A Distinctive γδ T Cell Repertoire in NOD Mice Weakens Immune Regulation and Favors Diabetic Disease
Abstract
:1. Introduction
2. Materials and Methods (Also See Supplemental Detailed Materials and Methods)
2.1. Mouse Strains
2.2. Genetic Screening of Mutant Mouse Strains
2.3. Diabetes Monitoring
2.4. Cell Preparation
2.5. Flow Cytometry
2.6. Statistics
3. Results
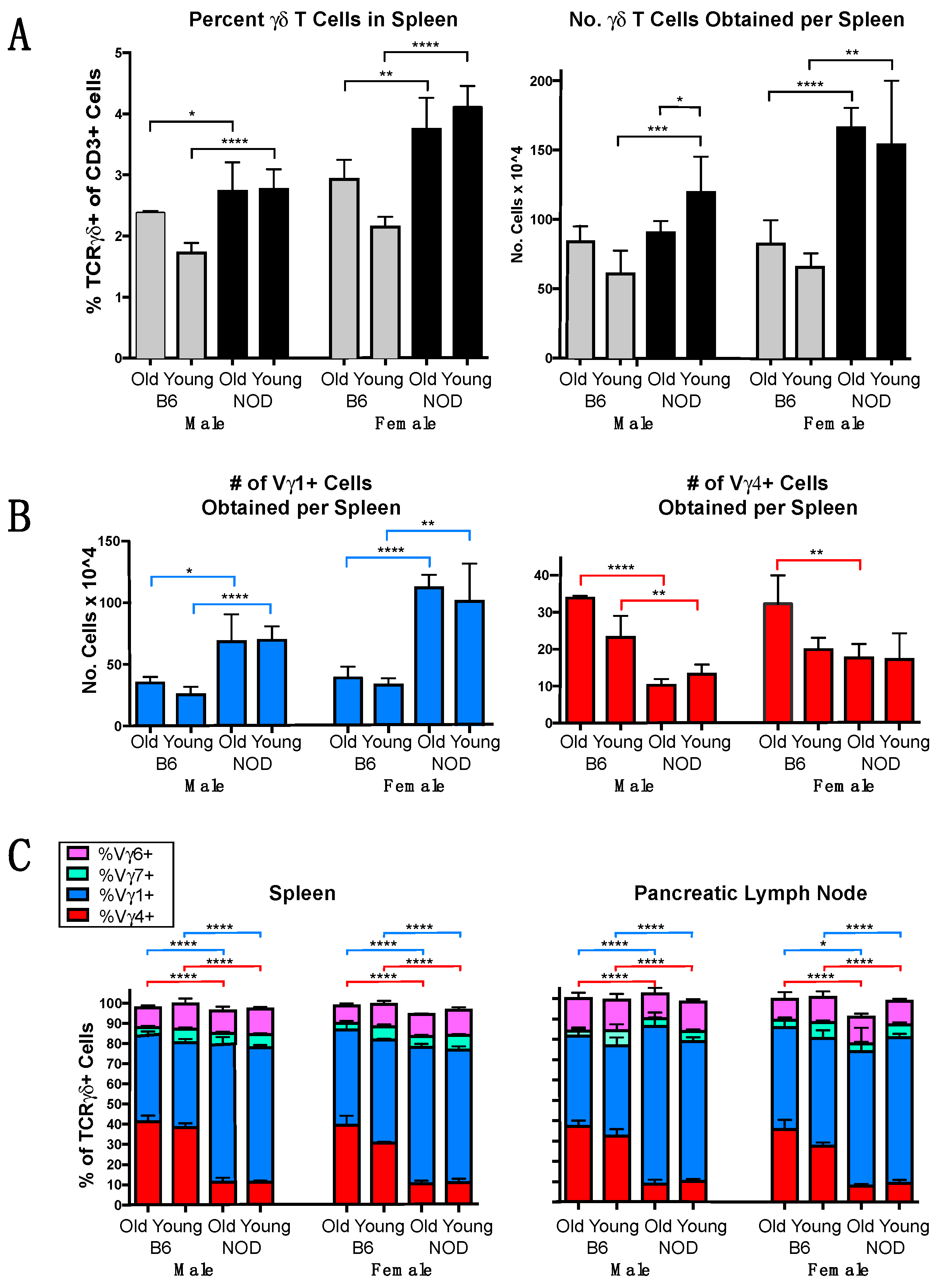
3.1. The γδ TCR Repertoire of NOD Mice Is Biased in Favor of Vγ1+ Cells
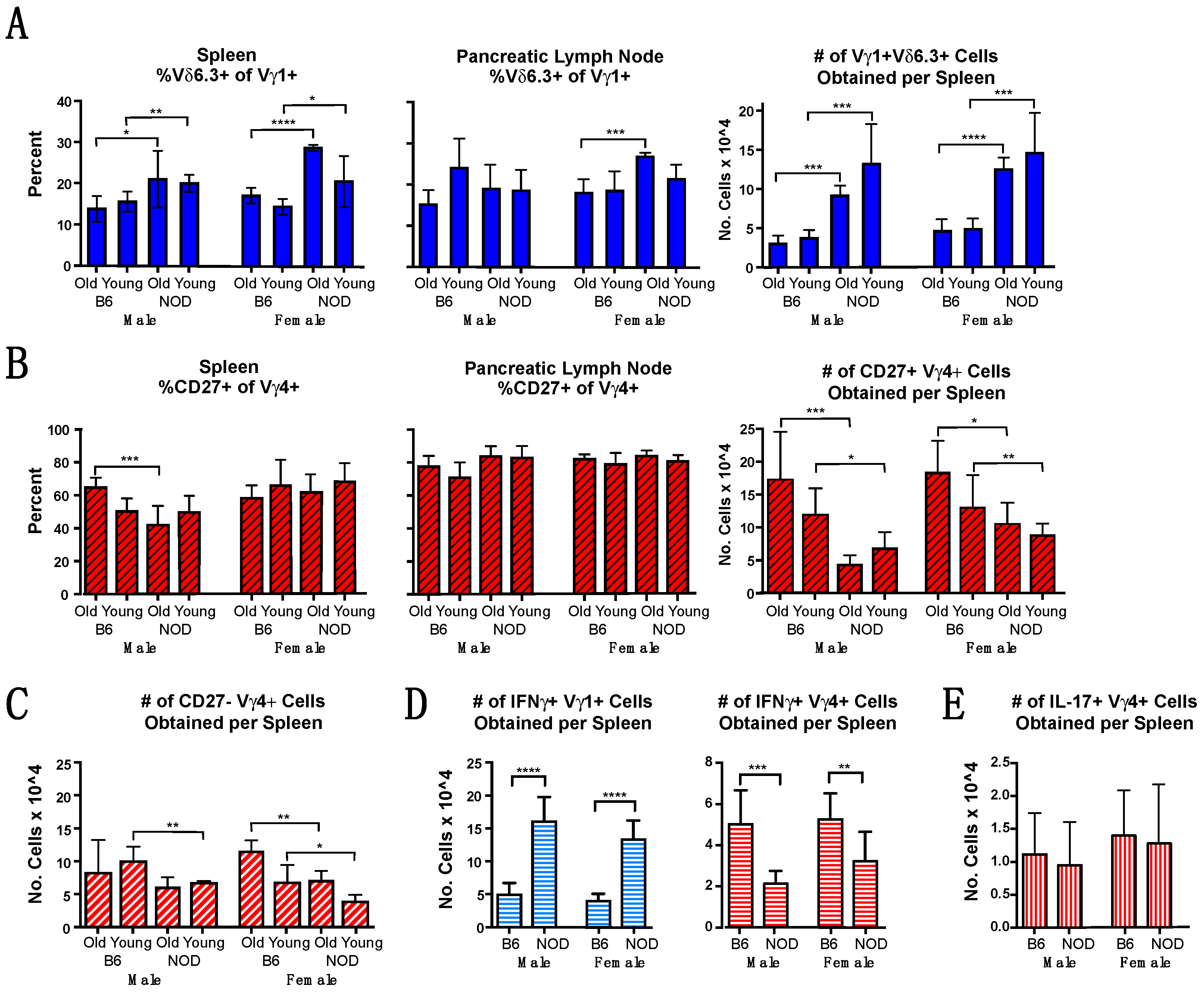
3.2. Vγ1+ and Vγ4+ γδ T Cell Subsets in NOD Mice Differ from Those in B6 Mice in Cellular Composition and Functional Bias
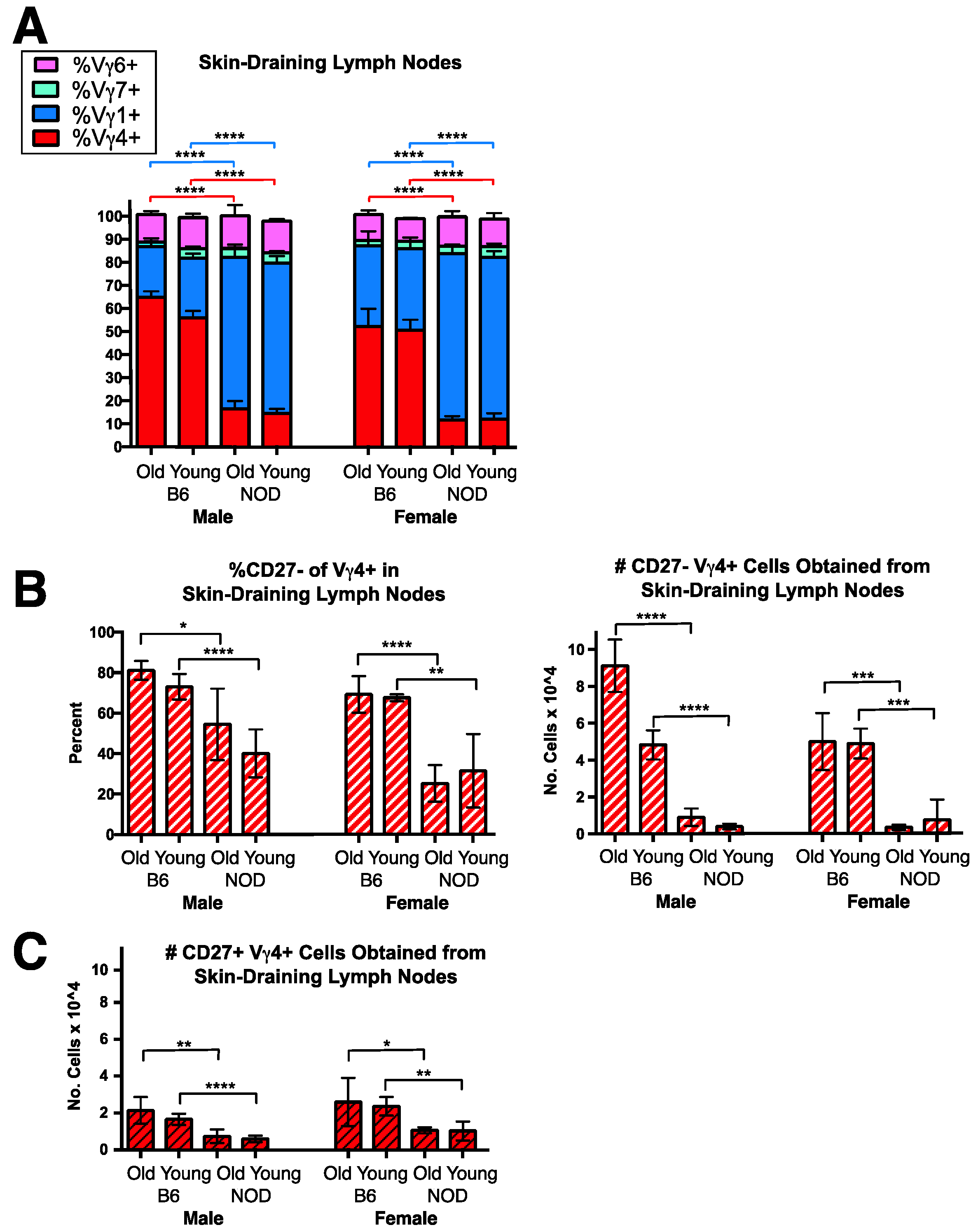
3.3. A Conspicuous Deficiency in NOD Mice of Vγ4+ IL-17-Biased Cells in Skin-Draining Lymph Nodes
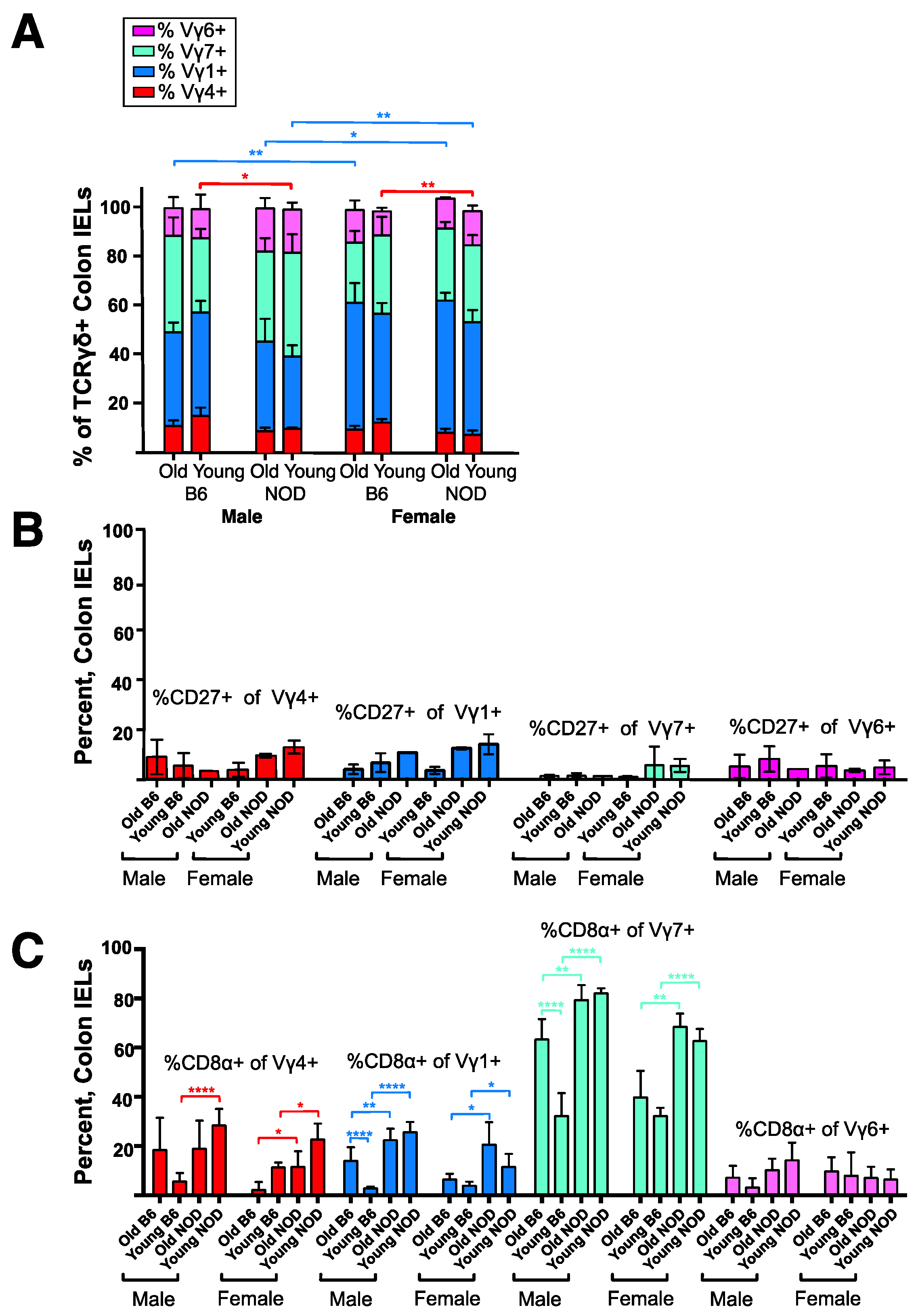
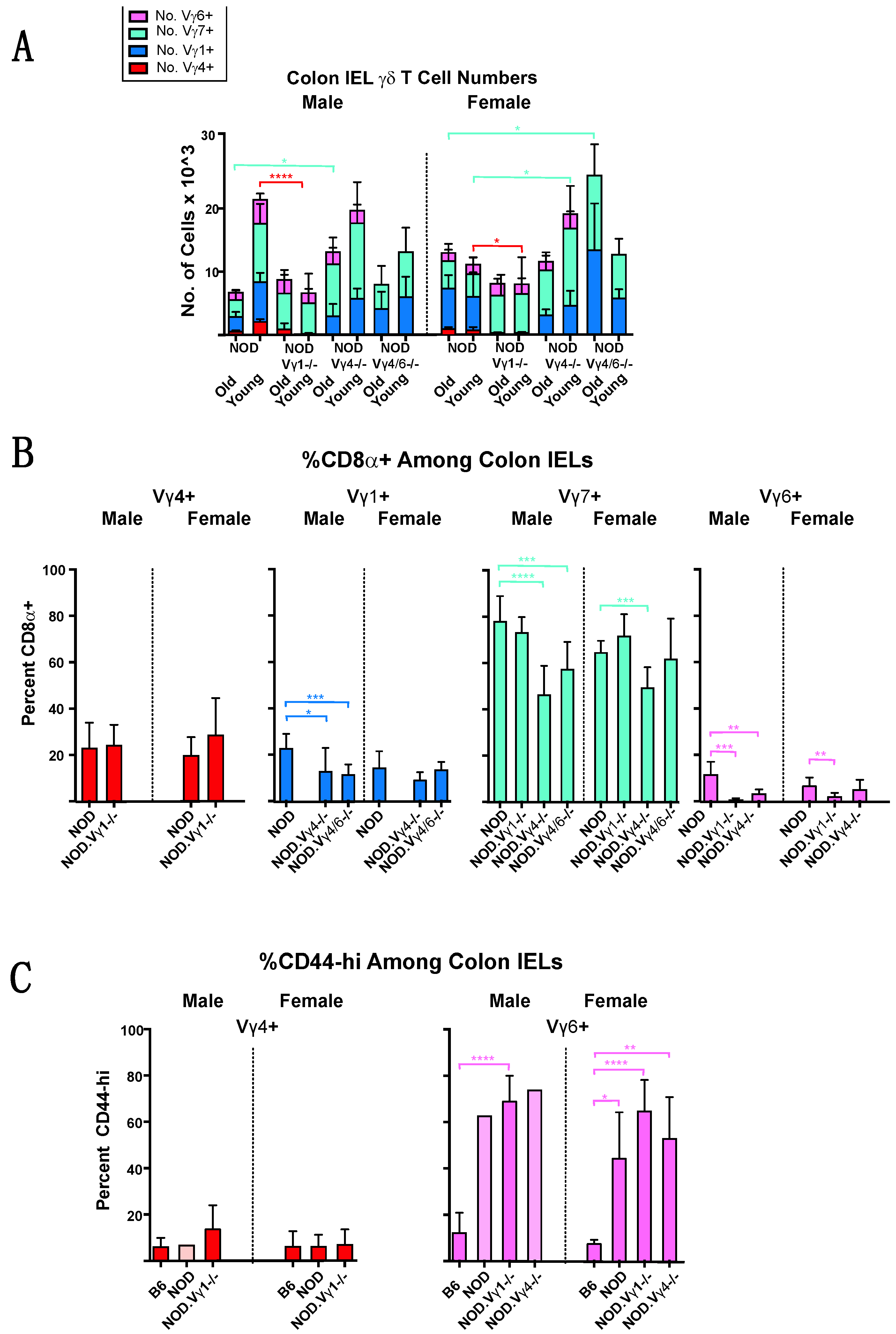
3.4. Intestinal Intraepithelial Lymphocyte (IEL) γδ T Cells Differ in CD8α Expression in NOD vs. B6 Mice
3.5. NOD Mice Lacking Vγ1+ Cells Are Less Prone to Develop Diabetes, Whereas Those Deficient in Vγ4+ Cells Develop Accelerated Disease
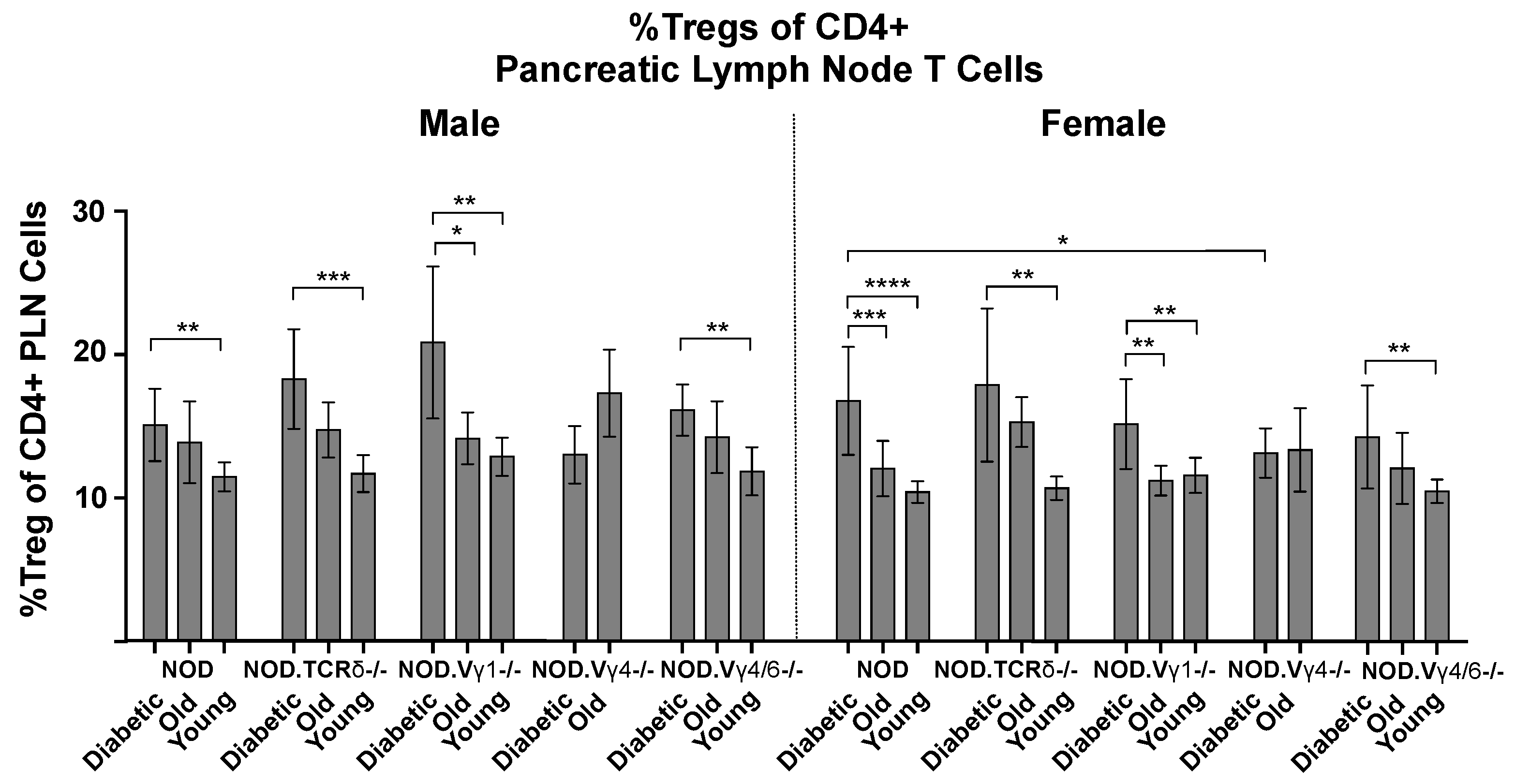
3.6. Vγ4+ γδ T Cells in NOD Mice May Provide Protection by Promoting CD4+ Tregs
3.7. Changes in NOD γδ T Cell Populations Associated with the Development of Diabetes, and Evidence for Crosstalk between γδ T Cell Subsets
4. Discussion
4.1. Protective γδ T Cells in NOD Diabetes
4.2. Pathogenic γδ T Cells in NOD Diabetes
4.3. Elimination of Particular γδ T Cell Subsets Precipitates Changes in Other T Cells
4.4. The Role of γδ T Cells in Human Type 1 Diabetes
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kachapati, K.; Adams, D.; Bednar, K.; Ridgway, W.M. The non-obese diabetic (NOD) mouse as a model of human type 1 diabetes. Methods Mol. Biol. 2012, 933, 3–16. [Google Scholar] [PubMed]
- Driver, J.P.; Chen, Y.G.; Mathews, C.E. Comparative genetics: Synergizing human and NOD mouse studies for identifying genetic causation of type 1 diabetes. Rev. Diabet. Stud. 2012, 9, 169–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, J.A.; Wong, F.S.; Wen, L. The importance of the Non Obese Diabetic (NOD) mouse model in autoimmune diabetes. J. Autoimmun. 2016, 66, 76–88. [Google Scholar] [CrossRef] [Green Version]
- Harrison, L.C.; Dempsey-Collier, M.; Kramer, D.R.; Takahashi, K. Aerosol insulin induced regulatory CD8 γδ T cells that prevent murine insulin-dependent diabetes. J. Exp. Med. 1996, 184, 2167–2174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, N.R.; Stankovic, S.; Funda, D.P.; Harrison, L.C. TCR γδ intraepithelial lymphocytes are required for self-tolerance. J. Immunol. 2006, 176, 6553–6559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, G.; Wang, R.; Chen, G.; Wang, J.; Xu, R.; Wang, L.; Feng, J.; Li, X.; Guo, R.; Fu, L.; et al. Interleukin-17-producing γδ T cells protect NOD mice from type 1 diabetes through a mechnism involving transforming gowth factor-β. Immunology 2009, 129, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Markle, J.G.; Mortin-Toth, S.; Wong, A.S.; Geng, L.; Hayday, A.; Danska, J.S. γδ T cells are essential effectors of type 1 diabetes in the nonobese diabetic mouse model. J. Immunol. 2013, 190, 5392–5401. [Google Scholar] [CrossRef] [Green Version]
