Synergies of Single Molecule Fluorescence and NMR for the Study of Intrinsically Disordered Proteins
Abstract
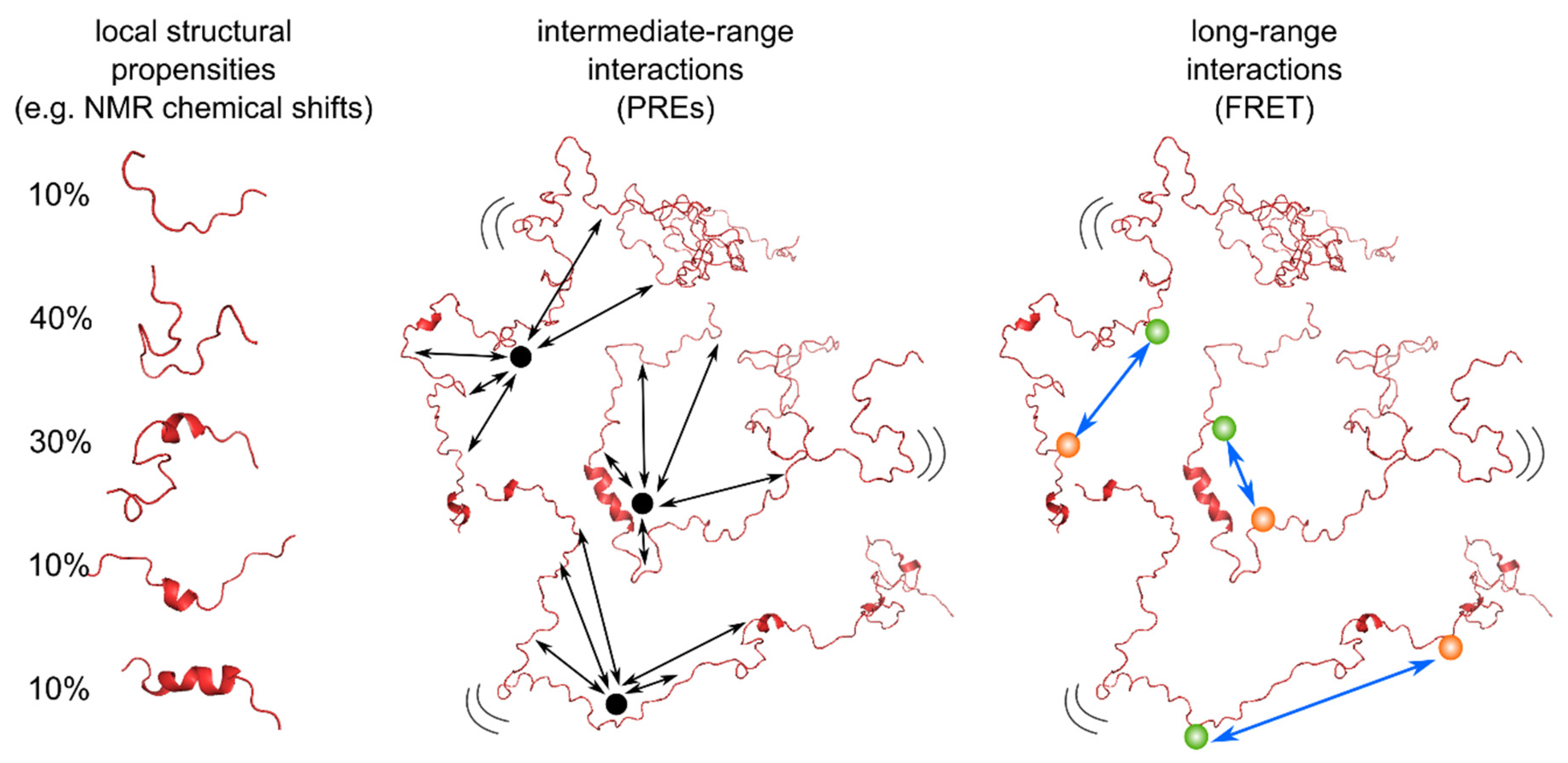
1. Introduction
2. Folded Domains Connected by Intrinsically Disordered Linkers
3. Large Intrinsically Disordered Proteins
3.1. Partially Folded Proteins
3.2. Disordered Protein Complexes
4. Liquid–Liquid Phase Separation
4.1. Ensemble Fluorescence Combined with NMR
4.2. Single Molecule FRET Combined with NMR
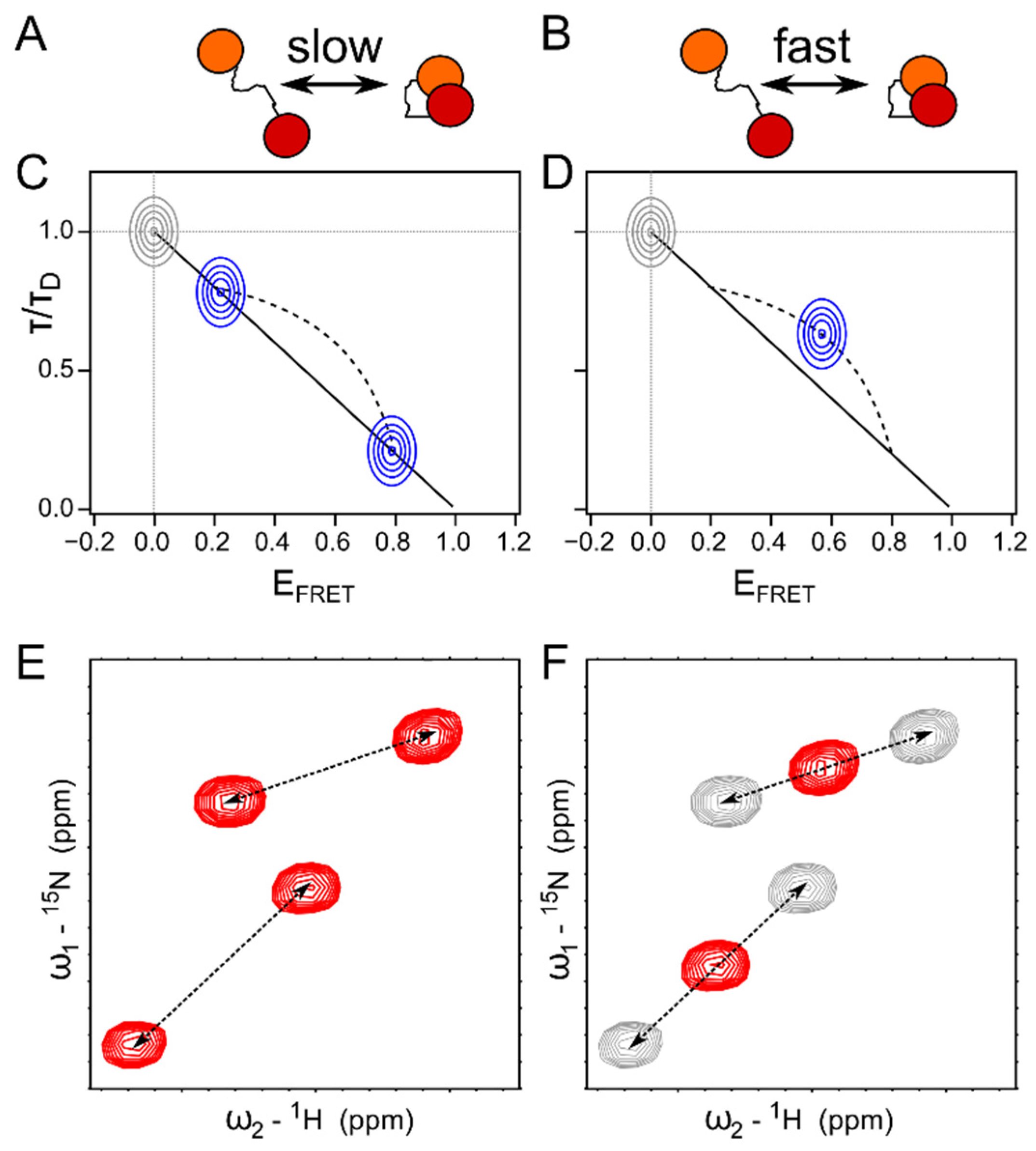
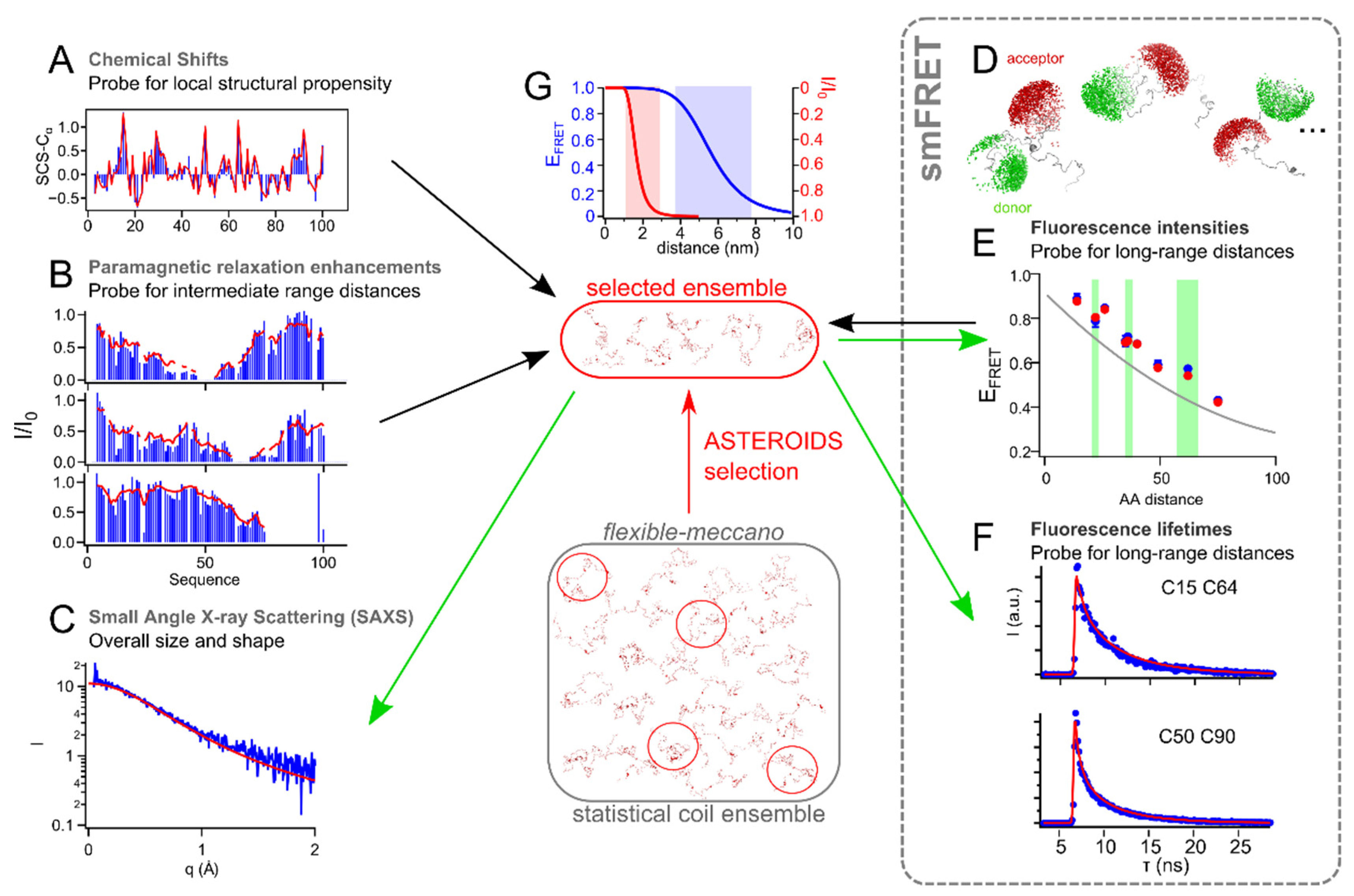
5. Towards a Quantitative Combination of smFRET and NMR
6. Conclusions and Perspectives for the Combined Use of Single Molecule Fluorescence and NMR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tompa, P. Intrinsically Disordered Proteins: A 10-Year Recap. Trends Biochem. Sci. 2012, 37, 509–516. [Google Scholar] [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically Unstructured Proteins and Their Functions. Nat. Rev. Mol. Cell Biol. 2005, 6, 197–208. [Google Scholar] [CrossRef]
- Arai, M. Unified Understanding of Folding and Binding Mechanisms of Globular and Intrinsically Disordered Proteins. Biophys. Rev. 2018, 10, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Davey, N.E.; Cowan, J.L.; Shields, D.C.; Gibson, T.J.; Coldwell, M.J.; Edwards, R.J. SLiMPrints: Conservation-Based Discovery of Functional Motif Fingerprints in Intrinsically Disordered Protein Regions. Nucleic Acids Res. 2012, 40, 10628–10641. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Birol, M.; Rhoades, E. IDPs in Macromolecular Complexes: The Roles of Multivalent Interactions in Diverse Assemblies. Curr. Opin. Struct. Biol. 2018, 49, 36–43. [Google Scholar] [CrossRef]
- Schneider, R.; Blackledge, M.; Jensen, M.R. Elucidating Binding Mechanisms and Dynamics of Intrinsically Disordered Protein Complexes Using NMR Spectroscopy. Curr. Opin. Struct. Biol. 2019, 54, 10–18. [Google Scholar] [CrossRef]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting Intrinsic Disorder in Proteins: An Overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B. Computational and Theoretical Advances in Studies of Intrinsically Disordered Proteins. Curr. Opin. Struct. Biol. 2017, 42, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Matysiak, S.; Mittal, J. Looking at the Disordered Proteins through the Computational Microscope. ACS Cent. Sci. 2018, 4, 534–542. [Google Scholar] [CrossRef]
- Schuler, B.; Soranno, A.; Hofmann, H.; Nettels, D. Single-Molecule FRET Spectroscopy and the Polymer Physics of Unfolded and Intrinsically Disordered Proteins. Annu. Rev. Biophys. 2016, 45, 207–231. [Google Scholar] [CrossRef]
- Jensen, M.R.; Zweckstetter, M.; Huang, J.; Blackledge, M. Exploring Free-Energy Landscapes of Intrinsically Disordered Proteins at Atomic Resolution Using NMR Spectroscopy. Chem. Rev. 2014, 114, 6632–6660. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Salvi, N.; Blackledge, M.; Jensen, M.R. Characterization of Intrinsically Disordered Proteins and Their Dynamic Complexes: From in Vitro to Cell-like Environments. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 109, 79–100. [Google Scholar] [CrossRef]
- Bernadó, P.; Svergun, D.I. Analysis of Intrinsically Disordered Proteins by Small-Angle X-Ray Scattering. Methods Mol. Biol. 2012, 896, 107–122. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Maurin, D.; Communie, G.; Kragelj, J.; Hansen, D.F.; Ruigrok, R.W.H.; Jensen, M.R.; Blackledge, M. Visualizing the Molecular Recognition Trajectory of an Intrinsically Disordered Protein Using Multinuclear Relaxation Dispersion NMR. J. Am. Chem. Soc. 2015, 137, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Soranno, A.; Borgia, A.; Gast, K.; Nettels, D.; Schuler, B. Polymer Scaling Laws of Unfolded and Intrinsically Disordered Proteins Quantified with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 16155–16160. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.R.; Salmon, L.; Nodet, G.; Blackledge, M. Defining Conformational Ensembles of Intrinsically Disordered and Partially Folded Proteins Directly from Chemical Shifts. J. Am. Chem. Soc. 2010, 132, 1270–1272. [Google Scholar] [CrossRef]
- Zheng, W.; Best, R.B. An Extended Guinier Analysis for Intrinsically Disordered Proteins. J. Mol. Biol. 2018, 430, 2540–2553. [Google Scholar] [CrossRef]
- Soranno, A.; Buchli, B.; Nettels, D.; Cheng, R.R.; Müller-Späth, S.; Pfeil, S.H.; Hoffmann, A.; Lipman, E.A.; Makarov, D.E.; Schuler, B. Quantifying Internal Friction in Unfolded and Intrinsically Disordered Proteins with Single-Molecule Spectroscopy. Proc. Natl. Acad. Sci. USA 2012, 109, 17800–17806. [Google Scholar] [CrossRef] [PubMed]
