Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi
Abstract
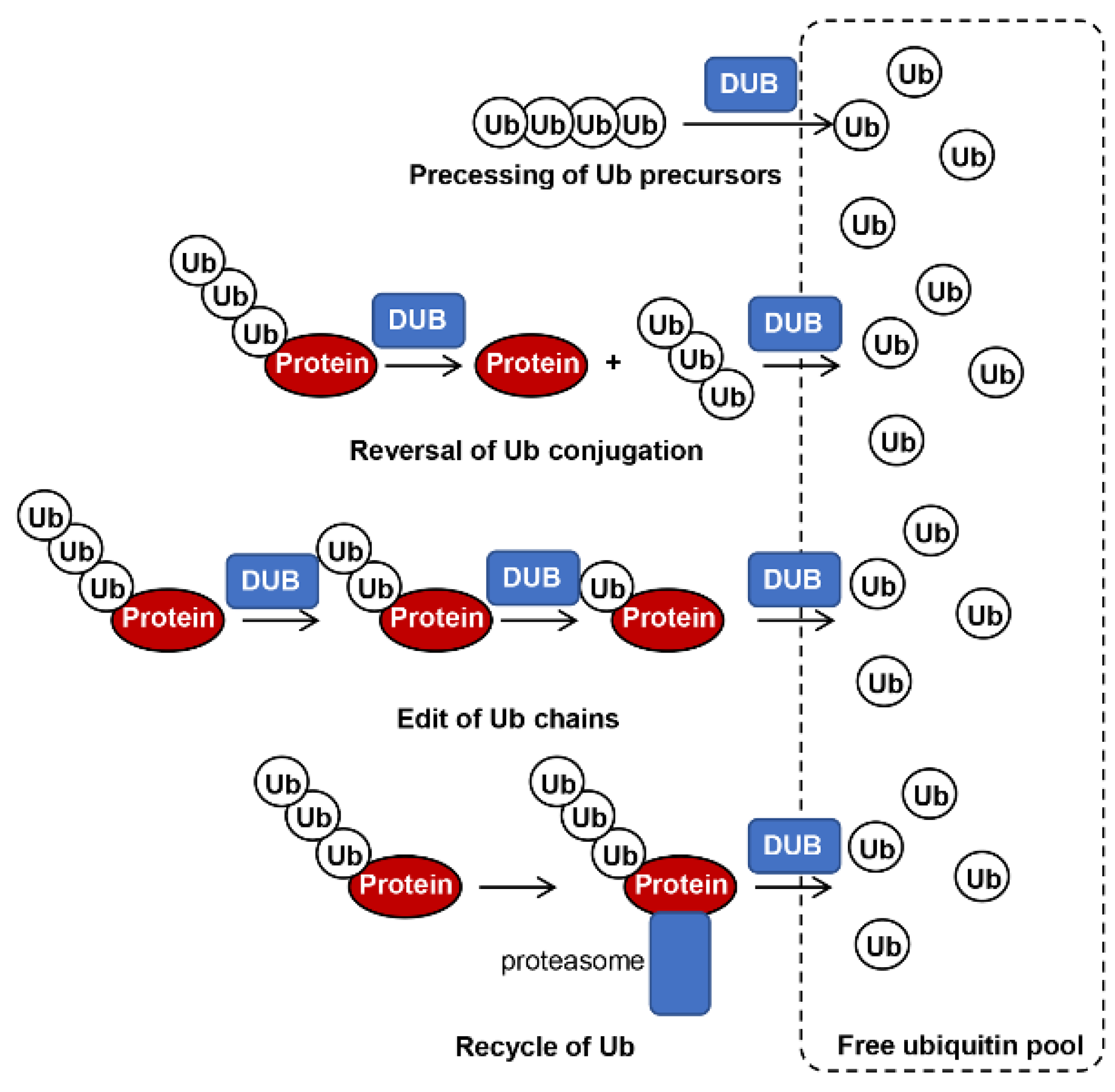
:1. Introduction
2. Summary of DUB Studies in Different Human and Plant Pathogenic Fungi
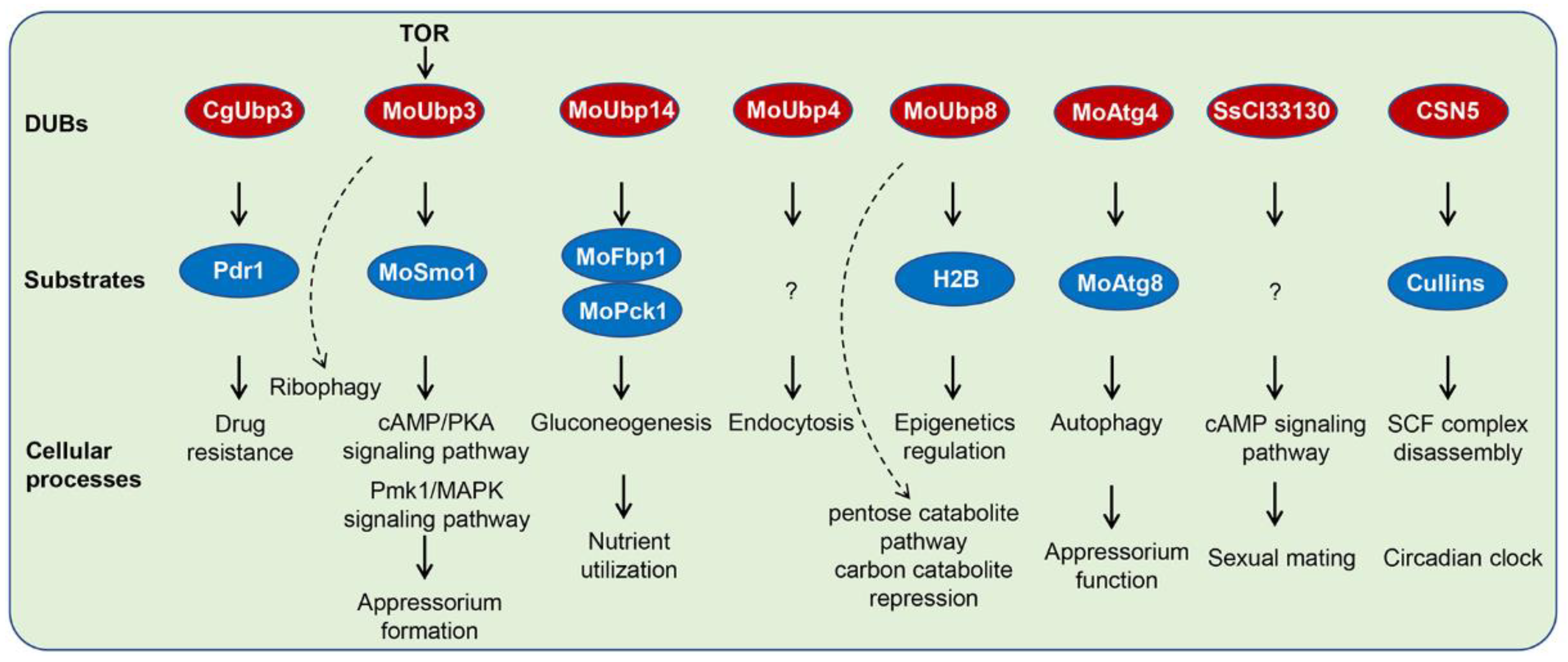
3. Biological Functions of DUBs in Different Pathogenic Fungi
3.1. Overview of Roles of Ubiquitination and DUBs in Different Pathogenic Fungi
3.2. DUBs in Fungal Growth
3.3. DUBs in Fungal Spore Production
3.4. DUBs in Fungal Sexual Reproduction
3.5. DUBs in Fungal Stress Response
3.6. DUBs in Fungal Nutrient Utilization
3.7. DUBs in Fungal Virulence
4. Regulatory Mechanisms Mediated by DUBs in Different Pathogenic Fungi
4.1. DUBs and Cellular Signaling Pathways
4.2. DUBs and Autophagy
4.3. DUBs and Fungal Carbon Source Utilization
4.4. DUBs and Histone Modification
4.5. DUBs and Endocytosis
4.6. DUBs and Circadian Clock
4.7. DUBs and Drug Resistance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grabbe, C.; Husnjak, K.; Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011, 12, 295–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 2014, 12, e0174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Hochstrasser, M. Origin and function of ubiquitin-like proteins. Nature 2009, 458, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Mevissen, T.E.T.; Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.H.; Baek, K.H. Deubiquitinating enzymes as therapeutic targets in cancer. Curr. Pharm. Des. 2013, 19, 4039–4052. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Vlasschaert, C.; Cook, D.; Xia, X.; Gray, D.A. The evolution and functional diversification of the deubiquitinating enzyme superfamily. Genome Biol. Evol. 2017, 9, 558–573. [Google Scholar] [CrossRef] [Green Version]
- Hutchins, A.P.; Liu, S.; Diez, D.; Miranda-Saavedra, D. The repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol. Biol. Evol. 2013, 30, 1172–1187. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.H.; Joo, J.Y.; Baek, K.H. The potential roles of deubiquitinating enzymes in brain diseases. Ageing Res. Rev. 2020, 61, 101088. [Google Scholar] [CrossRef]
- Suresh, B.; Lee, J.; Hong, S.H.; Kim, K.S.; Ramakrishna, S. The role of deubiquitinating enzymes in spermatogenesis. Cell. Mol. Life Sci. 2015, 72, 4711–4720. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Zhang, Z.; Wu, L.; Zhang, L.; Zhou, F. The functional deubiquitinating enzymes in control of innate antiviral immunity. Adv. Sci. 2020, 8, 2002484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Li, R.; Liu, X.; Qu, Y.Q. Advances in deubiquitinating enzymes in lung adenocarcinoma. J. Cancer 2021, 12, 5573–5582. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Cope, G.A.; Suh, G.S.; Aravind, L.; Schwarz, S.E.; Zipursky, S.L.; Koonin, E.V.; Deshaies, R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 2002, 298, 608–611. [Google Scholar] [CrossRef] [Green Version]