- Huber, S.A.; Graveline, D.; Newell, M.K.; Born, W.K.; O’Brien, R.L. Vγ1+ T cells suppress and Vγ4+ T cells promote susceptibility to coxsackievirus B3-induced myocarditis in mice. J. Immunol. 2000, 165, 4174–4181. [Google Scholar] [CrossRef] [Green Version]
- Hahn, Y.S.; Taube, C.; Jin, N.; Sharp, L.; Wands, J.M.; Aydintug, M.K.; Lahn, M.; Huber, S.A.; O’Brien, R.L.; Gelfand, E.W.; et al. Different potential of γδ T cell subsets in regulating airway responsiveness: Vγ1+ cells, but not Vγ4+ cells, promote airway hyperreactivity, Th-2 cytokines and airway inflammation. J. Immunol. 2004, 172, 12894–12902. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Hao, J.; Dong, S.; Gao, Y.; Tao, J.; Chi, H.; Flavell, R.; O’Brien, R.L.; Born, W.K.; Craft, J.; et al. Naturally activated Vγ4 γδ T cells play a protective role in tumor immunity through expression of eomesodermin. J. Immunol. 2010, 185, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Narayan, K.; Sylvia, K.E.; Malhotra, N.; Yin, C.C.; Martens, G.; Vallerskog, T.; Kornfeld, H.; Xiong, N.; Cohen, N.R.; Brenner, M.B.; et al. Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat. Immunol. 2012, 13, 511–518. [Google Scholar] [CrossRef] [Green Version]
- Blink, S.E.; Caldis, M.W.; Goings, G.E.; Harp, C.T.; Malissen, B.; Prinz, I.; Xu, D.; Miller, S.D. γδ T cell subsets play opposing roles in regulating experimental autoimmune encephalomyelitis. Cell. Immunol. 2014, 290, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, N.; Vegh, P.; Rothenberg, E.V.; Yui, M.A. Lineage divergence at the first TCR-dependent checkpoint: Preferential γδ and impaired αβ T cell development in nonobese diabetic mice. J. Immunol. 2011, 186, 826–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilig, J.S.; Tonegawa, S. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature 1986, 322, 836–840. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, R.L.; Yin, X.; Huber, S.A.; Ikuta, K.; Born, W.K. Depletion of a γδ T cell subset can increase host resistance to a bacterial infection. J. Immunol. 2000, 165, 6472–6479. [Google Scholar] [CrossRef] [Green Version]
- Roark, C.L.; French, J.D.; Taylor, M.A.; Bendele, A.M.; Born, W.K.; O’Brien, R.L. Exacerbation of collagen-induced arthritis by oligoclonal, IL-17-producing γδ T cells. J. Immunol. 2007, 179, 5576–5583. [Google Scholar] [CrossRef] [Green Version]
- Hao, J.; Dong, S.; Xia, S.; He, W.; Jia, H.; Zhang, S.; Wei, J.; O’Brien, R.L.; Born, W.K.; Wu, Z.; et al. Regulatory role of Vγ1 γδ T cells in tumor immunity through IL-4 production. J. Immunol. 2011, 187, 4979–4986. [Google Scholar] [CrossRef] [Green Version]
- Itohara, S.; Mombaerts, P.; Lafaille, J.; Iacomini, J.; Nelson, A.; Clarke, A.R.; Hooper, M.L.; Farr, A.; Tonegawa, S. T cell receptor δ gene mutant mice: Independent generation of αβ T cells and programmed rearrangements of γδ TCR genes. Cell 1993, 72, 337–348. [Google Scholar] [CrossRef]