- Abyzov, A.; Salvi, N.; Schneider, R.; Maurin, D.; Ruigrok, R.W.H.; Jensen, M.R.; Blackledge, M. Identification of Dynamic Modes in an Intrinsically Disordered Protein Using Temperature-Dependent NMR Relaxation. J. Am. Chem. Soc. 2016, 138, 6240–6251. [Google Scholar] [CrossRef]
- Krzeminski, M.; Marsh, J.A.; Neale, C.; Choy, W.-Y.; Forman-Kay, J.D. Characterization of Disordered Proteins with ENSEMBLE. Bioinformatics 2013, 29, 398–399. [Google Scholar] [CrossRef]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Mercadante, D.; Koehler, C.; Kachala, M.; Estrada Girona, G.; Milles, S.; Mishra, A.; et al. Decoupling of Size and Shape Fluctuations in Heteropolymeric Sequences Reconciles Discrepancies in SAXS vs. FRET Measurements. Proc. Natl. Acad. Sci. USA 2017, 114, E6342–E6351. [Google Scholar] [CrossRef]
- Milles, S.; Lemke, E.A. Mapping Multivalency and Differential Affinities within Large Intrinsically Disordered Protein Complexes with Segmental Motion Analysis. Angew. Chem. Int. Ed. 2014, 53, 7364–7367. [Google Scholar] [CrossRef] [PubMed]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of Coupled Folding and Binding of an Intrinsically Disordered Protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Bouvignies, G.; Pelupessy, P.; Walrant, A.; Marquant, R.; Kozlov, M.; De Ioannes, P.; Bolik-Coulon, N.; Sagan, S.; Cortes, P.; et al. Structure and Dynamics of an Intrinsically Disordered Protein Region That Partially Folds upon Binding by Chemical-Exchange NMR. J. Am. Chem. Soc. 2017, 139, 12219–12227. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.N.; Charlier, C.; Augustyniak, R.; Salvi, N.; Déjean, V.; Bodenhausen, G.; Lequin, O.; Pelupessy, P.; Ferrage, F. Distribution of Pico- and Nanosecond Motions in Disordered Proteins from Nuclear Spin Relaxation. Biophys. J. 2015, 109, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Nodet, G.; Salmon, L.; Ozenne, V.; Meier, S.; Jensen, M.R.; Blackledge, M. Quantitative Description of Backbone Conformational Sampling of Unfolded Proteins at Amino Acid Resolution from NMR Residual Dipolar Couplings. J. Am. Chem. Soc. 2009, 131, 17908–17918. [Google Scholar] [CrossRef]
- Kashtanov, S.; Borcherds, W.; Wu, H.; Daughdrill, G.W.; Ytreberg, F.M. Using Chemical Shifts to Assess Transient Secondary Structure and Generate Ensemble Structures of Intrinsically Disordered Proteins. Methods Mol. Biol. 2012, 895, 139–152. [Google Scholar] [CrossRef]
- Choy, W.-Y.; Forman-Kay, J.D. Calculation of Ensembles of Structures Representing the Unfolded State of an SH3 Domain11Edited by P. E. Wright. J. Mol. Biol. 2001, 308, 1011–1032. [Google Scholar] [CrossRef]
- Bernadó, P.; Blanchard, L.; Timmins, P.; Marion, D.; Ruigrok, R.W.H.; Blackledge, M. A Structural Model for Unfolded Proteins from Residual Dipolar Couplings and Small-Angle x-Ray Scattering. Proc. Natl. Acad. Sci. USA 2005, 102, 17002–17007. [Google Scholar] [CrossRef]
- Vallurupalli, P.; Bouvignies, G.; Kay, L.E. Studying “Invisible” Excited Protein States in Slow Exchange with a Major State Conformation. J. Am. Chem. Soc. 2012, 134, 8148–8161. [Google Scholar] [CrossRef] [PubMed]
- Kragelj, J.; Palencia, A.; Nanao, M.H.; Maurin, D.; Bouvignies, G.; Blackledge, M.; Jensen, M.R. Structure and Dynamics of the MKK7-JNK Signaling Complex. Proc. Natl. Acad. Sci. USA 2015, 112, 3409–3414. [Google Scholar] [CrossRef] [PubMed]
- Sisamakis, E.; Valeri, A.; Kalinin, S.; Rothwell, P.J.; Seidel, C.A.M. Accurate Single-Molecule FRET Studies Using Multiparameter Fluorescence Detection. Methods Enzymol. 2010, 475, 455–514. [Google Scholar] [CrossRef]
- Felekyan, S.; Kalinin, S.; Sanabria, H.; Valeri, A.; Seidel, C.A.M. Filtered FCS: Species Auto- and Cross-Correlation Functions Highlight Binding and Dynamics in Biomolecules. ChemPhysChem 2012, 13, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Aznauryan, M.; Delgado, L.; Soranno, A.; Nettels, D.; Huang, J.-R.; Labhardt, A.M.; Grzesiek, S.; Schuler, B. Comprehensive Structural and Dynamical View of an Unfolded Protein from the Combination of Single-Molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA. 2016, 113, E5389–E5398. [Google Scholar] [CrossRef] [PubMed]
- Rezaei-Ghaleh, N.; Parigi, G.; Soranno, A.; Holla, A.; Becker, S.; Schuler, B.; Luchinat, C.; Zweckstetter, M. Local and Global Dynamics in Intrinsically Disordered Synuclein. Angew. Chem. Int. Ed. 2018, 57, 15262–15266. [Google Scholar] [CrossRef]
