High-Resolution Ribosome Profiling Reveals Gene-Specific Details of UGA Re-Coding in Selenoprotein Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Model Validation and Cell Culture
2.2. Cell Transfection and Luciferase Assay
2.3. Western Blot
2.4. [75. Se] Labeling
2.5. TXNRD Activity Assay
2.6. Ribosome Profiling and RNA Sequencing
2.7. Quality Control and Preprocessing of Deep-sequencing Data
3. Results
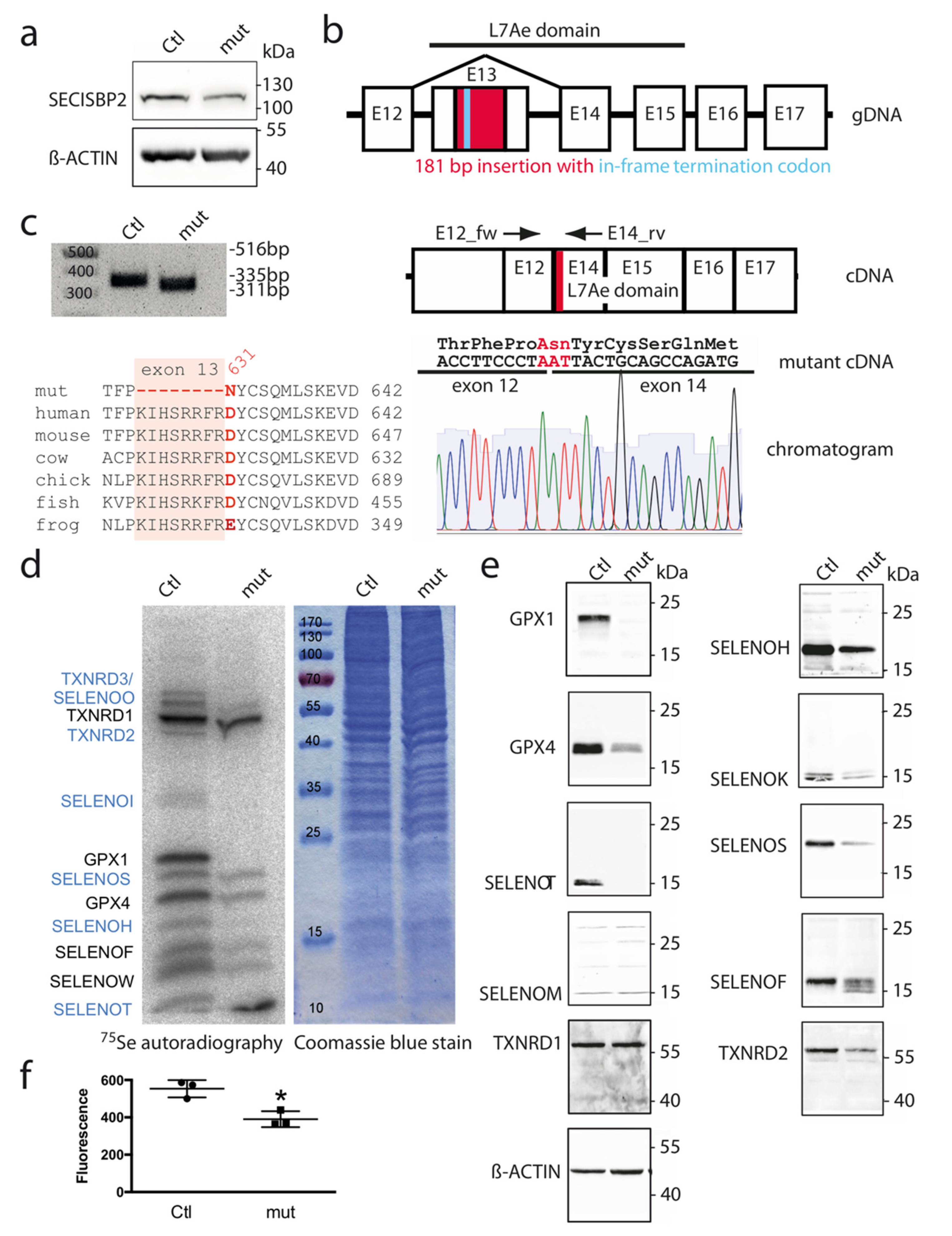
3.1. Deletion of 8 Amino Acids in the L7Ae Domain of SECISBP2 Greatly Reduces Expression of Selenoproteins
3.2. Ribosomal Profiling of Selenoprotein Transcripts in HAP1 Cells
3.3. Established Readouts of Ribosomal Profiling Underscore the Role of SECISBP2 in UGA Recoding
3.4. RPF Containing the UGA/Sec Codon in the Ribosomal A-Site
3.5. RPF Containing the UGA/Sec Codon in the Ribosomal P-Site Reflect UGA Re-Definition Efficiency
3.6. The Choices of the Ribosome at a UGA Codon
3.7. Evidence for Frameshifting at UGA Codons in Selenoprotein Translation
4. Discussion
4.1. Mutated SECISBP2 and UGA/Sec Re-Coding
4.2. SECISBP2 Opposes Termination
4.3. Does SECISBP2 Help Recruit Sec-tRNASec:EEFSEC?
4.4. pUGA as a Measure for UGA/Sec Redefinition Efficiency
4.5. Frameshifting, a New Process at UGA/Sec
4.6. Potential Technical Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


| Selenoprotein | URE Wild-Type | URE Mutant | ΔURE ± SD |
|---|---|---|---|
| GPX1 | 0.30 | 0.35 | 1.12 ± 0.02 a |
| 1.04 | 1.13 | 1.08 ± 0.29 b | |
| 1.84 | 1.82 | 0.99 ± 0.06 c | |
| GPX4 | 0.47 | 0.11 | 0.24 ± 0.01 a |
| 2.07 0.69 | 0.62 0.31 | 0.30 ± 0.02 b 0.45 ± 0.01 c | |
| SELENOF | 1.41 | 0.40 | 0.29 ± 0.01 a |
| 0.25 | 0.14 | 0.58 ± 0.17 b | |
| 0.34 | 0.04 | 0.12 ± 0.01 c | |
| SELENOH | 0.49 | 0.45 | 0.93 ± 0.04 a |
| 0.59 | 0.90 | 1.51 ± 0.03 b | |
| 0.40 | 0.22 | 0.55 ± 0.00 c | |
| SELENOT | 0.19 | 0.23 | 1.19 ± 0.08 a |
| 1.39 | 1.56 | 1.12 ± 0.15 b | |
| 2.00 | 0.60 | 0.30 ± 0.05 c | |
| SELENOW | 0.50 | 0.40 | 0.80 ± 0.27 a |
| 0.28 | 0.37 | 1.29 ± 0.02 b | |
| 2.59 | 1.67 | 0.64 ± 0.13 c | |
| SEPHS2 | 2.69 | 0.87 | 0.32 ± 0.01 a |
| 0.62 | 0.48 | 0.77 ± 0.06 b | |
| 2.02 | 0.52 | 0.26 ± 0.01 c |


References