- Mombaerts, P.; Arnoldi, J.; Russ, F.; Tonegawa, S.; Kaufmann, S.H.E. Different roles of αβ and γδ T cells in immunity against an intracellular bacterial pathogen. Nature 1993, 365, 53–56. [Google Scholar] [CrossRef]
- Andrew, E.M.; Newton, D.J.; Dalton, J.E.; Egan, C.E.; Goodwin, S.J.; Tramonti, D.; Scott, P.; Carding, S.R. Delineation of the function of a major γδ T cell subset during infection. J. Immunol. 2005, 175, 1741–1750. [Google Scholar] [CrossRef] [Green Version]
- Sunaga, S.; Maki, K.; Komagata, Y.; Miyazaki, J.-I.; Ikuta, K. Developmentally ordered V-J recombination in mouse T cell receptor γ locus is not perturbed by targeted deletion of the Vγ4 gene. J. Immunol. 1997, 158, 4223–4228. [Google Scholar] [PubMed]
- French, J.D.; Roark, C.L.; Born, W.K.; O’Brien, R.L. γδ T lymphocyte homeostasis is negatively regulated by β2-microglobulin. J. Immunol. 2009, 182, 1892–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigmann, B.; Tubbe, I.; Seidel, D.; Nicolaev, A.; Becker, C.; Neurath, M.F. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2007, 2, 2307–2311. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.; Gerber, D.; Huang, S.Y.; Tonegawa, S. Ontogenic development and tissue distribution of Vγ1-expressing γ/δ T lymphocytes in normal mice. J. Exp. Med. 1995, 182, 1921–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, P.; Boucontet, L. Rates of recombination and chain pair biases greatly influence the primary γδ TCR repertoire in the thymus of adult mice. J. Immunol. 2004, 173, 3261–3270. [Google Scholar] [CrossRef] [Green Version]
- Dent, A.L.; Matis, L.A.; Hooshmand, F.; Widacki, S.M.; Bluestone, J.A.; Hedrick, S.M. Self-reactive γδ cells are eliminated in the thymus. Nature 1990, 343, 714–719. [Google Scholar] [CrossRef]
- Havran, W.L.; Grell, S.; Duwe, G.; Kimura, J.; Wilson, A.; Kruisbeek, A.M.; O’Brien, R.L.; Born, W.; Tigelaar, R.E.; Allison, J.P. Limited diversity of TCR γ-chain expression of murine Thy-1+ dendritic epidermal cells revealed by Vγ3-specific monoclonal antibody. Proc. Natl. Acad. Sci. USA 1989, 86, 4185–4189. [Google Scholar] [CrossRef] [Green Version]
- Goodman, T.; LeCorre, R.; Lefrancois, L. A T-cell receptor γδ-specific monoclonal antibody detects a Vγ5 region polymorphism. Immunogenetics 1992, 35, 65–68. [Google Scholar] [CrossRef]
- Hatano, S.; Tun, X.; Noguchi, N.; Yue, D.; Yamada, H.; Sun, X.; Matsumoto, M.; Yoshikai, Y. Development of a new monoclonal antibody specific to mouse Vγ6 chain. Life Sci. Alliance 2019, 2, e201900363. [Google Scholar] [CrossRef] [Green Version]
- Pereira, P.; Gerber, D.; Regnault, A.; Huang, S.Y.; Hermitte, V.; Coutinho, A.; Tonegawa, S. Rearrangement and expression of Vγ1, Vγ2 and Vγ3 TCR γ genes in C57BL/6 mice. Internat. Immunol. 1996, 8, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Asarnow, D.M.; Kuziel, W.A.; Bonyhadi, M.; Tigelaar, R.E.; Tucker, P.W.; Allison, J.P. Limited diversity of γδ antigen receptor genes of Thy-1+ dendritic epidermal cells. Cell 1988, 55, 837–847. [Google Scholar] [CrossRef]
