Micro RNAs—The Small Big Players in Hepatitis E Virus Infection: A Comprehensive Review
Abstract
:1. Introduction
2. Hepatitis E Virus
2.1. Taxonomy and Etiology
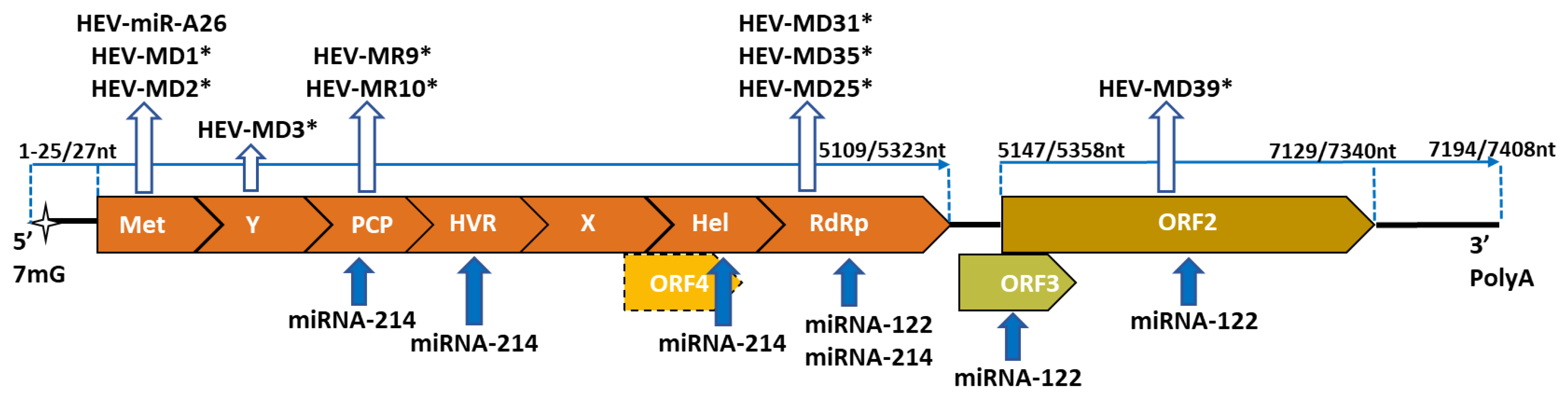
2.2. Genome Organization
2.3. Tropism
3. miRNA and the Viral Hepatitis
4. Hepatitis E Virus miRNA
4.1. HEV-miR-A26
4.2. HEV-miRNAs
5. Host miRNA Affecting HEV Life Cycle
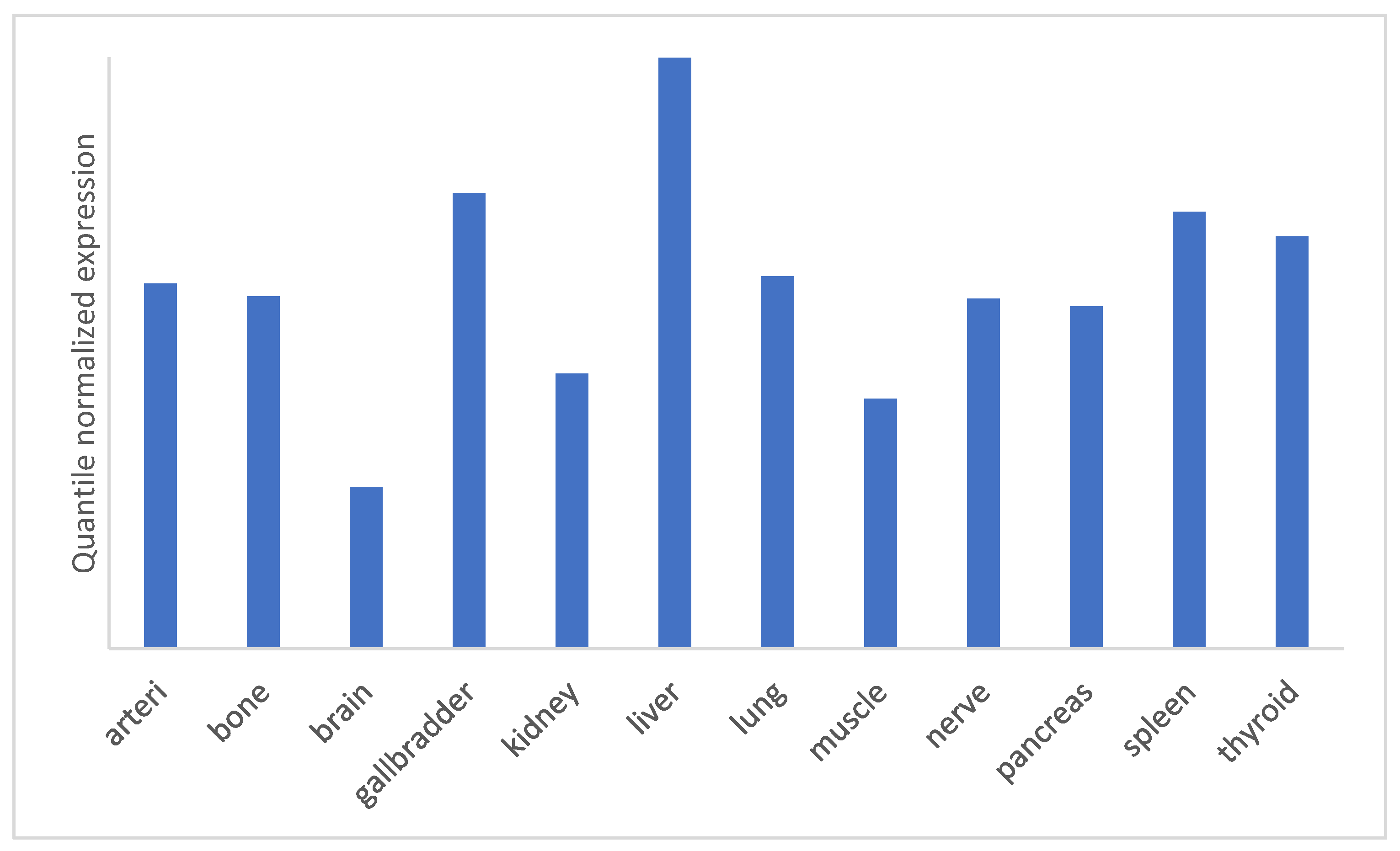
5.1. miRNA-122
5.2. miRNA-214
6. miRNA Pattern during HEV Infection
7. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pfeffer, S.; Zavolan, M.; Grässer, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Furuyama, W.; Lin, Z. RNA Virus-Encoded miRNAs: Current Insights and Future Challenges. Front. Microbiol. 2021, 12, 679210. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.-J.; Norder, H.; Okamoto, H.; van der Poel, W.H.M.; Reuter, G.; et al. Update: Proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698. [Google Scholar] [CrossRef]