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Forchhammer, K.; Leinfelder, W.; Böck, A. Identification of a Novel Translation Factor Necessary for the Incorporation of Selenocysteine into Protein. Nature 1989, 342, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.J.; Banu, L.; Chen, Y.Y.; Mandel, S.J.; Kieffer, J.D.; Harney, J.W.; Larsen, P.R. Recognition of UGA as a Selenocysteine Codon in Type I Deiodinase Requires Sequences in the 3’ Untranslated Region. Nature 1991, 353, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.T.; Aggarwal, G.; Anderson, C.B.; Khatri, S.; Flanigan, K.M.; Atkins, J.F. Recoding Elements Located Adjacent to a Subset of Eukaryal Selenocysteine-Specifying UGA Codons. EMBO J. 2005, 24, 1596–1607. [Google Scholar] [CrossRef]
- Howard, M.T.; Moyle, M.W.; Aggarwal, G.; Carlson, B.A.; Anderson, C.B. A Recoding Element That Stimulates Decoding of UGA Codons by Sec TRNA[Ser]Sec. RNA 2007, 13, 912–920. [Google Scholar] [CrossRef]
- Mariotti, M.; Shetty, S.; Baird, L.; Wu, S.; Loughran, G.; Copeland, P.R.; Atkins, J.F.; Howard, M.T. Multiple RNA Structures Affect Translation Initiation and UGA Redefinition Efficiency during Synthesis of Selenoprotein P. Nucleic Acids Res. 2017, 45, 13004–13015. [Google Scholar] [CrossRef]
- Shetty, S.P.; Copeland, P.R. The Selenium Transport Protein, Selenoprotein P, Requires Coding Sequence Determinants to Promote Efficient Selenocysteine Incorporation. J. Mol. Biol. 2018, 430, 5217–5232. [Google Scholar] [CrossRef]
- Cockman, E.M.; Narayan, V.; Willard, B.; Shetty, S.P.; Copeland, P.R.; Driscoll, D.M. Identification of the Selenoprotein S Positive UGA Recoding (SPUR) Element and Its Position-Dependent Activity. RNA Biol. 2019, 1682–1696. [Google Scholar] [CrossRef]
- Fagegaltier, D.; Hubert, N.; Yamada, K.; Mizutani, T.; Carbon, P.; Krol, A. Characterization of MSelB, a Novel Mammalian Elongation Factor for Selenoprotein Translation. EMBO J. 2000, 19, 4796–4805. [Google Scholar] [CrossRef]
- Tujebajeva, R.M.; Copeland, P.R.; Xu, X.M.; Carlson, B.A.; Harney, J.W.; Driscoll, D.M.; Hatfield, D.L.; Berry, M.J. Decoding Apparatus for Eukaryotic Selenocysteine Insertion. EMBO Rep. 2000, 1, 158–163. [Google Scholar] [CrossRef]
- Copeland, P.R.; Fletcher, J.E.; Carlson, B.A.; Hatfield, D.L.; Driscoll, D.M. A Novel RNA Binding Protein, SBP2, Is Required for the Translation of Mammalian Selenoprotein MRNAs. EMBO J. 2000, 19, 306–314. [Google Scholar] [CrossRef]
- Fischer, N.; Neumann, P.; Bock, L.V.; Maracci, C.; Wang, Z.; Paleskava, A.; Konevega, A.L.; Schroder, G.F.; Grubmuller, H.; Ficner, R.; et al. The Pathway to GTPase Activation of Elongation Factor SelB on the Ribosome. Nature 2016, 540, 80–85. [Google Scholar] [CrossRef]
- Hilal, T.; Killam, B.Y.; Grozdanović, M.; Dobosz-Bartoszek, M.; Loerke, J.; Bürger, J.; Mielke, T.; Copeland, P.R.; Simonović, M.; Spahn, C.M.T. Structure of the Mammalian Ribosome as It Decodes the Selenocysteine UGA Codon. Science 2022, 376, 1338–1343. [Google Scholar] [CrossRef]