- Havran, W.L.; Chien, Y.H.; Allison, J.P. Recognition of self antigens by skin-derived T cells with invariant γδ antigen receptors. Science 1991, 252, 1430–1432. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.J.; Azuara, V.; Levraud, J.P.; Huang, S.Y.; Lembezat, M.P.; Pereira, P. IL-4-producing γδ T cells that express a very restricted TCR repertoire are preferentially localized in liver and spleen. J. Immunol. 1999, 163, 3076–3082. [Google Scholar]
- Kreslavsky, T.; Savage, A.K.; Hobbs, R.; Gounari, F.; Bronson, R.; Pereira, P.; Pandolfi, P.P.; Bendelac, A.; von Boehmer, H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of γδ T cells with restricted TCR diversity. Proc. Natl. Acad. Sci. USA 2009, 106, 12453–12458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pereira, P.; Berthault, C.; Burlen-Defranoux, O.; Boucontet, L. Critical role of TCR specificity in the development of Vγ1Vδ6.3+ innate NKTγδ cells. J. Immunol. 2013, 191, 1716–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kisielow, J.; Kopf, M.; Karjalainen, K. SCART scavenger receptors identify a novel subset of adult γδ T cells. J. Immunol. 2008, 181, 1710–1716. [Google Scholar] [CrossRef] [Green Version]
- Hamada, S.; Umemura, M.; Shiono, T.; Tanaka, K.; Yahagi, A.; Begum, M.D.; Oshiro, K.; Okamoto, Y.; Watanabe, H.; Kawakami, K.; et al. IL-17A produced by γδ T cells plays a critical role in innate immunity against Listeria monocytogenes infection in the liver. J. Immunol. 2008, 181, 3456–3463. [Google Scholar] [CrossRef] [Green Version]
- Haas, J.D.; Gonzalez, F.H.; Schmitz, S.; Chennupati, V.; Fohse, L.; Kremmer, E.; Forster, R.; Prinz, I. CCR6 and NK1.1 distinguish between IL-17A and IFN-γ-producing γδ effector T cells. Eur. J. Immunol. 2009, 39, 3488–3497. [Google Scholar] [CrossRef]
- Okamoto-Yoshida, Y.; Umemura, M.; Yahagi, A.; O’Brien, R.L.; Ikuta, K.; Kishihara, K.; Hara, H.; Nakae, S.; Iwakura, Y.; Matsuzaki, G. Essential role of interleukin-17A in the formation of a mycobacterial infection-induced granuloma in the lung. J. Immunol. 2010, 184, 4414–4422. [Google Scholar] [CrossRef] [Green Version]
- Gray, E.E.; Suzuki, K.; Cyster, J.G. Cutting edge: Identification of a motile IL-17-producing γδ T cell population in the dermis. J. Immunol. 2011, 186, 6091–6095. [Google Scholar] [CrossRef] [Green Version]
- Roark, C.L.; Huang, Y.; Jin, N.; Aydintug, M.K.; Casper, T.; Sun, D.; Born, W.K.; O’Brien, R.L. A canonical Vγ4Vδ4+ γδ T cell population with distinct stimulation requirements which promotes the Th17 response. Immunol. Res. 2013, 55, 217–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashani, E.; Fohse, L.; Raha, S.; Sandrock, I.; Oberdorfer, L.; Koenecke, C.; Suerbaum, S.; Weiss, S.; Prinz, I. A clonotypic Vγ4Jγ1/Vδ5Dδ2Jδ1 innate γδ T-cell population restricted to the CCR6+ CD27-subset. Nat. Commun. 2015, 6, 6477. [Google Scholar] [CrossRef] [Green Version]
- Ribot, J.C.; de Barros, A.; Fang, D.J.; Neves, J.F.; Peperzak, V.; Roberts, S.J.; Girardi, M.; Borst, J.; Hayday, A.C.; Pennington, D.J.; et al. CD27 is a thymic determinant of the balance between interferon-γ and interleukin 17-producing γδ T cell subsets. Nat. Immunol. 2009, 10, 427–436. [Google Scholar] [CrossRef] [Green Version]