- Daniels, S.M.; Gatignol, A. The Multiple Functions of TRBP, at the Hub of Cell Responses to Viruses, Stress, and Cancer. Microbiol. Mol. Biol. Rev. 2012, 76, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Nettels, D.; Clark, J.; Borgia, A.; Radford, S.E.; Clarke, J.; Schuler, B. Quantifying Heterogeneity and Conformational Dynamics from Single Molecule FRET of Diffusing Molecules: Recurrence Analysis of Single Particles (RASP). Phys. Chem. Chem. Phys. 2011, 13, 1857–1871. [Google Scholar] [CrossRef] [PubMed]
- Masliah, G.; Maris, C.; König, S.L.; Yulikov, M.; Aeschimann, F.; Malinowska, A.L.; Mabille, J.; Weiler, J.; Holla, A.; Hunziker, J.; et al. Structural Basis of SiRNA Recognition by TRBP Double-Stranded RNA Binding Domains. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Boivin, S.; Cusack, S.; Ruigrok, R.W.H.; Hart, D.J. Influenza A Virus Polymerase: Structural Insights into Replication and Host Adaptation Mechanisms. J. Biol. Chem. 2010, 285, 28411–28417. [Google Scholar] [CrossRef]
- Tarendeau, F.; Boudet, J.; Guilligay, D.; Mas, P.J.; Bougault, C.M.; Boulo, S.; Baudin, F.; Ruigrok, R.W.H.; Daigle, N.; Ellenberg, J.; et al. Structure and Nuclear Import Function of the C-Terminal Domain of Influenza Virus Polymerase PB2 Subunit. Nat. Struct. Mol. Biol. 2007, 14, 229–233. [Google Scholar] [CrossRef]
- Tarendeau, F.; Crepin, T.; Guilligay, D.; Ruigrok, R.W.H.; Cusack, S.; Hart, D.J. Host Determinant Residue Lysine 627 Lies on the Surface of a Discrete, Folded Domain of Influenza Virus Polymerase PB2 Subunit. PLOS Pathog. 2008, 4, e1000136. [Google Scholar] [CrossRef] [PubMed]
- Delaforge, E.; Milles, S.; Bouvignies, G.; Bouvier, D.; Boivin, S.; Salvi, N.; Maurin, D.; Martel, A.; Round, A.; Lemke, E.A.; et al. Large-Scale Conformational Dynamics Control H5N1 Influenza Polymerase PB2 Binding to Importin α. J. Am. Chem. Soc. 2015, 137, 15122–15134. [Google Scholar] [CrossRef] [PubMed]
- Hengrung, N.; El Omari, K.; Serna Martin, I.; Vreede, F.T.; Cusack, S.; Rambo, R.P.; Vonrhein, C.; Bricogne, G.; Stuart, D.I.; Grimes, J.M.; et al. Crystal Structure of the RNA-Dependent RNA Polymerase from Influenza C Virus. Nature 2015, 527, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Kirui, J.; Bucci, M.D.; Poole, D.S.; Mehle, A. Conserved Features of the PB2 627 Domain Impact Influenza Virus Polymerase Function and Replication. J. Virol. 2014, 88, 5977–5986. [Google Scholar] [CrossRef] [PubMed]
- Thierry, E.; Guilligay, D.; Kosinski, J.; Bock, T.; Gaudon, S.; Round, A.; Pflug, A.; Hengrung, N.; El Omari, K.; Baudin, F.; et al. Influenza Polymerase Can Adopt an Alternative Configuration Involving a Radical Repacking of PB2 Domains. Mol. Cell 2016, 61, 125–137. [Google Scholar] [CrossRef]
- Huang, J.; Warner, L.R.; Sanchez, C.; Gabel, F.; Madl, T.; Mackereth, C.D.; Sattler, M.; Blackledge, M. Transient Electrostatic Interactions Dominate the Conformational Equilibrium Sampled by Multidomain Splicing Factor U2AF65: A Combined NMR and SAXS Study. J. Am. Chem. Soc. 2014, 136, 7068–7076. [Google Scholar] [CrossRef]
- Voith von Voithenberg, L.; Sánchez-Rico, C.; Kang, H.-S.; Madl, T.; Zanier, K.; Barth, A.; Warner, L.R.; Sattler, M.; Lamb, D.C. Recognition of the 3’ Splice Site RNA by the U2AF Heterodimer Involves a Dynamic Population Shift. Proc. Natl. Acad. Sci. USA 2016, 113, E7169–E7175. [Google Scholar] [CrossRef] [PubMed]
- Zuo, P.; Maniatis, T. The Splicing Factor U2AF35 Mediates Critical Protein-Protein Interactions in Constitutive and Enhancer-Dependent Splicing. Genes Dev. 1996, 10, 1356–1368. [Google Scholar] [CrossRef]
- Gopich, I.V.; Szabo, A. Theory of the Energy Transfer Efficiency and Fluorescence Lifetime Distribution in Single-Molecule FRET. Proc. Natl. Acad. Sci. USA 2012, 109, 7747–7752. [Google Scholar] [CrossRef]
- Ferreon, J.C.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Structural Basis for Subversion of Cellular Control Mechanisms by the Adenoviral E1A Oncoprotein. Proc. Natl. Acad. Sci. USA 2009, 106, 13260–13265. [Google Scholar] [CrossRef] [PubMed]
- Hošek, T.; Calçada, E.O.; Nogueira, M.O.; Salvi, M.; Pagani, T.D.; Felli, I.C.; Pierattelli, R. Structural and Dynamic Characterization of the Molecular Hub Early Region 1A (E1A) from Human Adenovirus. Chemistry 2016, 22, 13010–13013. [Google Scholar] [CrossRef] [PubMed]