- Schmaler, T.; Dubiel, W. Control of deneddylation by the COP9 signalosome. Subcell. Biochem. 2010, 54, 57–68. [Google Scholar]
- Cao, C.; Xue, C. More than just cleaning: Ubiquitin-mediated proteolysis in fungal pathogenesis. Front. Cell. Infect. Microbiol. 2021, 11, 774613. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, N.; Zheng, Y.; Yue, J.; Bhadauria, V.; Peng, Y.L.; Chen, Q. Ubiquitination in the rice blast fungus Magnaporthe oryzae: From development and pathogenicity to stress responses. Phytopathol. Res. 2022, 4, 1. [Google Scholar] [CrossRef]
- Roig, P.; Gozalbo, D. Depletion of polyubiquitin encoded by the UBI4 gene confers pleiotropic phenotype to Candida albicans cells. Fungal Genet. Biol. 2003, 39, 70–81. [Google Scholar] [CrossRef]
- Leach, M.D.; Stead, D.A.; Argo, E.; MacCallum, D.M.; Brown, A.J. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol. Microbiol. 2011, 79, 1574–1593. [Google Scholar] [CrossRef] [Green Version]
- Sandai, D.; Yin, Z.; Selway, L.; Stead, D.; Walker, J.; Leach, M.D.; Bohovych, I.; Ene, I.V.; Kastora, S.; Budge, S.; et al. The evolutionary rewiring of ubiquitination targets has reprogrammed the regulation of carbon assimilation in the pathogenic yeast Candida albicans. mBio 2012, 3, e00495-12. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, Y.; Fan, Y.; Yi, J.; Zhang, C.; Gu, Z.; Pan, W.; Gu, J.; Liao, W.; Fang, W. Ribosomal protein L40e fused with a ubiquitin moiety is essential for the vegetative growth, morphological homeostasis, cell cycle progression, and pathogenicity of Cryptococcus neoformans. Front. Microbiol. 2020, 11, 570269. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Li, Z.; Xing, J.; Yang, J.; Wang, Z.; Zhang, H.; Chen, D.; Peng, Y.L.; Chen, X.L. Global analysis of sumoylation function reveals novel insights into development and appressorium-mediated infection of the rice blast fungus. New Phytol. 2018, 219, 1031–1047. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.B.; Wang, Y.; Stukes, S.; Chen, Q.; Casadevall, A.; Xue, C. The F-Box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryot. Cell 2011, 10, 791–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, Y.; Franck, W.L.; Han, S.O.; Shows, A.; Gokce, E.; Muddiman, D.C.; Dean, R.A. Polyubiquitin is required for growth, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. PLoS ONE 2012, 7, e42868. [Google Scholar] [CrossRef] [Green Version]
- Miguel-Rojas, C.; Hera, C. Proteomic identification of potential target proteins regulated by the SCF(F) (bp1) -mediated proteolysis pathway in Fusarium oxysporum. Mol. Plant Pathol. 2013, 14, 934–945. [Google Scholar] [CrossRef]