- Sumaria, N.; Roediger, B.; Ng, L.G.; Qin, J.; Pinto, R.; Cavanagh, L.L.; Shklovskaya, E.; Fazekas de St Groth, B.; Triccas, J.A.; Weninger, W. Cutaneous immunosurveillance by self-renewing dermal γδ T cells. J. Exp. Med. 2011, 208, 505–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Shen, X.; Ding, C.; Qi, C.; Li, K.; Li, X.; Jala, V.R.; Zhang, H.G.; Wang, T.; Zheng, J.; et al. Pivotal role of dermal IL-17-producing γδ T cells in skin inflammation. Immunity 2011, 35, 596–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez-Valle, F.; Gray, E.E.; Cyster, J.G. Inflammation induces dermal Vγ4+ γδ T17 memory-like cells that travel to distant skin and accelerate secondary IL-17-driven responses. Proc. Natl. Acad. Sci. USA 2015, 112, 8046–8051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, L.; Sandrock, I.; Odak, I.; Aizenbud, Y.; Wilharm, A.; Barros-Martins, J.; Tabib, Y.; Borchers, A.; Amado, T.; Gangoda, L.; et al. Single-cell transcriptomics identifies the adaptation of Scart1(+) Vγ6(+) T cells to skin residency as activated effector cells. Cell Rep. 2019, 27, 3657–3671.e3654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegel, M.A.; Sefik, E.; Hill, J.A.; Wu, H.J.; Benoist, C.; Mathis, D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 11548–11553. [Google Scholar] [CrossRef] [Green Version]
- Markle, J.G.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [CrossRef] [Green Version]
- Sorini, C.; Cosorich, I.; Lo Conte, M.; De Giorgi, L.; Facciotti, F.; Lucianò, R.; Rocchi, M.; Ferrarese, R.; Sanvito, F.; Canducci, F.; et al. Loss of gut barrier integrity triggers activation of islet-reactive T cells and autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2019, 116, 15140–15149. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Tao, W.; Zhu, S. T lymphocytes in the intestinal mucosa: Defense and tolerance. Cell. Mol. Immunol. 2019, 16, 216–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Heiser, R.A.; Detanico, T.O.; Getahun, A.; Kirchenbaum, G.A.; Casper, T.L.; Aydintug, M.K.; Carding, S.R.; Ikuta, K.; Huang, H.; et al. γδ T cells affect IL-4 production and B-cell tolerance. Proc. Natl. Acad. Sci. USA 2015, 112, E39–E48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Getahun, A.; Heiser, R.A.; Detanico, T.O.; Aviszus, K.; Kirchenbaum, G.A.; Casper, T.L.; Huang, C.; Aydintug, M.K.; Carding, S.R.; et al. γδ T cells shape preimmune peripheral B cell populations. J. Immunol. 2016, 196, 217–231. [Google Scholar] [CrossRef] [Green Version]
- Daley, D.; Zambirinis, C.P.; Seifert, L.; Akkad, N.; Mohan, N.; Werba, G.; Barilla, R.; Torres-Hernandez, A.; Hundeyin, M.; Mani, V.R.; et al. γδ T cells support pancreatic oncogenesis by restraining αβ T cell activation. Cell 2016, 166, 1485–1499.e1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, A.E.; Freeman, G.J.; Mathis, D.; Benoist, C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 2004, 199, 1479–1489. [Google Scholar] [CrossRef]
- Spence, A.; Tang, Q. Restoring regulatory T cells in Type 1 Diabetes. Curr. Diab. Rep. 2016, 16, 110. [Google Scholar] [CrossRef] [Green Version]