- Glavina, J.; Román, E.A.; Espada, R.; de Prat-Gay, G.; Chemes, L.B.; Sánchez, I.E. Interplay between Sequence, Structure and Linear Motifs in the Adenovirus E1A Hub Protein. Virology 2018, 525, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, A.C.M.; Ferreon, J.C.; Wright, P.E.; Deniz, A.A. Modulation of Allostery by Protein Intrinsic Disorder. Nature 2013, 498, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Schuler, B. Single-Molecule FRET of Protein Structure and Dynamics—A Primer. J. Nanobiotechnol. 2013, 11, S2. [Google Scholar] [CrossRef] [PubMed]
- Murrali, M.G.; Felli, I.C.; Pierattelli, R. Adenoviral E1A Exploits Flexibility and Disorder to Target Cellular Proteins. Biomolecules 2020, 10, 1541. [Google Scholar] [CrossRef] [PubMed]
- Haberz, P.; Arai, M.; Martinez-Yamout, M.A.; Dyson, H.J.; Wright, P.E. Mapping the Interactions of Adenoviral E1A Proteins with the P160 Nuclear Receptor Coactivator Binding Domain of CBP. Protein Sci. 2016, 25, 2256–2267. [Google Scholar] [CrossRef] [PubMed]
- Tosatto, L.; Horrocks, M.H.; Dear, A.J.; Knowles, T.P.J.; Dalla Serra, M.; Cremades, N.; Dobson, C.M.; Klenerman, D. Single-Molecule FRET Studies on Alpha-Synuclein Oligomerization of Parkinson’s Disease Genetically Related Mutants. Sci. Rep. 2015, 5, 16696. [Google Scholar] [CrossRef]
- Gambin, Y.; Schug, A.; Lemke, E.A.; Lavinder, J.J.; Ferreon, A.C.M.; Magliery, T.J.; Onuchic, J.N.; Deniz, A.A. Direct Single-Molecule Observation of a Protein Living in Two Opposed Native Structures. Proc. Natl. Acad. Sci. USA 2009, 106, 10153–10158. [Google Scholar] [CrossRef]
- Nath, A.; Sammalkorpi, M.; DeWitt, D.C.; Trexler, A.J.; Elbaum-Garfinkle, S.; O’Hern, C.S.; Rhoades, E. The Conformational Ensembles of α-Synuclein and Tau: Combining Single-Molecule FRET and Simulations. Biophys. J. 2012, 103, 1940–1949. [Google Scholar] [CrossRef]
- Melo, A.M.; Coraor, J.; Alpha-Cobb, G.; Elbaum-Garfinkle, S.; Nath, A.; Rhoades, E. A Functional Role for Intrinsic Disorder in the Tau-Tubulin Complex. Proc. Natl Acad Sci USA 2016, 113, 14336–14341. [Google Scholar] [CrossRef]
- Seuring, C.; Verasdonck, J.; Gath, J.; Ghosh, D.; Nespovitaya, N.; Wälti, M.A.; Maji, S.K.; Cadalbert, R.; Güntert, P.; Meier, B.H.; et al. The Three-Dimensional Structure of Human β-Endorphin Amyloid Fibrils. Nat. Struct. Mol. Biol. 2020, 27, 1178–1184. [Google Scholar] [CrossRef]
- Tuttle, M.D.; Comellas, G.; Nieuwkoop, A.J.; Covell, D.J.; Berthold, D.A.; Kloepper, K.D.; Courtney, J.M.; Kim, J.K.; Barclay, A.M.; Kendall, A.; et al. Solid-State NMR Structure of a Pathogenic Fibril of Full-Length Human α-Synuclein. Nat. Struct. Mol. Biol. 2016, 23, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Kollmer, M.; Close, W.; Funk, L.; Rasmussen, J.; Bsoul, A.; Schierhorn, A.; Schmidt, M.; Sigurdson, C.J.; Jucker, M.; Fändrich, M. Cryo-EM Structure and Polymorphism of Aβ Amyloid Fibrils Purified from Alzheimer’s Brain Tissue. Nat. Commun. 2019, 10, 4760. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Babinchak, W.M.; Surewicz, W.K. Cryo-EM Structure of Amyloid Fibrils Formed by the Entire Low Complexity Domain of TDP-43. Nat. Commun. 2021, 12, 1620. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.A.; Jeffrey, P.D.; Patten, A.K.; Massagué, J.; Pavletich, N.P. Crystal Structure of the P27Kip1 Cyclin-Dependent-Kinase Inhibitor Bound to the Cyclin A-Cdk2 Complex. Nature 1996, 382, 325–331. [Google Scholar] [CrossRef]
- Lacy, E.R.; Filippov, I.; Lewis, W.S.; Otieno, S.; Xiao, L.; Weiss, S.; Hengst, L.; Kriwacki, R.W. P27 Binds Cyclin-CDK Complexes through a Sequential Mechanism Involving Binding-Induced Protein Folding. Nat. Struct. Mol. Biol. 2004, 11, 358–364. [Google Scholar] [CrossRef]
- Bloom, J.; Pagano, M. Deregulated Degradation of the Cdk Inhibitor P27 and Malignant Transformation. Semin. Cancer Biol. 2003, 13, 41–47. [Google Scholar] [CrossRef]
- Grimmler, M.; Wang, Y.; Mund, T.; Cilensek, Z.; Keidel, E.-M.; Waddell, M.B.; Jäkel, H.; Kullmann, M.; Kriwacki, R.W.; Hengst, L. Cdk-Inhibitory Activity and Stability of P27Kip1 Are Directly Regulated by Oncogenic Tyrosine Kinases. Cell 2007, 128, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Chu, I.; Sun, J.; Arnaout, A.; Kahn, H.; Hanna, W.; Narod, S.; Sun, P.; Tan, C.-K.; Hengst, L.; Slingerland, J. P27 Phosphorylation by Src Regulates Inhibition of Cyclin E-Cdk2. Cell 2007, 128, 281–294. [Google Scholar] [CrossRef]
- Tsytlonok, M.; Sanabria, H.; Wang, Y.; Felekyan, S.; Hemmen, K.; Phillips, A.H.; Yun, M.-K.; Waddell, M.B.; Park, C.-G.; Vaithiyalingam, S.; et al. Dynamic Anticipation by Cdk2/Cyclin A-Bound P27 Mediates Signal Integration in Cell Cycle Regulation. Nat. Commun. 2019, 10, 1676. [Google Scholar] [CrossRef]