- Nelson, K.E.; Labrique, A.B.; Kmush, B.L. Epidemiology of Genotype 1 and 2 Hepatitis E Virus Infections. Cold Spring Harb. Perspect. Med. 2019, 9, a031732. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wu, X.; Xia, J. Hepatitis E virus infection during pregnancy. Virol. J. 2020, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Mirazo, S.; Ramos, N.; Mainardi, V.; Arbiza, J.; Gerona, S. Transmission, diagnosis, and management of hepatitis E: An update. Hepatic Med. Evid. Res. 2014, 6, 45–59. [Google Scholar] [CrossRef]
- Purdy, M.A.; Khudyakov, Y.E. Evolutionary history and population dynamics of hepatitis E virus. PLoS ONE 2010, 5, e14376. [Google Scholar] [CrossRef] [PubMed]
- Papp, C.-P.; Biedermann, P.; Harms, D.; Wang, B.; Kebelmann, M.; Choi, M.; Helmuth, J.; Corman, V.M.; Thürmer, A.; Altmann, B.; et al. Advanced sequencing approaches detected insertions of viral and human origin in the viral genome of chronic hepatitis E virus patients. Sci. Rep. 2022, 12, 1720. [Google Scholar] [CrossRef]
- Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202103665 (accessed on 19 September 2022).