- Aydintug, M.K.; Zhang, L.; Wang, C.; Liang, D.; Wands, J.M.; Michels, A.W.; Hirsch, B.; Day, B.J.; Zhang, G.; Sun, D.; et al. γδ T cells recognize the insulin B: 9–23 peptide antigen when it is dimerized through thiol oxidation. Mol. Immunol. 2014, 60, 116–128. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Xue, F.; Fleming, C.; Yang, J.; Ding, C.; Ma, Y.; Liu, M.; Zhang, H.G.; Zheng, J.; Xiong, N.; et al. Differential developmental requirement and peripheral regulation for dermal Vγ4 and Vγ6 T17 cells in health and inflammation. Nat. Commun. 2014, 5, 3986. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Cao, X.; Zhang, X.; Kovalovsky, D. PLZF controls the development of fetal-derived IL-17+Vγ6+ γδ T cells. J. Immunol. 2015, 195, 4273–4281. [Google Scholar] [CrossRef] [Green Version]
- Nikoopour, E.; Schwartz, J.A.; Huszarik, K.; Sandrock, C.; Krougly, O.; Lee-Chan, E.; Singh, B. Th17 polarized cells from nonobese diabetic mice following mycobacterial adjuvant immunotherapy delay type 1 diabetes. J. Immunol. 2010, 184, 4779–4788. [Google Scholar] [CrossRef]
- Lau, K.; Benitez, P.; Ardissone, A.; Wilson, T.D.; Collins, E.L.; Lorca, G.; Li, N.; Sankar, D.; Wasserfall, C.; Neu, J.; et al. Inhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J. Immunol. 2011, 186, 3538–3546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bellemore, S.M.; Nikoopour, E.; Schwartz, J.A.; Krougly, O.; Lee-Chan, E.; Singh, B. Preventative role of interleukin-17 producing regulatory T helper type 17 (Treg 17) cells in type 1 diabetes in non-obese diabetic mice. Clin. Exp. Immunol. 2015, 182, 261–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnan, S.; Prise, I.E.; Wemyss, K.; Schenck, L.P.; Bridgeman, H.M.; McClure, F.A.; Zangerle-Murray, T.; O’Boyle, C.; Barbera, T.A.; Mahmood, F.; et al. Amphiregulin-producing γδ T cells are vital for safeguarding oral barrier immune homeostasis. Proc. Natl. Acad. Sci. USA 2018, 115, 10738–10743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.J.; Dash, P.; Crawford, J.C.; Allen, E.K.; Zamora, A.E.; Boyd, D.F.; Duan, S.; Bajracharya, R.; Awad, W.A.; Apiwattanakul, N.; et al. Lung γδ T cells mediate protective responses during neonatal influenza infection that are associated with Type 2 immunity. Immunity 2018, 49, 531–544.e536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlgruber, A.C.; Gal-Oz, S.T.; LaMarche, N.M.; Shimazaki, M.; Duquette, D.; Nguyen, H.N.; Mina, A.I.; Paras, T.; Tavakkoli, A.; von Andrian, U.; et al. γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol. 2018, 19, 464–474. [Google Scholar] [CrossRef]
- Zaiss, D.M.; van Loosdregt, J.; Gorlani, A.; Bekker, C.P.; Gröne, A.; Sibilia, M.; van Bergen en Henegouwen, P.M.; Roovers, R.C.; Coffer, P.J.; Sijts, A.J. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity 2013, 38, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, C.; Rezende, R.M.; da Cunha, A.P.; Valette, F.; Quintana, F.J.; Chatenoud, L.; Weiner, H.L. Mucosal administration of CD3-specific monoclonal antibody inhibits diabetes in NOD mice and in a preclinical mouse model transgenic for the CD3 epsilon chain. J. Autoimmun. 2017, 76, 115–122. [Google Scholar] [CrossRef]