- Chen, Q.; Li, Y.; Wang, J.; Li, R.; Chen, B. cpubi4 is essential for development and virulence in chestnut blight fungus. Front. Microbiol. 2018, 9, 1286. [Google Scholar] [CrossRef]
- Marblestone, J.G.; Butt, S.; McKelvey, D.M.; Sterner, D.E.; Mattern, M.R.; Nicholson, B.; Eddins, M.J. Comprehensive ubiquitin E2 profiling of ten ubiquitin E3 ligases. Cell Biochem. Biophys. 2013, 67, 161–167. [Google Scholar] [CrossRef]
- Leng, P.; Sudbery, P.E.; Brown, A.J. Rad6p represses yeast-hypha morphogenesis in the human fungal pathogen Candida albicans. Mol. Microbiol. 2000, 35, 1264–1275. [Google Scholar] [CrossRef]
- Shi, H.B.; Chen, G.Q.; Chen, Y.P.; Dong, B.; Lu, J.P.; Liu, X.H.; Lin, F.C. MoRad6-mediated ubiquitination pathways are essential for development and pathogenicity in Magnaporthe oryzae. Environ. Microbiol. 2016, 18, 4170–4187. [Google Scholar] [CrossRef]
- Liu, T.B.; Xue, C. The ubiquitin-proteasome system and F-box proteins in pathogenic fungi. Mycobiology 2011, 39, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Butler, D.K.; All, O.; Goffena, J.; Loveless, T.; Wilson, T.; Toenjes, K.A. The GRR1 gene of Candida albicans is involved in the negative control of pseudohyphal morphogenesis. Fungal Genet. Biol. 2006, 43, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.K.; Kim, M.D.; Lee, S.H.; Yun, S.H.; Lee, Y.W. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol. Microbiol. 2007, 63, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Chen, N.; Zhu, X.M.; Liang, S.; Li, L.; Wang, J.Y.; Lu, J.P.; Lin, F.C.; Liu, X.H. F-box proteins MoFwd1, MoCdc4 and MoFbx15 regulate development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2019, 21, 3027–3045. [Google Scholar] [CrossRef] [PubMed]
- Shieh, J.C.; White, A.; Cheng, Y.C.; Rosamond, J. Identification and functional characterization of Candida albicans CDC4. J. Biomed. Sci. 2005, 12, 913–924. [Google Scholar] [CrossRef]
- Chin, C.; Lai, W.C.; Lee, T.L.; Tseng, T.L.; Shieh, J.C. Dissection of the Candida albicans Cdc4 protein reveals the involvement of domains in morphogenesis and cell flocculation. J. Biomed. Sci. 2013, 20, 97. [Google Scholar] [CrossRef] [Green Version]
- Han, L.T.; Wu, Y.J.; Liu, T.B. The F-box protein fbp1 regulates virulence of Cryptococcus neoformans through the putative zinc-binding protein Zbp1. Front. Cell. Infect. Microbiol. 2021, 11, 794661. [Google Scholar] [CrossRef]
- Trunk, K.; Gendron, P.; Nantel, A.; Lemieux, S.; Roemer, T.; Raymond, M. Depletion of the cullin Cdc53p induces morphogenetic changes in Candida albicans. Eukaryot. Cell. 2009, 8, 756–767. [Google Scholar] [CrossRef] [Green Version]