- Baha, S.; Behloul, N.; Liu, Z.; Wei, W.; Shi, R.; Meng, J. Comprehensive analysis of genetic and evolutionary features of the hepatitis E virus. BMC Genom. 2019, 20, 790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.-H.; Tan, B.-H.; Teo, E.C.-Y.; Lim, S.-G.; Dan, Y.-Y.; Wee, A.; Aw, P.P.K.; Zhu, Y.; Hibberd, M.; Tan, C.-K.; et al. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 2016, 150, 355–357. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Seth, R.; Gupta, A.; Nayak, B.; Acharya, S.K.; Das, P. Chronic hepatitis E-an emerging disease in an immunocompromised host. Gastroenterol. Rep. 2018, 6, 152–155. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Zhang, H.; Huang, W.; Harrison, T.J.; Geng, K.; Li, Z.; Wang, Y. Persistent hepatitis E virus genotype 4 infection in a child with acute lymphoblastic leukemia. Hepat. Mon. 2014, 14, e15618. [Google Scholar] [CrossRef] [Green Version]
- Nair, V.P.; Anang, S.; Subramani, C.; Madhvi, A.; Bakshi, K.; Srivastava, A.; Nayak, B.; Ranjith Kumar, C.T.; Surjit, M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepa-titis E Virus. PLoS Pathog. 2016, 12, e1005521. [Google Scholar] [CrossRef]
- Wang, B.; Meng, X.-J. Structural and molecular biology of hepatitis E virus. Comput. Struct. Biotechnol. J. 2021, 19, 1907–1916. [Google Scholar] [CrossRef]
- Kenney, S.P.; Meng, X.-J. Hepatitis E Virus Genome Structure and Replication Strategy. Cold Spring Harb. Perspect. Med. 2019, 9, a031724. [Google Scholar] [CrossRef]
- LeDesma, R.; Nimgaonkar, I.; Ploss, A. Hepatitis E Virus Replication. Viruses 2019, 11, 719. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Qu, C.; Yu, P.; Ou, X.; Pan, Q.; Wang, W. The Interplay between Host Innate Immunity and Hepatitis E Virus. Viruses 2019, 11, 541. [Google Scholar] [CrossRef] [Green Version]
- Haldipur, B.; Bhukya, P.L.; Arankalle, V.; Lole, K. Positive Regulation of Hepatitis E Virus Replication by MicroRNA-122. J. Virol. 2018, 92, e01999-17. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Lemon, S.M. Innate Immunity to Enteric Hepatitis Viruses. Cold Spring Harb. Perspect. Med. 2019, 9, a033464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nan, Y.; Yu, Y.; Ma, Z.; Khattar, S.K.; Fredericksen, B.; Zhang, Y.-J. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J. Virol. 2014, 88, 11924–11932. [Google Scholar] [CrossRef]
- Lhomme, S.; Abravanel, F.; Dubois, M.; Sandres-Saune, K.; Mansuy, J.M.; Rostaing, L.; Kamar, N.; Izopet, J. Characterization of the polyproline region of the hepatitis E virus in immunocompromised patients. J. Virol. 2014, 88, 12017–12025. [Google Scholar] [CrossRef] [Green Version]
- Van Tong, H.; Hoan, N.X.; Wang, B.; Wedemeyer, H.; Bock, C.T.; Velavan, T.P. Hepatitis E Virus Mutations: Functional and Clinical Relevance. EBioMedicine 2016, 11, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Fousekis, F.S.; Mitselos, I.V.; Christodoulou, D.K. Extrahepatic manifestations of hepatitis E virus: An overview. Clin. Mol. Hepatol. 2020, 26, 16–23. [Google Scholar] [CrossRef]