- Rezende, R.M.; da Cunha, A.P.; Kuhn, C.; Rubino, S.; M’Hamdi, H.; Gabriely, G.; Vandeventer, T.; Liu, S.; Cialic, R.; Pinheiro-Rosa, N.; et al. Identification and characterization of latency-associated peptide-expressing γδ T cells. Nat. Commun. 2015, 6, 8726. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Yang, Z.; Huang, C.; McGowan, J.; Casper, T.; Sun, D.; Born, W.K.; O’Brien, R.L. γδ T cell-dependent regulatory T cells prevent the development of autoimmune keratitis. J. Immunol. 2015, 195, 5572–5581. [Google Scholar] [CrossRef] [Green Version]
- Nagy, N.; Kuipers, H.F.; Marshall, P.L.; Wang, E.; Kaber, G.; Bollyky, P.L. Hyaluronan in immune dysregulation and autoimmune diseases. Matrix Biol. 2019, 78–79, 292–313. [Google Scholar] [CrossRef]
- Li, C.R.; Mueller, E.E.; Bradley, L.M. Targeting CD44 augments the efficacy of Tregs in autoimmune diabetes. Immunol. Lett. 2015, 163, 199–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollyky, P.L.; Bogdani, M.; Bollyky, J.B.; Hull, R.L.; Wight, T.N. The role of hyaluronan and the extracellular matrix in islet inflammation and immune regulation. Curr. Diab. Rep. 2012, 12, 471–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, J.; Stanko, K.; Schliesser, U.; Appelt, C.; Sawitzki, B. Differences in CD44 surface expression levels and function discriminates IL-17 and IFN-γ producing helper T cells. PLoS ONE 2015, 10, e0132479. [Google Scholar]
- Tard, C.; Rouxel, O.; Lehuen, A. Regulatory role of natural killer T cells in diabetes. Biomed. J. 2015, 38, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Jiao, Y.; Xiong, Q.; Wang, C.Y.; Gerling, I.; Gu, W. Genetic and molecular basis of QTL of diabetes in mouse: Genes and polymorphisms. Curr. Genom. 2008, 9, 324–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Jin, N.; Nakayama, M.; O’Brien, R.L.; Eisenbarth, G.S.; Born, W.K. γδ T cell receptors confer autonomous responsiveness to the insulin-peptide B: 9–23. J. Autoimmun. 2010, 34, 478–484. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Nakayama, M.; Eisenbarth, G.S. Insulin as an autoantigen in NOD/human diabetes. Curr. Opin. Immunol. 2008, 20, 111–118. [Google Scholar] [CrossRef] [Green Version]
- Lang, F.P.; Pollock, B.H.; Riley, W.J.; Maclaren, N.K.; Barrett, D.J. The temporal association between γδ T cells and the natural history of insulin-dependent diabetes. J. Autoimmun. 1993, 6, 107–119. [Google Scholar] [CrossRef]
- Gyarmati, J.; Szekeres-Bartho, J.; Fischer, B.; Soltesz, G. Fetal type lymphocytes in insulin dependent diabetes mellitus. Autoimmunity 1999, 30, 63–69. [Google Scholar] [CrossRef]
- Kretowski, A.; Mysliwiec, J.; Szelachowska, M.; Turowski, D.; Wysocka, J.; Kowalska, I.; Kinalska, I. γδ T-cells alterations in the peripheral blood of high risk diabetes type 1 subjects with subclinical pancreatic B-cells impairment. Immunol. Lett. 1999, 68, 289–293. [Google Scholar] [CrossRef]
- Kretowski, A.; Mysliwiec, J.; Kinalska, I. Abnormal distribution of γδ T lymphocytes in Graves’ disease and insulin-dependent diabetes type 1. Arch. Immunol. Ther. Exp. 2000, 48, 39–42. [Google Scholar]