- Dumitrescu, A.M.; Liao, X.H.; Abdullah, M.S.; Lado-Abeal, J.; Majed, F.A.; Moeller, L.C.; Boran, G.; Schomburg, L.; Weiss, R.E.; Refetoff, S. Mutations in SECISBP2 Result in Abnormal Thyroid Hormone Metabolism. Nat. Genet. 2005, 37, 1247–1252. [Google Scholar] [CrossRef]
- Schoenmakers, E.; Chatterjee, K. Human Disorders Affecting the Selenocysteine Incorporation Pathway Cause Systemic Selenoprotein Deficiency. Antioxid. Redox Signal. 2020, 33, 481–497. [Google Scholar] [CrossRef]
- Azevedo, M.F.; Barra, G.B.; Naves, L.A.; Ribeiro Velasco, L.F.; Godoy Garcia, C.P.; de Castro, L.C.; Amato, A.A.; Miniard, A.; Driscoll, D.; Schomburg, L.; et al. Selenoprotein-Related Disease in a Young Girl Caused by Nonsense Mutations in the SBP2 Gene. J. Clin. Endocrinol. Metab. 2010, 95, 4066–4071. [Google Scholar] [CrossRef]
- Di Cosmo, C.; McLellan, N.; Liao, X.H.; Khanna, K.K.; Weiss, R.E.; Papp, L.; Refetoff, S. Clinical and Molecular Characterization of a Novel Selenocysteine Insertion Sequence-Binding Protein 2 (SBP2) Gene Mutation (R128X). J. Clin. Endocrinol. Metab. 2009, 94, 4003–4009. [Google Scholar] [CrossRef]
- Schoenmakers, E.; Agostini, M.; Mitchell, C.; Schoenmakers, N.; Papp, L.; Rajanayagam, O.; Padidela, R.; Ceron-Gutierrez, L.; Doffinger, R.; Prevosto, C.; et al. Mutations in the Selenocysteine Insertion Sequence-Binding Protein 2 Gene Lead to a Multisystem Selenoprotein Deficiency Disorder in Humans. J. Clin. Investig. 2010, 120, 4220–4235. [Google Scholar] [CrossRef]
- Caban, K.; Kinzy, S.A.; Copeland, P.R. The L7Ae RNA Binding Motif Is a Multifunctional Domain Required for the Ribosome-Dependent Sec Incorporation Activity of Sec Insertion Sequence Binding Protein 2. Mol. Cell. Biol. 2007, 27, 6350–6360. [Google Scholar] [CrossRef][Green Version]
- Donovan, J.; Copeland, P.R. Evolutionary History of Selenocysteine Incorporation from the Perspective of SECIS Binding Proteins. BMC Evol. Biol. 2009, 9, 229. [Google Scholar] [CrossRef]
- Donovan, J.; Copeland, P.R. Selenocysteine Insertion Sequence Binding Protein 2L Is Implicated as a Novel Post-Transcriptional Regulator of Selenoprotein Expression. PLoS ONE 2012, 7, e35581. [Google Scholar] [CrossRef]
- Kiledjian, N.T.; Shah, R.; Vetick, M.B.; Copeland, P.R. The Expression of Essential Selenoproteins during Development Requires SECIS-Binding Protein 2-Like. Life Sci. Alliance 2022, 5, e202101291. [Google Scholar] [CrossRef]