- Antonik, M.; Felekyan, S.; Gaiduk, A.; Seidel, C.A.M. Separating Structural Heterogeneities from Stochastic Variations in Fluorescence Resonance Energy Transfer Distributions via Photon Distribution Analysis. J. Phys. Chem. B 2006, 110, 6970–6978. [Google Scholar] [CrossRef] [PubMed]
- Tsytlonok, M.; Hemmen, K.; Hamilton, G.; Kolimi, N.; Felekyan, S.; Seidel, C.A.M.; Tompa, P.; Sanabria, H. Specific Conformational Dynamics and Expansion Underpin a Multi-Step Mechanism for Specific Binding of P27 with Cdk2/Cyclin, A. J. Mol. Biol. 2020, 432, 2998–3017. [Google Scholar] [CrossRef] [PubMed]
- Wälde, S.; Kehlenbach, R.H. The Part and the Whole: Functions of Nucleoporins in Nucleocytoplasmic Transport. Trends Cell Biol. 2010, 20, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Paci, G.; Caria, J.; Lemke, E.A. Cargo Transport through the Nuclear Pore Complex at a Glance. J. Cell Sci. 2021, 134. [Google Scholar] [CrossRef] [PubMed]
- Kubitscheck, U.; Grünwald, D.; Hoekstra, A.; Rohleder, D.; Kues, T.; Siebrasse, J.P.; Peters, R. Nuclear Transport of Single Molecules: Dwell Times at the Nuclear Pore Complex. J. Cell Biol. 2005, 168, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Gelles, J.; Musser, S.M. Imaging of Single-Molecule Translocation through Nuclear Pore Complexes. Proc. Natl. Acad. Sci. USA 2004, 101, 12887–12892. [Google Scholar] [CrossRef]
- Bayliss, R.; Littlewood, T.; Stewart, M. Structural Basis for the Interaction between FxFG Nucleoporin Repeats and Importin-Beta in Nuclear Trafficking. Cell 2000, 102, 99–108. [Google Scholar] [CrossRef]
- Bayliss, R.; Littlewood, T.; Strawn, L.A.; Wente, S.R.; Stewart, M. GLFG and FxFG Nucleoporins Bind to Overlapping Sites on Importin-Beta. J. Biol. Chem. 2002, 277, 50597–50606. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, R.; Leung, S.W.; Baker, R.P.; Quimby, B.B.; Corbett, A.H.; Stewart, M. Structural Basis for the Interaction between NTF2 and Nucleoporin FxFG Repeats. EMBO J. 2002, 21, 2843–2853. [Google Scholar] [CrossRef]
- Morrison, J.; Yang, J.-C.; Stewart, M.; Neuhaus, D. Solution NMR Study of the Interaction between NTF2 and Nucleoporin FxFG Repeats. J. Mol. Biol. 2003, 333, 587–603. [Google Scholar] [CrossRef]
- Isgro, T.A.; Schulten, K. Cse1p-Binding Dynamics Reveal a Binding Pattern for FG-Repeat Nucleoporins on Transport Receptors. Structure 2007, 15, 977–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Isgro, T.A.; Schulten, K. Association of Nuclear Pore FG-Repeat Domains to NTF2 Import and Export Complexes. J. Mol. Biol. 2007, 366, 330–345. [Google Scholar] [CrossRef] [PubMed]
- Isgro, T.A.; Schulten, K. Binding Dynamics of Isolated Nucleoporin Repeat Regions to Importin-Beta. Structure 2005, 13, 1869–1879. [Google Scholar] [CrossRef] [PubMed]
- Bednenko, J.; Cingolani, G.; Gerace, L. Importin β Contains a COOH-Terminal Nucleoporin Binding Region Important for Nuclear Transport. J. Cell Biol. 2003, 162, 391–401. [Google Scholar] [CrossRef]
- Ben-Efraim, I.; Gerace, L. Gradient of Increasing Affinity of Importin β for Nucleoporins along the Pathway of Nuclear Import. J. Cell Biol 2001, 152, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Tetenbaum-Novatt, J.; Hough, L.E.; Mironska, R.; McKenney, A.S.; Rout, M.P. Nucleocytoplasmic Transport: A Role for Nonspecific Competition in Karyopherin-Nucleoporin Interactions. Mol. Cell. Proteom. 2012, 11, 31–46. [Google Scholar] [CrossRef]
- Tu, L.-C.; Fu, G.; Zilman, A.; Musser, S.M. Large Cargo Transport by Nuclear Pores: Implications for the Spatial Organization of FG-Nucleoporins. EMBO J. 2013, 32, 3220–3230. [Google Scholar] [CrossRef]
- Milles, S.; Mercadante, D.; Aramburu, I.V.; Jensen, M.R.; Banterle, N.; Koehler, C.; Tyagi, S.; Clarke, J.; Shammas, S.L.; Blackledge, M.; et al. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell 2015, 163, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Hough, L.E.; Dutta, K.; Sparks, S.; Temel, D.B.; Kamal, A.; Tetenbaum-Novatt, J.; Rout, M.P.; Cowburn, D. The Molecular Mechanism of Nuclear Transport Revealed by Atomic-Scale Measurements. eLife 2015, 4, e10027. [Google Scholar] [CrossRef]
- Tan, P.S.; Aramburu, I.V.; Mercadante, D.; Tyagi, S.; Chowdhury, A.; Spitz, D.; Shammas, S.L.; Gräter, F.; Lemke, E.A. Two Differential Binding Mechanisms of FG-Nucleoporins and Nuclear Transport Receptors. Cell Rep. 2018, 22, 3660–3671. [Google Scholar] [CrossRef]
- Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/26489467/ (accessed on 6 August 2020).