- Lang, F.P.; Schatz, D.A.; Pollock, B.H.; Riley, W.J.; Maclaren, N.K.; Dumont-Driscoll, M.; Barrett, D.J. Increased T lymphocytes bearing the γδ T cell receptor in subjects at high risk for insulin dependent diabetes. J. Autoimmun. 1991, 4, 925–933. [Google Scholar] [CrossRef]
- Zubkiewicz-Kucharska, A.; Noczynska, A. Abnormal Distribution of γδ T lymphocytes and their subsets in Type 1 diabetes. Adv. Clin. Exp. Med. 2016, 25, 665–6671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishell, R.I.; Dutton, R.W. Immunization of dissociated spleen cell cultures from normal mice. J. Exp. Med. 1967, 67, 423–442. [Google Scholar] [CrossRef] [Green Version]
- Goodman, T.; Lefrancois, L. Intraepithelial lymphocytes: Anatomical site, not T cell receptor form, dictates phenotype and function. J. Exp. Med. 1989, 170, 1569–1581. [Google Scholar] [CrossRef] [Green Version]
- Belles, C.; Kuhl, A.L.; Donoghue, A.J.; Sano, Y.; O’Brien, R.L.; Born, W.; Bottomly, K.; Carding, S.R. Bias in the γδ T cell response to Listeria monocytogenes. Vδ6.3+ cells are a major component of the γδ T cell response to Listeria monocytogenes. J. Immunol. 1996, 156, 4280–4289. [Google Scholar]
- Dialynas, D.P.; Quan, Z.S.; Wall, K.A.; Pierres, A.; Quintans, J.; Loken, M.R.; Pierres, M.; Fitch, F.W. Characterization of the murine T cell surface molecule, designated L3T4, identified by monoclonal antibody GK1.5: Similarity of L3T4 to the human Leu-3/T4 molecule. J. Immunol. 1983, 131, 2445–2451. [Google Scholar]
- Unkeless, J.C. Characterization of a monoclonal antibody directed against mouse macrophage and lymphocyte Fc receptors. J. Exp. Med. 1979, 150, 580–588. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, R.L.; Fu, Y.-X.; Cranfill, R.; Dallas, A.; Reardon, C.; Lang, J.; Carding, S.R.; Kubo, R.; Born, W. Heat shock protein Hsp-60 reactive γδ cells: A large, diversified T lymphocyte subset with highly focused specificity. Proc. Natl. Acad. Sci. USA 1992, 89, 4348–4352. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Brien, R.L.; Matsuda, J.; Aydintug, M.K.; Jin, N.; Phalke, S.; Born, W.K. A Distinctive γδ T Cell Repertoire in NOD Mice Weakens Immune Regulation and Favors Diabetic Disease. Biomolecules 2022, 12, 1406. https://doi.org/10.3390/biom12101406
O’Brien RL, Matsuda J, Aydintug MK, Jin N, Phalke S, Born WK. A Distinctive γδ T Cell Repertoire in NOD Mice Weakens Immune Regulation and Favors Diabetic Disease. Biomolecules. 2022; 12(10):1406. https://doi.org/10.3390/biom12101406
Chicago/Turabian StyleO’Brien, Rebecca L., Jennifer Matsuda, M. Kemal Aydintug, Niyun Jin, Swati Phalke, and Willi K. Born. 2022. "A Distinctive γδ T Cell Repertoire in NOD Mice Weakens Immune Regulation and Favors Diabetic Disease" Biomolecules 12, no. 10: 1406. https://doi.org/10.3390/biom12101406
APA StyleO’Brien, R. L., Matsuda, J., Aydintug, M. K., Jin, N., Phalke, S., & Born, W. K. (2022). A Distinctive γδ T Cell Repertoire in NOD Mice Weakens Immune Regulation and Favors Diabetic Disease. Biomolecules, 12(10), 1406. https://doi.org/10.3390/biom12101406