- Dai, Z.-M.; Guo, W.; Yu, D.; Zhu, X.-J.; Sun, S.; Huang, H.; Jiang, M.; Xie, B.; Zhang, Z.; Qiu, M. SECISBP2L-Mediated Selenoprotein Synthesis Is Essential for Autonomous Regulation of Oligodendrocyte Differentiation. J. Neurosci. 2022, 42, 5860–5869. [Google Scholar] [CrossRef]
- Seeher, S.; Atassi, T.; Mahdi, Y.; Carlson, B.A.; Braun, D.; Wirth, E.K.; Klein, M.O.; Reix, N.; Miniard, A.C.; Schomburg, L.; et al. Secisbp2 Is Essential for Embryonic Development and Enhances Selenoprotein Expression. Antioxid. Redox Signal. 2014, 21, 835–849. [Google Scholar] [CrossRef]
- Seeher, S.; Carlson, B.A.; Miniard, A.C.; Wirth, E.K.; Mahdi, Y.; Hatfield, D.L.; Driscoll, D.M.; Schweizer, U. Impaired Selenoprotein Expression in Brain Triggers Striatal Neuronal Loss Leading to Co-Ordination Defects in Mice. Biochem. J. 2014, 462, 67–75. [Google Scholar] [CrossRef]
- Fradejas-Villar, N.; Seeher, S.; Anderson, C.B.; Doengi, M.; Carlson, B.A.; Hatfield, D.L.; Schweizer, U.; Howard, M.T. The RNA-Binding Protein Secisbp2 Differentially Modulates UGA Codon Reassignment and RNA Decay. Nucleic Acids Res. 2017, 45, 4094–4107. [Google Scholar] [CrossRef]
- Zhao, W.; Bohleber, S.; Schmidt, H.; Seeher, S.; Howard, M.T.; Braun, D.; Arndt, S.; Reuter, U.; Wende, H.; Birchmeier, C.; et al. Ribosome Profiling of Selenoproteins in Vivo Reveals Consequences of Pathogenic Secisbp2 Missense Mutations. J. Biol. Chem. 2019, 294, 14185–14200. [Google Scholar] [CrossRef]
- Howard, M.T.; Carlson, B.A.; Anderson, C.B.; Hatfield, D.L. Translational Redefinition of UGA Codons Is Regulated by Selenium Availability. J. Biol. Chem. 2013, 288, 19401–19413. [Google Scholar] [CrossRef]
- Gladyshev, V.N.; Arnér, E.S.; Berry, M.J.; Brigelius-Flohé, R.; Bruford, E.A.; Burk, R.F.; Carlson, B.A.; Castellano, S.; Chavatte, L.; Conrad, M.; et al. Selenoprotein Gene Nomenclature. J. Biol. Chem. 2016, 291, 24036–24040. [Google Scholar] [CrossRef]
- Mehta, A.; Rebsch, C.M.; Kinzy, S.A.; Fletcher, J.E.; Copeland, P.R. Efficiency of Mammalian Selenocysteine Incorporation. J. Biol. Chem. 2004, 279, 37852–37859. [Google Scholar] [CrossRef]
- Fradejas-Villar, N.; Zhao, W.; Reuter, U.; Doengi, M.; Ingold, I.; Bohleber, S.; Conrad, M.; Schweizer, U. Missense Mutation in Selenocysteine Synthase Causes Cardio-Respiratory Failure and Perinatal Death in Mice Which Can Be Compensated by Selenium-Independent GPX4. Redox Biol. 2021, 48, 102188. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FASTQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010. [Google Scholar]