- Jung, S.; Seo, D.J.; Yeo, D.; Wang, Z.; Min, A.; Zhao, Z.; Song, M.; Choi, I.-S.; Myoung, J.; Choi, C. Experimental infection of hepatitis E virus induces pancreatic necroptosis in miniature pigs. Sci. Rep. 2020, 10, 12022. [Google Scholar] [CrossRef]
- Soomro, M.H.; Shi, R.; She, R.; Yang, Y.; Hu, F.; Li, H. Antigen detection and apoptosis in mongolian gerbil’s kidney experimentally intraperitoneally infected by swine hepatitis e virus. Virus Res. 2016, 213, 343–352. [Google Scholar] [CrossRef]
- Lhomme, S.; Marion, O.; Abravanel, F.; Izopet, J.; Kamar, N. Clinical Manifestations, Pathogenesis and Treatment of Hepatitis E Virus Infections. J. Clin. Med. 2020, 9, 331. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Zhao, C.; Huang, W.; Harrison, T.J.; Zhang, H.; Geng, K.; Wang, Y.J. Detection and assessment of infectivity of hepatitis e virus in urine. J. Hepatol. 2016, 64, 37–43. [Google Scholar] [CrossRef]
- Shrivastava, S.; Steele, R.; Ray, R.; Ray, R.B. MicroRNAs: Role in Hepatitis C Virus pathogenesis. Genes Dis. 2015, 2, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Trobaugh, D.W.; Klimstra, W.B. MicroRNA Regulation of RNA Virus Replication and Pathogenesis. Trends Mol. Med. 2017, 23, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Chipman, L.B.; Pasquinelli, A.E. miRNA Targeting: Growing beyond the Seed. Trends Genet. 2019, 35, 215–222. [Google Scholar] [CrossRef]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Chen, X.M. MicroRNA signatures in liver diseases. World J. Gastroenterol. 2009, 15, 1665–1672. [Google Scholar] [CrossRef] [Green Version]
- Loureiro, D.; Tout, I.; Narguet, S.; Benazzouz, S.M.; Mansouri, A.; Asselah, T. miRNAs as Potential Biomarkers for Viral Hepatitis B and C. Viruses 2020, 12, 1440. [Google Scholar] [CrossRef]
- Hu, J.; Xu, Y.; Hao, J.; Wang, S.; Li, C.; Meng, S. MiR-122 in hepatic function and liver diseases. Protein Cell 2012, 3, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. MiR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [Green Version]
- Lefkowitch, J.H. Acute Viral Hepatitis. Scheuer’s Liver Biopsy Interpret. 2021, 89–107. [Google Scholar] [CrossRef]
- Walker, P.J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Dempsey, D.M.; Dutilh, B.E.; García, M.L.; Hendrickson, R.C.; et al. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch. Virol. 2022, 167, 2429–2440. [Google Scholar] [CrossRef]
- Tabata, K.; Neufeldt, C.J.; Bartenschlager, R. Hepatitis C Virus Replication. Cold Spring Harb. Perspect. Med. 2020, 10, a037093. [Google Scholar] [CrossRef]
- Kunden, R.D.; Khan, J.Q.; Ghezelbash, S.; Wilson, J.A. The Role of the Liver-Specific microRNA, miRNA-122 in the HCV Replication Cycle. Int. J. Mol. Sci. 2020, 21, 5677. [Google Scholar] [CrossRef]
- Schult, P.; Roth, H.; Adams, R.L.; Mas, C.; Imbert, L.; Orlik, C.; Ruggieri, A.; Pyle, A.M.; Lohmann, V. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018, 9, 2613. [Google Scholar] [CrossRef]
- Machlin, E.S.; Sarnow, P.; Sagan, S.M. Masking the 5’ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. USA 2011, 108, 3193–3198. [Google Scholar] [CrossRef] [Green Version]