- Jöhnk, B.; Bayram, Ö.; Abelmann, A.; Heinekamp, T.; Mattern, D.J.; Brakhage, A.A.; Jacobsen, I.D.; Valerius, O.; Braus, G.H. SCF ubiquitin ligase F-box protein Fbx15 controls nuclear co-repressor localization, stress response and virulence of the human pathogen Aspergillus fumigatus. PLoS Pathog. 2016, 12, e1005899. [Google Scholar] [CrossRef] [Green Version]
- Duyvesteijn, R.G.; van Wijk, R.; Boer, Y.; Rep, M.; Cornelissen, B.J.; Haring, M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005, 57, 1051–1063. [Google Scholar] [CrossRef]
- Chang, S.C.; Hsu, W.; Su, E.C.; Hung, C.S.; Ding, J.L. Human FBXL8 is a novel E3 ligase which promotes BRCA metastasis by stimulating pro-tumorigenic cytokines and inhibiting tumor suppressors. Cancers 2020, 12, 2210. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Hung, C.S.; Zhang, B.X.; Hsieh, T.H.; Hsu, W.; Ding, J.L. A novel signature of CCNF-associated E3 ligases collaborate and counter each other in breast cancer. Cancers 2021, 13, 2873. [Google Scholar] [CrossRef] [PubMed]
- Kahana, A. The deubiquitinating enzyme Dot4p is involved in regulating nutrient uptake. Biochem. Biophys. Res. Commun. 2001, 282, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Enyenihi, A.H.; Saunders, W.S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 2003, 163, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Auesukaree, C.; Damnernsawad, A.; Kruatrachue, M.; Pokethitiyook, P.; Boonchird, C.; Kaneko, Y.; Harashima, S. Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J. Appl. Genet. 2009, 50, 301–310. [Google Scholar] [CrossRef]
- Amerik, A.Y.; Li, S.J.; Hochstrasser, M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 2000, 381, 981–992. [Google Scholar] [CrossRef]
- Hämmerle, M.; Bauer, J.; Rose, M.; Szallies, A.; Thumm, M.; Düsterhus, S.; Mecke, D.; Entian, K.D.; Wolf, D.H. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 25000–25005. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, K.; Tanaka, T.; Ida, Y.; Furusawa, C.; Hirasawa, T.; Shimizu, H. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 2011, 28, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Wang, Z.; Hou, Y.; Liu, C.; Ahmed, H.; Xing, J.; Chen, X.L. Systematic characterization of the ubiquitin-specific proteases in Magnaporthe oryzae. Phytopathol. Res. 2020, 2, 8. [Google Scholar] [CrossRef]
- Cai, X.; Xiang, S.; He, W.; Tang, M.; Zhang, S.; Chen, D.; Zhang, X.; Liu, C.; Li, G.; Xing, J.; et al. Deubiquitinase Ubp3 regulates ribophagy and deubiquitinates Smo1 for appressorium-mediated infection by Magnaporthe oryzae. Mol. Plant Pathol. 2022, 23, 832–844. [Google Scholar] [CrossRef]