- Krueger, F. Trim Galore! A Wrapper Tool around Cutadapt and FastQC; Babraham Bioinformatics: Cambridge, UK, 2012. [Google Scholar]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser Data Retrieval Tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Giron, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Agric. Sci. 2018, 9, 8. [Google Scholar]
- Lin, H.C.; Ho, S.C.; Chen, Y.Y.; Khoo, K.H.; Hsu, P.H.; Yen, H.C. SELENOPROTEINS. CRL2 Aids Elimination of Truncated Selenoproteins Produced by Failed UGA/Sec Decoding. Science 2015, 349, 91–95. [Google Scholar] [CrossRef]
- Lin, H.C.; Yeh, C.W.; Chen, Y.F.; Lee, T.T.; Hsieh, P.Y.; Rusnac, D.V.; Lin, S.Y.; Elledge, S.J.; Zheng, N.; Yen, H.S. C-Terminal End-Directed Protein Elimination by CRL2 Ubiquitin Ligases. Mol. Cell 2018, 70, 602–613.e3. [Google Scholar] [CrossRef]
- Lareau, L.F.; Hite, D.H.; Hogan, G.J.; Brown, P.O. Distinct Stages of the Translation Elongation Cycle Revealed by Sequencing Ribosome-Protected MRNA Fragments. eLife 2014, 3, e01257. [Google Scholar] [CrossRef]
- Wu, C.C.; Zinshteyn, B.; Wehner, K.A.; Green, R. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 2019, 73, 959–970. [Google Scholar] [CrossRef]
- Renko, K.; Martitz, J.; Hybsier, S.; Heynisch, B.; Voss, L.; Everley, R.A.; Gygi, S.P.; Stoedter, M.; Wisniewska, M.; Kohrle, J.; et al. Aminoglycoside-Driven Biosynthesis of Selenium-Deficient Selenoprotein P. Sci. Rep. 2017, 7, 4391. [Google Scholar] [CrossRef]
- Lee, B.J.; Worland, P.J.; Davis, J.N.; Stadtman, T.C.; Hatfield, D.L. Identification of a Selenocysteyl-TRNA(Ser) in Mammalian Cells That Recognizes the Nonsense Codon, UGA. J. Biol. Chem. 1989, 264, 9724–9727. [Google Scholar] [CrossRef]
- Cridge, A.G.; Major, L.L.; Mahagaonkar, A.A.; Poole, E.S.; Isaksson, L.A.; Tate, W.P. Comparison of Characteristics and Function of Translation Termination Signals between and within Prokaryotic and Eukaryotic Organisms. Nucleic Acids Res. 2006, 34, 1959–1973. [Google Scholar] [CrossRef]
- Fixsen, S.M.; Howard, M.T. Processive Selenocysteine Incorporation during Synthesis of Eukaryotic Selenoproteins. J. Mol. Biol. 2010, 399, 385–396. [Google Scholar] [CrossRef]
- Moriarty, P.M.; Reddy, C.C.; Maquat, L.E. Selenium Deficiency Reduces the Abundance of MRNA for Se-Dependent Glutathione Peroxidase 1 by a UGA-Dependent Mechanism Likely to Be Nonsense Codon-Mediated Decay of Cytoplasmic MRNA. Mol. Cell. Biol. 1998, 18, 2932–2939. [Google Scholar] [CrossRef]