- Bugge, K.; Brakti, I.; Fernandes, C.B.; Dreier, J.E.; Lundsgaard, J.E.; Olsen, J.G.; Skriver, K.; Kragelund, B.B. Interactions by Disorder—A Matter of Context. Front. Mol. Biosci. 2020, 7, 110. [Google Scholar] [CrossRef]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The Liquid State of FG-Nucleoporins Mimics Permeability Barrier Properties of Nuclear Pore Complexes. J. Cell Biol. 2020, 219, e201907157. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Lemke, E.A. Single Molecule Study of the Intrinsically Disordered FG-Repeat Nucleoporin 153. Biophys. J. 2011, 101, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.; Görlich, D. A Saturated FG-Repeat Hydrogel Can Reproduce the Permeability Properties of Nuclear Pore Complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Bednar, J.; Garcia-Saez, I.; Boopathi, R.; Cutter, A.R.; Papai, G.; Reymer, A.; Syed, S.H.; Lone, I.N.; Tonchev, O.; Crucifix, C.; et al. Structure and Dynamics of a 197 Bp Nucleosome in Complex with Linker Histone H1. Mol. Cell 2017, 66, 384–397.e8. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhou, B.-R.; Bai, Y. Binding Affinity and Function of the Extremely Disordered Protein Complex Containing Human Linker Histone H1.0 and Its Chaperone ProTα. Biochemistry 2018, 57, 6645–6648. [Google Scholar] [CrossRef] [PubMed]
- Müller-Späth, S.; Soranno, A.; Hirschfeld, V.; Hofmann, H.; Rüegger, S.; Reymond, L.; Nettels, D.; Schuler, B. From the Cover: Charge Interactions Can Dominate the Dimensions of Intrinsically Disordered Proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 14609–14614. [Google Scholar] [CrossRef]
- Borgia, A.; Borgia, M.B.; Bugge, K.; Kissling, V.M.; Heidarsson, P.O.; Fernandes, C.B.; Sottini, A.; Soranno, A.; Buholzer, K.J.; Nettels, D.; et al. Extreme Disorder in an Ultrahigh-Affinity Protein Complex. Nature 2018, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Sottini, A.; Borgia, A.; Borgia, M.B.; Bugge, K.; Nettels, D.; Chowdhury, A.; Heidarsson, P.O.; Zosel, F.; Best, R.B.; Kragelund, B.B.; et al. Polyelectrolyte Interactions Enable Rapid Association and Dissociation in High-Affinity Disordered Protein Complexes. Nat. Commun. 2020, 11, 5736. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Jensen, M.R.; Lazert, C.; Guseva, S.; Ivashchenko, S.; Communie, G.; Maurin, D.; Gerlier, D.; Ruigrok, R.W.H.; Blackledge, M. An Ultraweak Interaction in the Intrinsically Disordered Replication Machinery Is Essential for Measles Virus Function. Sci. Adv. 2018, 4, eaat7778. [Google Scholar] [CrossRef]
- Taylor, N.O.; Wei, M.-T.; Stone, H.A.; Brangwynne, C.P. Quantifying Dynamics in Phase-Separated Condensates Using Fluorescence Recovery after Photobleaching. Biophys. J. 2019, 117, 1285–1300. [Google Scholar] [CrossRef]
- Guseva, S.; Milles, S.; Jensen, M.R.; Salvi, N.; Kleman, J.-P.; Maurin, D.; Ruigrok, R.W.H.; Blackledge, M. Measles Virus Nucleo- and Phosphoproteins Form Liquid-like Phase-Separated Compartments That Promote Nucleocapsid Assembly. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.-P.; Sun, Y.-C.; Qiu, D.-C.; Lin, Y.-H.; Chen, Y.-Q.; Kuo, J.-C.; Huang, J.-R. Liquid-Liquid Phase Separation and Extracellular Multivalent Interactions in the Tale of Galectin-3. Nat. Commun. 2020, 11, 1229. [Google Scholar] [CrossRef] [PubMed]
- Burke, K.A.; Janke, A.M.; Rhine, C.L.; Fawzi, N.L. Residue-by-Residue View of in Vitro FUS Granules That Bind the C-Terminal Domain of RNA Polymerase II. Mol. Cell 2015, 60, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Brady, J.P.; Farber, P.J.; Sekhar, A.; Lin, Y.-H.; Huang, R.; Bah, A.; Nott, T.J.; Chan, H.S.; Baldwin, A.J.; Forman-Kay, J.D.; et al. Structural and Hydrodynamic Properties of an Intrinsically Disordered Region of a Germ Cell-Specific Protein on Phase Separation. Proc. Natl. Acad. Sci. USA 2017, 114, E8194–E8203. [Google Scholar] [CrossRef]
- Reichheld, S.E.; Muiznieks, L.D.; Keeley, F.W.; Sharpe, S. Direct Observation of Structure and Dynamics during Phase Separation of an Elastomeric Protein. Proc. Natl. Acad. Sci. USA 2017, 114, E4408–E4415. [Google Scholar] [CrossRef]
- Wong, L.E.; Kim, T.H.; Muhandiram, D.R.; Forman-Kay, J.D.; Kay, L.E. NMR Experiments for Studies of Dilute and Condensed Protein Phases: Application to the Phase-Separating Protein CAPRIN1. J. Am. Chem. Soc. 2020, 142, 2471–2489. [Google Scholar] [CrossRef]
- Yuwen, T.; Brady, J.P.; Kay, L.E. Probing Conformational Exchange in Weakly Interacting, Slowly Exchanging Protein Systems via Off-Resonance R1ρ Experiments: Application to Studies of Protein Phase Separation. J. Am. Chem. Soc. 2018, 140, 2115–2126. [Google Scholar] [CrossRef]
- Yuwen, T.; Bah, A.; Brady, J.P.; Ferrage, F.; Bouvignies, G.; Kay, L.E. Measuring Solvent Hydrogen Exchange Rates by Multifrequency Excitation 15N CEST: Application to Protein Phase Separation. J. Phys. Chem. B 2018, 122, 11206–11217. [Google Scholar] [CrossRef]
- Murthy, A.C.; Fawzi, N.L. The (Un)Structural Biology of Biomolecular Liquid-Liquid Phase Separation Using NMR Spectroscopy. J. Biol. Chem. 2020, 295, 2375–2384. [Google Scholar] [CrossRef]
- Majumdar, A.; Dogra, P.; Maity, S.; Mukhopadhyay, S. Liquid–Liquid Phase Separation Is Driven by Large-Scale Conformational Unwinding and Fluctuations of Intrinsically Disordered Protein Molecules. J. Phys. Chem. Lett. 2019, 10, 3929–3936. [Google Scholar] [CrossRef] [PubMed]
- Martin, E.W.; Holehouse, A.S.; Peran, I.; Farag, M.; Incicco, J.J.; Bremer, A.; Grace, C.R.; Soranno, A.; Pappu, R.V.; Mittag, T. Valence and Patterning of Aromatic Residues Determine the Phase Behavior of Prion-like Domains. Science 2020, 367, 694–699. [Google Scholar] [CrossRef]