- Que, Y.; Xu, Z.; Wang, C.; Lv, W.; Yue, X.; Xu, L.; Tang, S.; Dai, H.; Wang, Z. The putative deubiquitinating enzyme MoUbp4 is required for infection-related morphogenesis and pathogenicity in the rice blast fungus Magnaporthe oryzae. Curr. Genet. 2020, 66, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, D.; Matar, K.; Zheng, T.; Zhao, Q.; Xie, Y.; Gao, X.; Li, M.; Wang, B.; Lu, G.D. The deubiquitinating enzyme MoUbp8 is required for infection-related development, pathogenicity, and carbon catabolite repression in Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2020, 104, 5081–5094. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, H.; Liu, C.; Xing, J.; Chen, X.L. A deubiquitinating enzyme Ubp14 is required for development, stress response, nutrient utilization, and pathogenesis of Magnaporthe oryzae. Front. Microbiol. 2018, 9, 769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, W.; Price, M.S.; Toffaletti, D.L.; Tenor, J.; Betancourt-Quiroz, M.; Price, J.L.; Pan, W.H.; Liao, W.Q.; Perfect, J.R. Pleiotropic effects of deubiquitinating enzyme Ubp5 on growth and pathogenesis of Cryptococcus neoformans. PLoS ONE 2012, 7, e38326. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Zhang, C.; Yi, J.; Zhou, Z.; Fa, Z.; Zhao, J.; Yang, Y.; Fang, W.; Wang, Y.; Liao, W.Q. Deubiquitinase Ubp5 Is Required for the Growth and Pathogenicity of Cryptococcus gattii. PLoS ONE 2016, 11, e0153219. [Google Scholar] [CrossRef] [Green Version]
- Paul, S.; McDonald, W.H.; Moye-Rowley, W.S. Negative regulation of Candida glabrata Pdr1 by the deubiquitinase subunit Bre5 occurs in a ubiquitin independent manner. Mol. Microbiol. 2018, 110, 309–323. [Google Scholar] [CrossRef]
- Li, H.; Cai, Y.; Deng, Q.; Bao, H.; Chen, J.; Shen, W. Ovarian tumor domain-containing proteases-deubiquitylation enzyme gene SsCI33130 involved in the regulation of mating/filamentation and pathogenicity in Sporisorium scitamineum. Front. Microbiol. 2021, 12, 746550. [Google Scholar] [CrossRef]
- Wang, M.; Yang, X.; Ruan, R.; Fu, H.; Li, H. Csn5 is required for the conidiogenesis and pathogenesis of the Alternaria alternata tangerine pathotype. Front. Microbiol. 2018, 9, 508. [Google Scholar] [CrossRef] [Green Version]
- Liu, O.W.; Chun, C.D.; Chow, E.D.; Chen, C.; Madhani, H.D.; Noble, S.M. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 2008, 135, 174–188. [Google Scholar] [CrossRef] [Green Version]