- Schuller, A.P.; Green, R. Roadblocks and Resolutions in Eukaryotic Translation. Nat. Rev. Mol. Cell. Biol. 2018. [Google Scholar] [CrossRef]
- Taguchi, T.; Kurata, M.; Onishi, I.; Kinowaki, Y.; Sato, Y.; Shiono, S.; Ishibashi, S.; Ikeda, M.; Yamamoto, M.; Kitagawa, M.; et al. SECISBP2 Is a Novel Prognostic Predictor That Regulates Selenoproteins in Diffuse Large B-Cell Lymphoma. Lab. Investig. 2021, 101, 218–227. [Google Scholar] [CrossRef]
- Cheng, Q.; Roveri, A.; Cozza, G.; Bordin, L.; Rohn, I.; Schwerdtle, T.; Kipp, A.; Ursini, F.; Maiorino, M.; Miotto, G.; et al. Production and Purification of Homogenous Recombinant Human Selenoproteins Reveals a Unique Codon Skipping Event in E. Coli and GPX4-Specific Affinity to Bromosulfophthalein. Redox Biol. 2021, 46, 102070. [Google Scholar] [CrossRef]
- Rodnina, M.V.; Korniy, N.; Klimova, M.; Karki, P.; Peng, B.Z.; Senyushkina, T.; Belardinelli, R.; Maracci, C.; Wohlgemuth, I.; Samatova, E.; et al. Translational Recoding: Canonical Translation Mechanisms Reinterpreted. Nucleic Acids Res. 2020, 48, 1056–1067. [Google Scholar] [CrossRef]
- Farabaugh, P.J.; Zhao, H.; Vimaladithan, A. A Novel Programed Frameshift Expresses the POL3 Gene of Retrotransposon Ty3 of Yeast: Frameshifting without TRNA Slippage. Cell 1993, 74, 93–103. [Google Scholar] [CrossRef]
- Dinman, J.D. Mechanisms and Implications of Programmed Translational Frameshifting. Wiley Interdiscip. Rev. RNA 2012, 3, 661–673. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohleber, S.; Fradejas-Villar, N.; Zhao, W.; Reuter, U.; Schweizer, U. High-Resolution Ribosome Profiling Reveals Gene-Specific Details of UGA Re-Coding in Selenoprotein Biosynthesis. Biomolecules 2022, 12, 1504. https://doi.org/10.3390/biom12101504
Bohleber S, Fradejas-Villar N, Zhao W, Reuter U, Schweizer U. High-Resolution Ribosome Profiling Reveals Gene-Specific Details of UGA Re-Coding in Selenoprotein Biosynthesis. Biomolecules. 2022; 12(10):1504. https://doi.org/10.3390/biom12101504
Chicago/Turabian StyleBohleber, Simon, Noelia Fradejas-Villar, Wenchao Zhao, Uschi Reuter, and Ulrich Schweizer. 2022. "High-Resolution Ribosome Profiling Reveals Gene-Specific Details of UGA Re-Coding in Selenoprotein Biosynthesis" Biomolecules 12, no. 10: 1504. https://doi.org/10.3390/biom12101504
APA StyleBohleber, S., Fradejas-Villar, N., Zhao, W., Reuter, U., & Schweizer, U. (2022). High-Resolution Ribosome Profiling Reveals Gene-Specific Details of UGA Re-Coding in Selenoprotein Biosynthesis. Biomolecules, 12(10), 1504. https://doi.org/10.3390/biom12101504