- Wei, M.-T.; Elbaum-Garfinkle, S.; Holehouse, A.S.; Chen, C.C.-H.; Feric, M.; Arnold, C.B.; Priestley, R.D.; Pappu, R.V.; Brangwynne, C.P. Phase Behaviour of Disordered Proteins Underlying Low Density and High Permeability of Liquid Organelles. Nat. Chem. 2017, 9, 1118–1125. [Google Scholar] [CrossRef]
- Choi, J.-M.; Holehouse, A.S.; Pappu, R.V. Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu. Rev. Biophys. 2020, 49, 107–133. [Google Scholar] [CrossRef]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.-H.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The Disordered P Granule Protein LAF-1 Drives Phase Separation into Droplets with Tunable Viscosity and Dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin Integrates within the Nucleolus via Multi-Modal Interactions with Proteins Displaying R-Rich Linear Motifs and RRNA. eLife 2016, 5, e13571. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Cika, J.A.; Stanley, C.B.; Nourse, A.; Onuchic, P.L.; Banerjee, P.R.; Phillips, A.H.; Park, C.-G.; Deniz, A.A.; Kriwacki, R.W. Self-Interaction of NPM1 Modulates Multiple Mechanisms of Liquid-Liquid Phase Separation. Nat. Commun. 2018, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.R.; Mitrea, D.M.; Kriwacki, R.W.; Deniz, A.A. Asymmetric Modulation of Protein Order–Disorder Transitions by Phosphorylation and Partner Binding. Angew. Chem. Int. Ed. 2016, 55, 1675–1679. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, D.M.; Grace, C.R.; Buljan, M.; Yun, M.-K.; Pytel, N.J.; Satumba, J.; Nourse, A.; Park, C.-G.; Madan Babu, M.; White, S.W.; et al. Structural Polymorphism in the N-Terminal Oligomerization Domain of NPM1. Proc. Natl. Acad. Sci. USA 2014, 111, 4466–4471. [Google Scholar] [CrossRef]
- Jensen, M.R.; Communie, G.; Ribeiro, E.A.; Martinez, N.; Desfosses, A.; Salmon, L.; Mollica, L.; Gabel, F.; Jamin, M.; Longhi, S.; et al. Intrinsic Disorder in Measles Virus Nucleocapsids. Proc. Natl. Acad. Sci. USA 2011, 108, 9839–9844. [Google Scholar] [CrossRef]
- Wells, M.; Tidow, H.; Rutherford, T.J.; Markwick, P.; Jensen, M.R.; Mylonas, E.; Svergun, D.I.; Blackledge, M.; Fersht, A.R. Structure of Tumor Suppressor P53 and Its Intrinsically Disordered N-Terminal Transactivation Domain. Proc. Natl. Acad. Sci. USA 2008, 105, 5762–5767. [Google Scholar] [CrossRef]
- Kubáň, V.; Srb, P.; Štégnerová, H.; Padrta, P.; Zachrdla, M.; Jaseňáková, Z.; Šanderová, H.; Vítovská, D.; Krásný, L.; Koval’, T.; et al. Quantitative Conformational Analysis of Functionally Important Electrostatic Interactions in the Intrinsically Disordered Region of Delta Subunit of Bacterial RNA Polymerase. J. Am. Chem. Soc. 2019, 141, 16817–16828. [Google Scholar] [CrossRef]
- Ziv, G.; Haran, G. Protein Folding, Protein Collapse, and Tanford’s Transfer Model: Lessons from Single-Molecule FRET. J. Am. Chem. Soc. 2009, 131, 2942–2947. [Google Scholar] [CrossRef] [PubMed]
- Borgia, A.; Zheng, W.; Buholzer, K.; Borgia, M.B.; Schüler, A.; Hofmann, H.; Soranno, A.; Nettels, D.; Gast, K.; Grishaev, A.; et al. Consistent View of Polypeptide Chain Expansion in Chemical Denaturants from Multiple Experimental Methods. J. Am. Chem. Soc. 2016, 138, 11714–11726. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zheng, W.; Borgia, A.; Buholzer, K.; Borgia, M.B.; Hofmann, H.; Soranno, A.; Nettels, D.; Gast, K.; Grishaev, A.; et al. Comment on “Innovative Scattering Analysis Shows That Hydrophobic Disordered Proteins Are Expanded in Water”. Science 2018, 361, eaar7101. [Google Scholar] [CrossRef] [PubMed]
- Fuertes, G.; Banterle, N.; Ruff, K.M.; Chowdhury, A.; Pappu, R.V.; Svergun, D.I.; Lemke, E.A. Comment on “Innovative Scattering Analysis Shows That Hydrophobic Disordered Proteins Are Expanded in Water”. Science 2018, 361, eaau8230. [Google Scholar] [CrossRef]
- Clore, G.M.; Iwahara, J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and Their Complexes. Chem. Rev. 2009, 109, 4108–4139. [Google Scholar] [CrossRef]
- Schwieters, C.D.; Kuszewski, J.J.; Tjandra, N.; Clore, G.M. The Xplor-NIH NMR Molecular Structure Determination Package. J. Magn. Reson. 2003, 160, 65–73. [Google Scholar] [CrossRef]
- Meier, S.; Grzesiek, S.; Blackledge, M. Mapping the Conformational Landscape of Urea-Denatured Ubiquitin Using Residual Dipolar Couplings. J. Am. Chem. Soc. 2007, 129, 9799–9807. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Strohmeier, M.; Blackledge, M.; Grzesiek, S. Direct Observation of Dipolar Couplings and Hydrogen Bonds across a β-Hairpin in 8 M Urea. J. Am. Chem. Soc. 2007, 129, 754–755. [Google Scholar] [CrossRef]
- Huang, J.; Grzesiek, S. Ensemble Calculations of Unstructured Proteins Constrained by RDC and PRE Data: A Case Study of Urea-Denatured Ubiquitin. J. Am. Chem. Soc. 2010, 132, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Vajpai, N.; Gentner, M.; Huang, J.-R.; Blackledge, M.; Grzesiek, S. Side-Chain Chi(1) Conformations in Urea-Denatured Ubiquitin and Protein G from (3)J Coupling Constants and Residual Dipolar Couplings. J. Am. Chem. Soc. 2010, 132, 3196–3203. [Google Scholar] [CrossRef] [PubMed]
- Gabel, F.; Jensen, M.R.; Zaccaï, G.; Blackledge, M. Quantitative Modelfree Analysis of Urea Binding to Unfolded Ubiquitin Using a Combination of Small Angle X-Ray and Neutron Scattering. J. Am. Chem. Soc. 2009, 131, 8769–8771. [Google Scholar] [CrossRef] [PubMed]
- McCarney, E.R.; Werner, J.H.; Bernstein, S.L.; Ruczinski, I.; Makarov, D.E.; Goodwin, P.M.; Plaxco, K.W. Site-Specific Dimensions across a Highly Denatured Protein; a Single Molecule Study. J. Mol. Biol. 2005, 352, 672–682. [Google Scholar] [CrossRef]