- Ostapenko, D.; Burton, J.L.; Solomon, M.J. The Ubp15 deubiquitinase promotes timely entry into S phase in Saccharomyces cerevisiae. Mol. Biol. Cell 2015, 26, 2205–2216. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Zhang, X.; Li, W.; Liu, G.; Wang, S.; Yan, X.; Zou, H.; Yin, W.B. COP9 signalosome subunit PfCsnE regulates secondary metabolism and conidial formation in Pestalotiopsis fici. Sci. China Life Sci. 2017, 60, 656–664. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hu, Q.; Chen, H.; Zhou, Z.; Li, W.; Wang, Y.; Li, S.; He, Q. Role of individual subunits of the Neurospora crassa CSN complex in regulation of deneddylation and stability of cullin proteins. PLoS Genet. 2010, 6, e1001232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Wang, Y.; Cai, G.; He, Q. Neurospora COP9 signalosome integrity plays major roles for hyphal growth, conidial development, and circadian function. PLoS Genet. 2012, 8, e1002712. [Google Scholar] [CrossRef]
- Nahlik, K.; Dumkow, M.; Bayram, O.; Helmstaedt, K.; Busch, S.; Valerius, O.; Gerke, J.; Hoppert, M.; Schwier, E.; Opitz, L.; et al. The COP9 signalosome mediates transcriptional and metabolic response to hormones, oxidative stress protection and cell wall rearrangement during fungal development. Mol. Microbiol. 2010, 78, 964–979. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, X. Cryptococcus neoformans: Sex, morphogenesis, and virulence. Infect. Genet. Evol. 2021, 89, 104731. [Google Scholar] [CrossRef]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Liu, T.B.; Liu, X.H.; Lu, J.P.; Zhang, L.; Min, H.; Lin, F.C. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010, 6, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, H. Regulation of autophagy by mTOR signaling pathway. Adv. Exp. Med. Biol. 2019, 1206, 67–83. [Google Scholar]
- Marroquin-Guzman, M.; Wilson, R.A. GATA- dependent glutaminolysis drives appressorium formation in Magnaporthe oryzae by suppressing TOR inhibition of cAMP/PKA signaling. PLoS Pathog. 2015, 11, e1004851. [Google Scholar] [CrossRef]
- Müller, M.; Kötter, P.; Behrendt, C.; Walter, E.; Scheckhuber, C.Q.; Entian, K.D.; Reichert, A.S. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep. 2015, 10, 1215–1225. [Google Scholar] [CrossRef] [Green Version]
- Kershaw, M.J.; Basiewicz, M.; Soanes, D.M.; Yan, X.; Ryder, L.S.; Csukai, M.; Oses-Ruiz, M.; Valent, B.; Talbot, N.J. Conidial morphogenesis and septin-mediated plant infection require Smo1, a Ras GTPase-activating protein in Magnaporthe oryzae. Genetics 2019, 211, 151–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Abreu, S.; Kriegenburg, F.; Gómez-Sánchez, R.; Mari, M.; Sánchez-Wandelmer, J.; Skytte Rasmussen, M.; Soares Guimarães, R.; Zens, B.; Schuschnig, M.; Hardenberg, R.; et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2017, 18, 765–780. [Google Scholar] [CrossRef] [Green Version]
- Hollenstein, D.M.; Kraft, C. Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 2020, 65, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Stanojevic, N.; Duan, Z.; Sen, P.; Bhaumik, S.R. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell Biol. 2006, 26, 3339–3352. [Google Scholar] [CrossRef] [Green Version]