- Kunden, R.D.; Ghezelbash, S.; Khan, J.Q.; Wilson, J.A. Location specific annealing of miR-122 and other small RNAs defines an Hepatitis C Virus 5’ UTR regulatory element with distinct impacts on virus translation and genome stability. Nucleic Acids Res. 2020, 48, 9235–9249. [Google Scholar] [CrossRef]
- Li, H.C.; Yang, C.H.; Lo, S.Y. Roles of microRNAs in Hepatitis C Virus Replication and Patho-genesis. Viruses 2022, 14, 1776. [Google Scholar] [CrossRef]
- Lamontagne, J.; Steel, L.F.; Bouchard, M.J. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J. Gastroenterol. 2015, 21, 7375–7399. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Sun, H.; Fan, H.; Hu, Y.; Liu, M.; Li, X.; Tang, H. Hepatitis B Virus-Encoded MicroRNA Controls Viral Replication. J. Virol. 2017, 91, e01919-16. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.M. Host RNA circles and the origin of hepatitis delta virus. World J. Gastroenterol. 2014, 20, 2971–2978. [Google Scholar] [CrossRef]
- Chang, J.; Provost, P.; Taylor, J.M. Resistance of human hepatitis delta virus RNAs to dicer activity. J. Virol. 2003, 77, 11910–11917. [Google Scholar] [CrossRef] [Green Version]
- McKnight, K.L.; Lemon, S.M. Hepatitis A Virus Genome Organization and Replication Strategy. Cold Spring Harb. Perspect. Med. 2018, 8, a033480. [Google Scholar] [CrossRef]
- Shi, J.; Sun, J.; Wang, B.; Wu, M.; Zhang, J.; Duan, Z.; Wang, H.; Hu, N.; Hu, Y. Novel microRNA-like viral small regulatory RNAs arising during human hepatitis A virus infection. FASEB J. 2014, 28, 4381–4393. [Google Scholar] [CrossRef]
- Shi, J.; Sun, J.; Wu, M.; Hu, N.; Hu, Y. Hepatitis A virus-encoded miRNAs attenuate the accumulation of viral genomic RNAs in infected cells. Virus Genes 2016, 52, 317–324. [Google Scholar] [CrossRef]
- Shi, J.; Sun, J.; Wu, M.; Wang, H.; Hu, N.; Hu, Y. Comprehensive profiling and characterization of cellular miRNAs in response to hepatitis A virus infection in human fibroblasts. Infect. Genet. Evol. 2016, 45, 176–186. [Google Scholar] [CrossRef]
- Xu, L.-D.; Zhang, F.; Peng, L.; Luo, W.-T.; Chen, C.; Xu, P.; Huang, Y.-W. Stable Expression of a Hepatitis E Virus (HEV) RNA Replicon in Two Mammalian Cell Lines to Assess Mechanism of Innate Immunity and Antiviral Response. Front. Microbiol. 2020, 11, 603699. [Google Scholar] [CrossRef]
- Qian, Z.; Yang, C.; Xu, L.; Mickael, H.K.; Chen, S.; Zhang, Y.; Xia, Y.; Li, T.; Yu, W.; Huang, F. Hepatitis E virus-encoded microRNA promotes viral replication by inhibiting type I interferon. FASEB J. 2022, 36, e22104. [Google Scholar] [CrossRef]
- Baruah, V.; Bose, S. Computational identification of hepatitis E virus-encoded microRNAs and their targets in human. J. Med. Virol. 2019, 91, 1545–1552. [Google Scholar] [CrossRef]
- Chaikuad, A.; Keates, T.; Vincke, C.; Kaufholz, M.; Zenn, M.; Zimmermann, B.; Gutiérrez, C.; Zhang, R.; Hatzos-Skintges, C.; Joachimiak, A.; et al. Structure of cyclin G-associated kinase (GAK) trapped in different conformations using nanobodies. Biochem. J. 2014, 459, 59–69. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Feng, Z. Hepatitis E Virus Entry. Viruses 2019, 11, 883. [Google Scholar] [CrossRef] [Green Version]
