanner | Equipment | Light microscope |
| New era | Research purposes | Gold standard |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bencze, J.; Szarka, M.; Kóti, B.; Seo, W.; Hortobágyi, T.G.; Bencs, V.; Módis, L.V.; Hortobágyi, T. Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry. Biomolecules 2022, 12, 19. https://doi.org/10.3390/biom12010019
Bencze J, Szarka M, Kóti B, Seo W, Hortobágyi TG, Bencs V, Módis LV, Hortobágyi T. Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry. Biomolecules. 2022; 12(1):19. https://doi.org/10.3390/biom12010019
Chicago/Turabian StyleBencze, János, Máté Szarka, Balázs Kóti, Woosung Seo, Tibor G. Hortobágyi, Viktor Bencs, László V. Módis, and Tibor Hortobágyi. 2022. "Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry" Biomolecules 12, no. 1: 19. https://doi.org/10.3390/biom12010019
APA StyleBencze, J., Szarka, M., Kóti, B., Seo, W., Hortobágyi, T. G., Bencs, V., Módis, L. V., & Hortobágyi, T. (2022). Comparison of Semi-Quantitative Scoring and Artificial Intelligence Aided Digital Image Analysis of Chromogenic Immunohistochemistry. Biomolecules, 12(1), 19. https://doi.org/10.3390/biom12010019