- Gardner, R.G.; Nelson, Z.W.; Gottschling, D.E. Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: Distinct roles in telomeric silencing and general chromatin. Mol. Cell Biol. 2005, 25, 6123–6139. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Fan, X.; Zhao, Q.; Xu, Y.; Wang, X.; Chen, J. Bre1 and Ubp8 regulate H2B mono-ubiquitination and the reversible yeast-hyphae transition in Candida albicans. Mol. Microbiol. 2021, 115, 332–343. [Google Scholar] [CrossRef]
- Swaminathan, S.; Amerik, A.Y.; Hochstrasser, M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol. Biol. Cell 1999, 10, 2583–2594. [Google Scholar] [CrossRef] [Green Version]
- Buysse, D.; Pfitzner, A.K.; West, M.; Roux, A.; Odorizzi, G. The ubiquitin hydrolase Doa4 directly binds Snf7 to inhibit recruitment of ESCRT-III remodeling factors in S. cerevisiae. J. Cell Sci. 2020, 133, jcs241455. [Google Scholar] [CrossRef]
- Cohen, M.; Stutz, F.; Belgareh, N.; Haguenauer-Tsapis, R.; Dargemont, C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 2003, 5, 661–667. [Google Scholar] [CrossRef]
- Schauer, N.J.; Magin, R.S.; Liu, X.; Doherty, L.M.; Buhrlage, S.J. Advances in discovering deubiquitinating enzyme (DUB) inhibitors. J. Med. Chem. 2020, 63, 2731–2750. [Google Scholar] [CrossRef] [PubMed]
- Nanduri, B.; Suvarnapunya, A.E.; Venkatesan, M.; Edelmann, M.J. Deubiquitinating enzymes as promising drug targets for infectious diseases. Curr. Pharm. Des. 2013, 19, 3234–3247. [Google Scholar] [CrossRef] [PubMed]
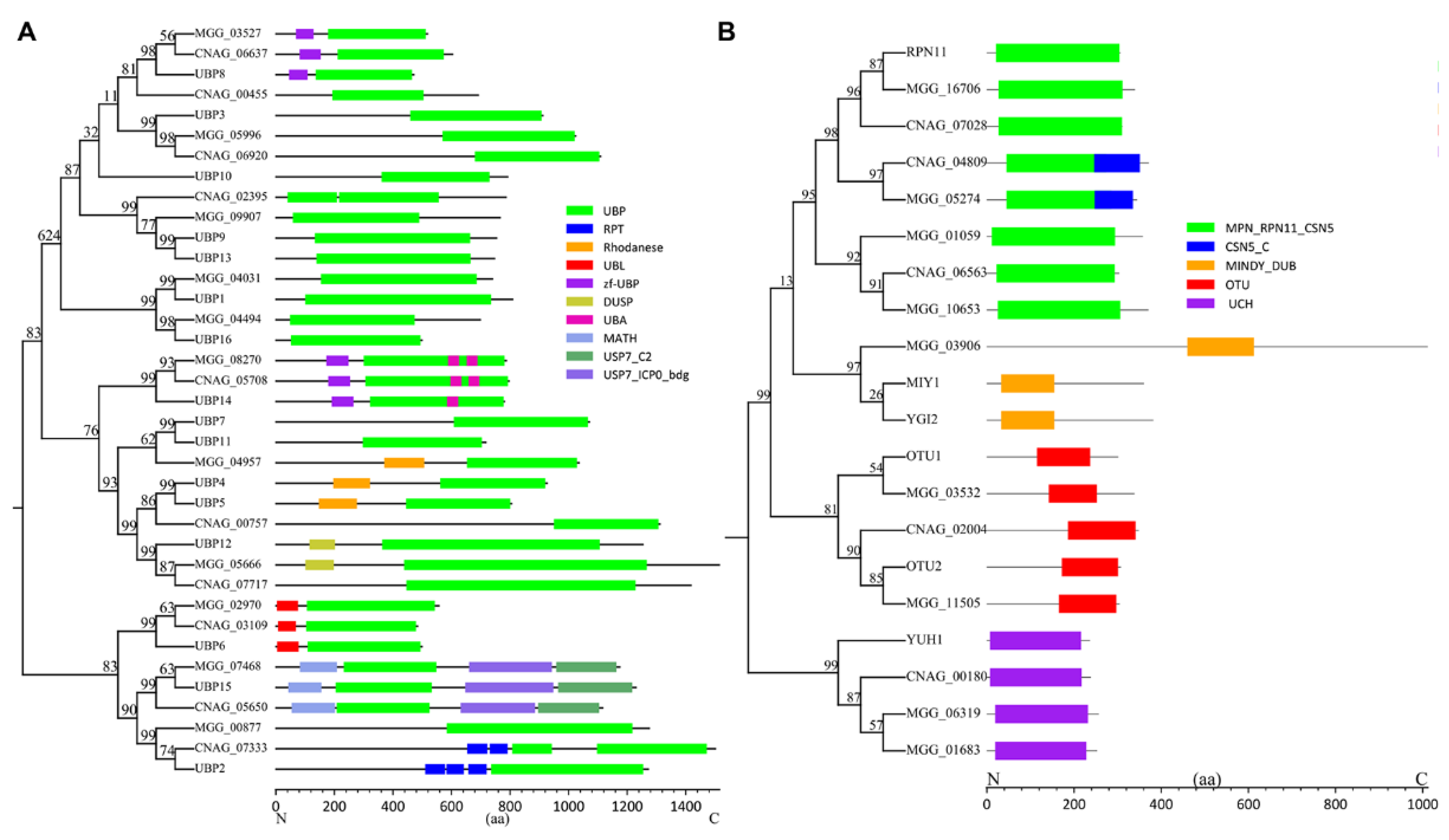
| S. cerevisiae | C. neoformans | M. oryzae | |
|---|---|---|---|
| UBPs | UBP1 | - | MGG_04031 |
| UBP2 | CNAG_07333 | MGG_00877 | |
| UBP3 | CNAG_06920 | MGG_05996 | |
| UBP4 | CNAG_00757 | MGG_04957 | |
| UBP5 | - | ||
| UBP6 | CNAG_03109 | MGG_02970 | |
| UBP7 | - | - | |
| UBP8 | CNAG_06637 | MGG_03527 | |
| UBP9 | CNAG_02395 | - | |
| UBP10 | CNAG_00455 | - | |
| UBP11 | - | - | |
| UBP12 | CNAG_07717 | MGG_05666 | |
| UBP13 | CNAG_02395 | MGG_09907 | |
| UBP14 | CNAG_05708 | MGG_08270 | |
| UBP15 | CNAG_05650 | MGG_07468 | |
| UBP16 | - | MGG_04494 | |
| UCHs | YUH1 | CNAG_00180 | MGG_06319 |
| MGG_01683 | |||
| OTUs | OTU1 | - | MGG_03532 |
| OTU2 | CNAG_02004 | MGG_11505 | |
| MINDY | MIY1 | - | MGG_03906 |
| YGI2 | - | MGG_05274 | |
| JAMM | RPN11 | CNAG_07028 | MGG_16706 |
| CNAG_04809 | MGG_05274 | ||
| CNAG_06563 | MGG_01059 | ||
| MGG_10653 |
| Fungal DUBs | Biological Functions | Mechanisms | Ref. |
|---|---|---|---|
| Magnaporthe oryzae Ubp1 | Growth, conidiation, virulence, stress response | [49] | |
| M. oryzae UBP2 | No evident function was found | [49] | |