- Haldipur, B.; Arankalle, V. Circulating miR-122 levels in self-recovering hepatitis E patients. ExRNA 2019, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Harms, D.; Choi, M.; Allers, K.; Wang, B.; Pietsch, H.; Papp, C.-P.; Hanisch, L.; Kurreck, J.; Hofmann, J.; Bock, C.-T. Specific circulating microRNAs during hepatitis E infection can serve as indicator for chronic hepatitis E. Sci. Rep. 2020, 10, 5337. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Xu, S.J.; Xie, S.J.; Zhang, Y.; Yang, J.H.; Zhang, W.Q.; Zheng, M.-N.; Zhou, H.; Qu, L.-H. MicroRNA-122 supports robust innate immunity in hepatocytes by targeting the RTKs/STAT3 signaling pathway. eLife 2019, 8, e41159. [Google Scholar] [CrossRef]
- Li, C.; Deng, M.; Hu, J.; Li, X.; Chen, L.; Ju, Y.; Hao, J.; Meng, S. Chronic inflammation contributes to the development of hepatocellular carcinoma by decreasing miR-122 levels. Oncotarget 2016, 7, 17021–17034. [Google Scholar] [CrossRef]
- Penna, E.; Orso, F.; Taverna, D. miR-214 as a key hub that controls cancer networks: Small player, multiple functions. J. Investig. Dermatol. 2015, 135, 960–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, M.M.J.; Trevelyan, C.J.; Turner, N.A. MicroRNA-214 in Health and Disease. Cells 2021, 10, 3274. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; Borentain, P.; Gérolami, R. Hepatitis E virus as an agent of hepatocellular carcinoma. Int. J. Infect. Dis. 2019, 80, 62–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, R.N.; Karpe, Y.A. Uncovering the Roles of miR-214 in Hepatitis E Virus Replication. J. Mol. Biol. 2020, 432, 5322–5342. [Google Scholar] [CrossRef]
- Kanade, G.D.; Pingale, K.D.; Karpe, Y.A. Activities of Thrombin and Factor Xa Are Essential for Replication of Hepatitis E Virus and Are Possibly Implicated in ORF1 Polyprotein Processing. J. Virol. 2018, 92, e01853-17. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhang, H.; Hao, H.; Zhang, X.; Song, H.; He, B.; Wang, Y.; Zhou, Y.; Zhu, Z.; Hu, Y.; et al. Thrombin induces morphological and inflammatory astrocytic responses via activation of PAR1 receptor. Cell Death Discov. 2022, 8, 189. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- McGowan, K.; Simpson, K.J.; Petrik, J. Expression Profiles of Exosomal MicroRNAs from HEV- and HCV-Infected Blood Donors and Patients: A Pilot Study. Viruses 2020, 12, 833. [Google Scholar] [CrossRef]
- Trehanpati, N.; Sehgal, R.; Patra, S.; Vyas, A.; Vasudevan, M.; Khosla, R.; Khanam, A.; Kumar, G.; Maiwall, R.; Ramakrishna, G.; et al. miRNA signatures can predict acute liver failure in hepatitis E infected pregnant females. Heliyon 2017, 3, e00287. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golkocheva-Markova, E. Micro RNAs—The Small Big Players in Hepatitis E Virus Infection: A Comprehensive Review. Biomolecules 2022, 12, 1543. https://doi.org/10.3390/biom12111543
Golkocheva-Markova E. Micro RNAs—The Small Big Players in Hepatitis E Virus Infection: A Comprehensive Review. Biomolecules. 2022; 12(11):1543. https://doi.org/10.3390/biom12111543
Chicago/Turabian StyleGolkocheva-Markova, Elitsa. 2022. "Micro RNAs—The Small Big Players in Hepatitis E Virus Infection: A Comprehensive Review" Biomolecules 12, no. 11: 1543. https://doi.org/10.3390/biom12111543