- Guinier, A. La diffraction des rayons X aux très petits angles: Application à l’étude de phénomènes ultramicroscopiques. Ann. Phys. 1939, 11, 161–237. [Google Scholar] [CrossRef]
- Pérez, J.; Vachette, P.; Russo, D.; Desmadril, M.; Durand, D. Heat-Induced Unfolding of Neocarzinostatin, a Small All-Beta Protein Investigated by Small-Angle X-Ray Scattering. J. Mol. Biol. 2001, 308, 721–743. [Google Scholar] [CrossRef]
- Riback, J.A.; Bowman, M.A.; Zmyslowski, A.M.; Knoverek, C.R.; Jumper, J.M.; Hinshaw, J.R.; Kaye, E.B.; Freed, K.F.; Clark, P.L.; Sosnick, T.R. Innovative Scattering Analysis Shows That Hydrophobic Disordered Proteins Are Expanded in Water. Science 2017, 358, 238–241. [Google Scholar] [CrossRef] [PubMed]
- Bernadó, P.; Mylonas, E.; Petoukhov, M.V.; Blackledge, M.; Svergun, D.I. Structural Characterization of Flexible Proteins Using Small-Angle X-Ray Scattering. J Am. Chem. Soc. 2007, 129, 5656–5664. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, S.; Peulen, T.; Sindbert, S.; Rothwell, P.J.; Berger, S.; Restle, T.; Goody, R.S.; Gohlke, H.; Seidel, C.A.M. A Toolkit and Benchmark Study for FRET-Restrained High.h-Precision Structural Modeling. Nat. Methods 2012, 9, 1218–1225. [Google Scholar] [CrossRef] [PubMed]
- Dimura, M.; Peulen, T.-O.; Sanabria, H.; Rodnin, D.; Hemmen, K.; Hanke, C.A.; Seidel, C.A.M.; Gohlke, H. Automated and Optimally FRET-Assisted Structural Modeling. Nat. Commun. 2020, 11, 5394. [Google Scholar] [CrossRef]
- Dingfelder, F.; Benke, S.; Nettels, D.; Schuler, B. Mapping an Equilibrium Folding Intermediate of the Cytolytic Pore Toxin ClyA with Single-Molecule FRET. J. Phys. Chem. B 2018, 122, 11251–11261. [Google Scholar] [CrossRef] [PubMed]
- Mercier, E.; Wang, X.; Maiti, M.; Wintermeyer, W.; Rodnina, M.V. Lateral Gate Dynamics of the Bacterial Translocon during Cotranslational Membrane Protein Insertion. Proc. Natl. Acad. Sci. USA 2021, 118, e2100474118. [Google Scholar] [CrossRef] [PubMed]
- Fijen, C.; Mahmoud, M.M.; Kronenberg, M.; Kaup, R.; Fontana, M.; Towle-Weicksel, J.B.; Sweasy, J.B.; Hohlbein, J. Using Single-Molecule FRET to Probe the Nucleotide-Dependent Conformational Landscape of Polymerase β-DNA Complexes. J. Biol. Chem. 2020, 295, 9012–9020. [Google Scholar] [CrossRef] [PubMed]
- Gomes, G.-N.W.; Krzeminski, M.; Namini, A.; Martin, E.W.; Mittag, T.; Head-Gordon, T.; Forman-Kay, J.D.; Gradinaru, C.C. Conformational Ensembles of an Intrinsically Disordered Protein Consistent with NMR, SAXS and Single-Molecule FRET. J. Am. Chem. Soc. 2020, 142, 15697–15710. [Google Scholar] [CrossRef]
- Ozenne, V.; Bauer, F.; Salmon, L.; Huang, J.-R.; Jensen, M.R.; Segard, S.; Bernadó, P.; Charavay, C.; Blackledge, M. Flexible-Meccano: A Tool for the Generation of Explicit Ensemble Descriptions of Intrinsically Disordered Proteins and Their Associated Experimental Observables. Bioinformatics 2012, 28, 1463–1470. [Google Scholar] [CrossRef]
- Salmon, L.; Nodet, G.; Ozenne, V.; Yin, G.; Jensen, M.R.; Zweckstetter, M.; Blackledge, M. NMR Characterization of Long-Range Order in Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010, 132, 8407–8418. [Google Scholar] [CrossRef]
- Naudi-Fabra, S.; Tengo, M.; Jensen, M.R.; Blackledge, M.; Milles, S. Quantitative Description of Intrinsically Disordered Proteins Using Single-Molecule FRET, NMR, and SAXS. J. Am. Chem. Soc. 2021, 143, 20109–20121. [Google Scholar] [CrossRef]
- Walczewska-Szewc, K.; Deplazes, E.; Corry, B. Comparing the Ability of Enhanced Sampling Molecular Dynamics Methods To Reproduce the Behavior of Fluorescent Labels on Proteins. J. Chem. Theory Comput. 2015, 11, 3455–3465. [Google Scholar] [CrossRef] [PubMed]
- Piana, S.; Donchev, A.G.; Robustelli, P.; Shaw, D.E. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. J. Phys. Chem. B 2015, 119, 5113–5123. [Google Scholar] [CrossRef]
- König, I.; Soranno, A.; Nettels, D.; Schuler, B. Impact of In-Cell and In-Vitro Crowding on the Conformations and Dynamics of an Intrinsically Disordered Protein. Angew. Chem. Int. Ed. 2021, 60, 10724–10729. [Google Scholar] [CrossRef]
- König, I.; Zarrine-Afsar, A.; Aznauryan, M.; Soranno, A.; Wunderlich, B.; Dingfelder, F.; Stüber, J.C.; Plückthun, A.; Nettels, D.; Schuler, B. Single-Molecule Spectroscopy of Protein Conformational Dynamics in Live Eukaryotic Cells. Nat. Methods 2015, 12, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Sakon, J.J.; Weninger, K.R. Detecting the Conformation of Individual Proteins in Live Cells. Nat. Methods 2010, 7, 203–205. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naudi-Fabra, S.; Blackledge, M.; Milles, S. Synergies of Single Molecule Fluorescence and NMR for the Study of Intrinsically Disordered Proteins. Biomolecules 2022, 12, 27. https://doi.org/10.3390/biom12010027
Naudi-Fabra S, Blackledge M, Milles S. Synergies of Single Molecule Fluorescence and NMR for the Study of Intrinsically Disordered Proteins. Biomolecules. 2022; 12(1):27. https://doi.org/10.3390/biom12010027
Chicago/Turabian StyleNaudi-Fabra, Samuel, Martin Blackledge, and Sigrid Milles. 2022. "Synergies of Single Molecule Fluorescence and NMR for the Study of Intrinsically Disordered Proteins" Biomolecules 12, no. 1: 27. https://doi.org/10.3390/biom12010027
APA StyleNaudi-Fabra, S., Blackledge, M., & Milles, S. (2022). Synergies of Single Molecule Fluorescence and NMR for the Study of Intrinsically Disordered Proteins. Biomolecules, 12(1), 27. https://doi.org/10.3390/biom12010027