| M. oryzae UBP3 | Growth, conidiation, virulence, stress response | Regulate ribophagy and GTPase-activating protein Smo1, cAMP, and Pmk1-MAPK signaling | [49,50] |
| M. oryzae UBP4 | Growth, conidiation, virulence, stress response, nutrient utilization | Recycle Ub at the late endosome, endocytosis | [49,51] |
| M. oryzae UBP6 | Conidiation, virulence, stress response | [49] | |
| M. oryzae UBP8 | Growth, conidiation, virulence, stress response | Component of SAGA complex, regulates transcriptional activation by deubiquitinating H2B | [49,52] |
| M. oryzae UBP12 | Growth, conidiation, virulence, stress response | [49] | |
| M. oryzae UBP13 | Conidiation, virulence, stress response | [49] | |
| M. oryzae UBP14 | growth, conidiation, virulence, stress response, nutrient utilization | Regulate gluconeogenesis key enzymes MoFBP1 and MoPCK1 | [49,53] |
| M. oryzae UBP15 | Virulence, stress response | [49] | |
| M. oryzae UBP16 | Conidiation | [49] | |
| Cryptococcus neoformans DOA4 | Sporulation, pigment production, capsule formation, virulence, stress response, sexual reproduction | [54] | |
| C. neoformans UBP13 | Stress response, pigment production | [54] | |
| C. neoformans UBP14 | Stress response, pigment production | [54] | |
| C. neoformans UBP5 | Growth, sporulation, melanization, capsule formation, virulence, stress response, sexual reproduction | Major deubiquitinating enzyme for stress response | [54] |
| Candida gattii UBP5 | Growth, stress response, virulence | [55] | |
| Candida glabrata BRE5 | Azole resistance | Association with UBP3, negatively regulate transcription of PDR1 | [56] |
| Sporisorium scitamineum SsCI33130 | Sexual mating and pathogenicity | Regulate the synthesis of the small-molecule signaling substances (cAMP or tryptophol) | [57] |
| Alternaria alternata CSN5 | Light response, sexual development, virulence, and secondary metabolism | Components of COP9 signalosome, regulate activity of cullin–RING ubiquitin E3 ligases | [58] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Cai, X.; Chen, X.-L. Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi. Biomolecules 2022, 12, 1424. https://doi.org/10.3390/biom12101424
Wang W, Cai X, Chen X-L. Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi. Biomolecules. 2022; 12(10):1424. https://doi.org/10.3390/biom12101424
Chicago/Turabian StyleWang, Weixiang, Xuan Cai, and Xiao-Lin Chen. 2022. "Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi" Biomolecules 12, no. 10: 1424. https://doi.org/10.3390/biom12101424
APA StyleWang, W., Cai, X., & Chen, X. -L. (2022). Recent Progress of Deubiquitinating Enzymes in Human and Plant Pathogenic Fungi. Biomolecules, 12(10), 1424. https://doi.org/10.3390/biom12101424







