Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis
Abstract
1. Introduction
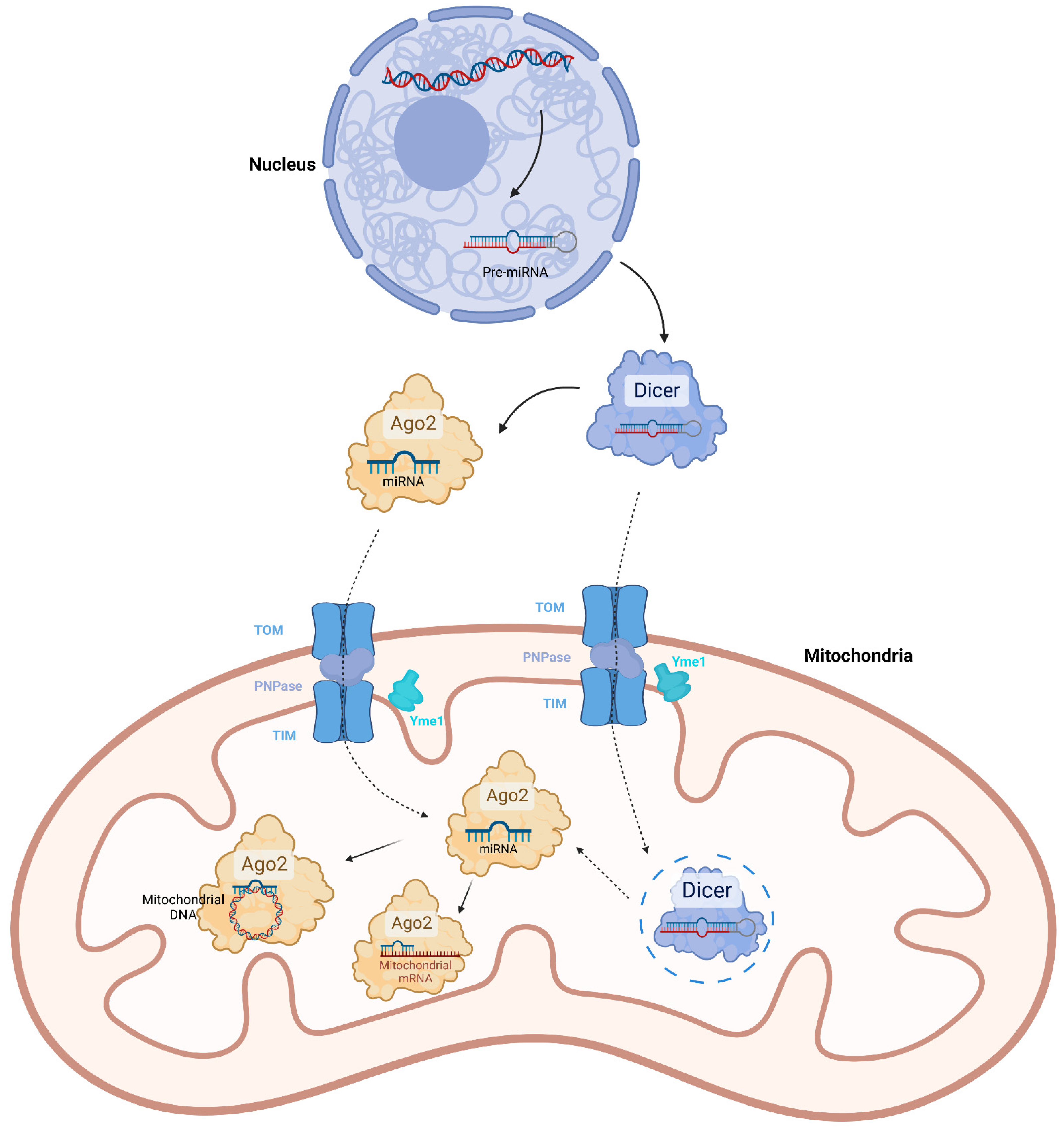
2. Mitochondrial miRNAs
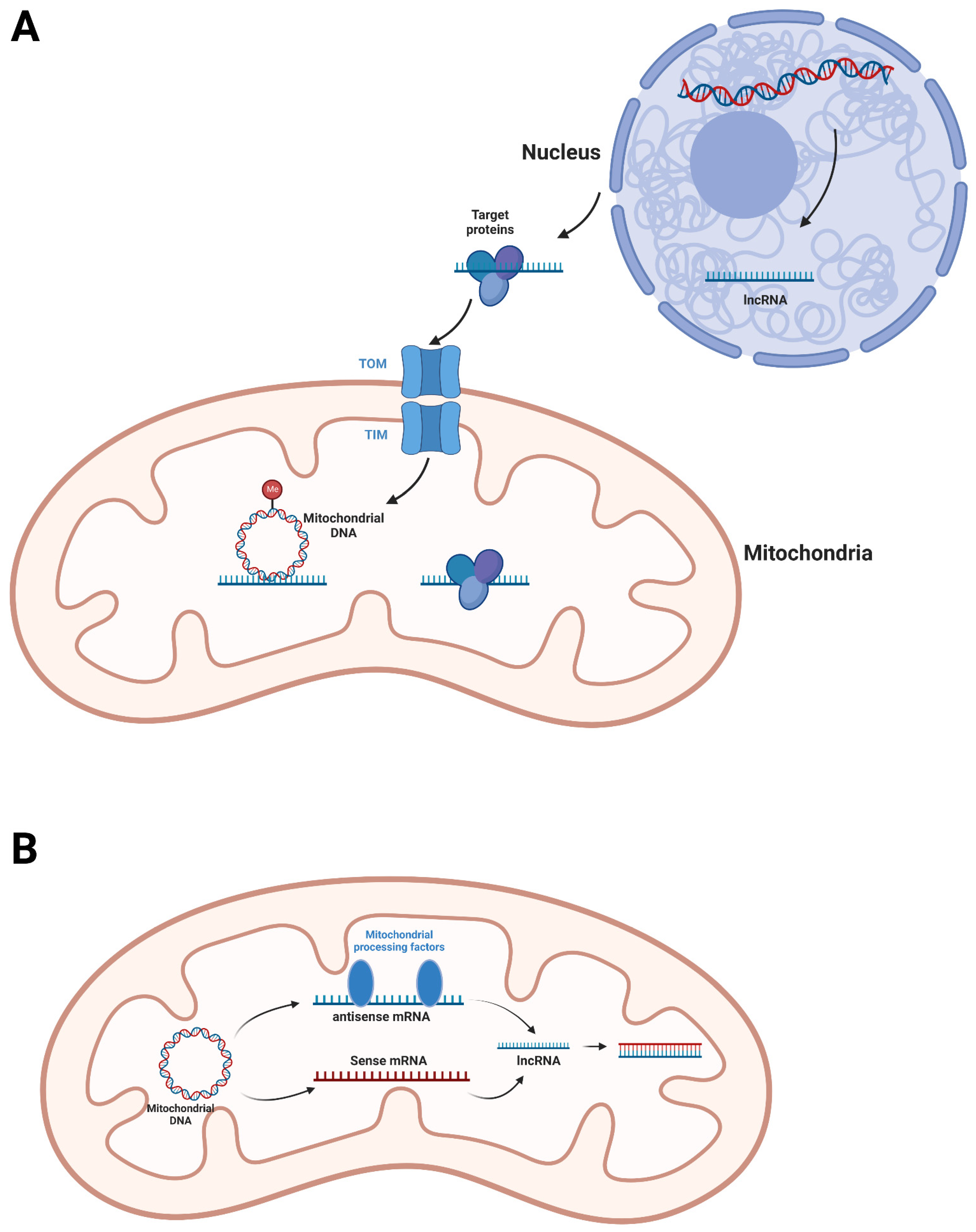
3. Mitochondrial lncRNA
4. Mitochondrial Piwi-Interacting RNAs
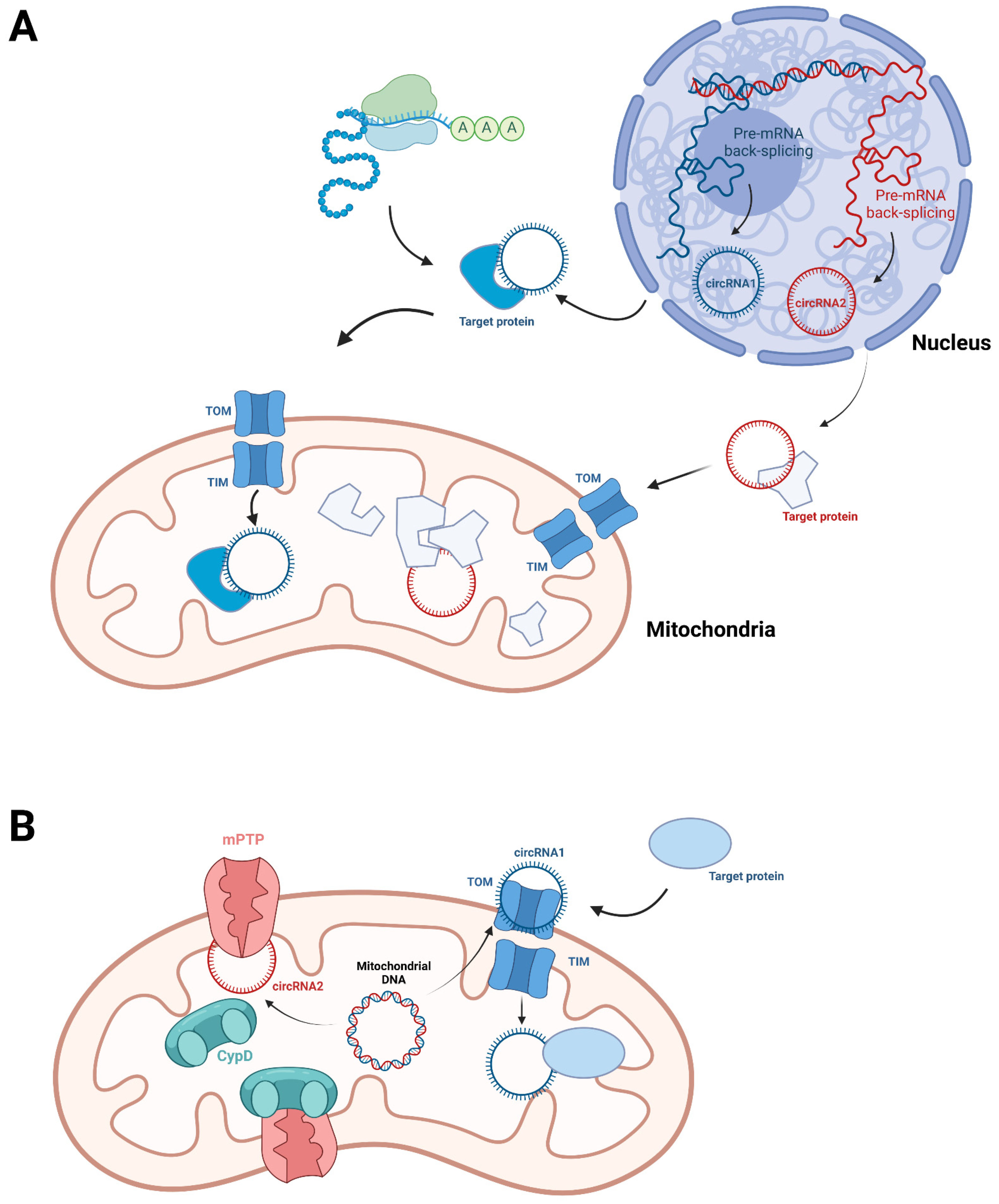
5. Mitochondrial circRNA
6. Mitochondrial ncRNAs Mediate Mitochondrial Homeostasis
7. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 4, a011403. [Google Scholar] [CrossRef] [PubMed]
- Tao, M.; You, C.P.; Zhao, R.R.; Liu, S.-J.; Zhang, Z.-H.; Zhang, C.; Liu, Y. Animal mitochondria: Evolution, function, and disease. Curr. Mol. Med. 2014, 14, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Garbincius, J.F.; Elrod, J.W. Mitochondrial calcium exchange in physiology and disease. Physiol. Rev. 2022, 102, 893–992. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, O.; Quirós, P.M.; Auwerx, J. Mitochondria and Epigenetics—Crosstalk in Homeostasis and Stress. Trends Cell Biol. 2017, 27, 453–463. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; De Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Virciglio, C.; Abel, Y.; Rederstorff, M. Regulatory Non-Coding RNAs: An Overview. Methods Mol. Biol. 2021, 2300, 3–9. [Google Scholar] [CrossRef]
- Kren, B.T.; Wong, P.Y.; Sarver, A.; Zhang, X.; Zeng, Y.; Steer, C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009, 6, 65–72. [Google Scholar] [CrossRef]
- Bian, Z.; Li, L.M.; Tang, R.; Hou, D.-X.; Chen, X.; Zhang, C.-Y.; Zen, K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010, 20, 1076–1078. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.-M.J.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Bandiera, S.; Rüberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chrétien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef] [PubMed]
- Sripada, L.; Tomar, D.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: Detailed analysis of mitochondrial associated miRNA. PLoS ONE 2012, 7, e44873. [Google Scholar] [CrossRef] [PubMed]
- Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Stricker, J.C.; Croston, T.L.; Baseler, W.A.; Lewis, S.E.; Martinez, I.; Hollander, J.M. Translational Regulation of the Mitochondrial Genome Following Redistribution of Mitochondrial MicroRNA in the Diabetic Heart. Circ. Cardiovasc. Genet. 2015, 8, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Visavadiya, N.P.; Pandya, J.D.; Nelson, P.T.; Sullivan, P.G.; Springer, J.E. Mitochondria-associated microRNAs in rat hippocampus following traumatic brain injury. Exp. Neurol. 2015, 265, 84–93. [Google Scholar] [CrossRef]
- Wang, X.; Song, C.; Zhou, X.; Han, X.; Li, J.; Wang, Z.; Shang, H.; Liu, Y.; Cao, H. Mitochondria Associated MicroRNA Expression Profiling of Heart Failure. Biomed. Res. Int. 2017, 2017, 4042509. [Google Scholar] [CrossRef]
- Sang, L.; Ju, H.Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 2021, 3, 90–106. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.; Li, H.; Sun, T.; Wen, X.; Li, X.; Meng, Y.; Li, Y.; Liu, M.; Liu, S.; et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol. Ther. Nucleic Acids 2020, 23, 264–276. [Google Scholar] [CrossRef]
- Kwon, C.; Tak, H.; Rho, M.; Chang, H.R.; Kim, Y.H.; Kim, K.T.; Balch, C.; Lee, E.K.; Nam, S. Detection of PIWI and piRNAs in the mitochondria of mammalian cancer cells. Biochem. Biophys. Res. Commun. 2014, 446, 218–223. [Google Scholar] [CrossRef]
- Larriba, E.; Rial, E.; Del Mazo, J. The landscape of mitochondrial small non-coding RNAs in the PGCs of male mice, spermatogonia, gametes and in zygotes. BMC Genom. 2018, 19, 634. [Google Scholar] [CrossRef] [PubMed]
- Barreñada, O.; Larriba, E.; Fernández-Pérez, D.; Brieño-Enríquez, M.Á.; Del Mazo Martínez, J. Unraveling mitochondrial piRNAs in mouse embryonic gonadal cells. Sci. Rep. 2022, 12, 10730. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, X.; Li, J.; Hu, S.; Deng, Y.; Yin, H.; Bao, X.; Zhang, Q.C.; Wang, G.; Wang, B.; et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 2020, 63, 1429–1449. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Lung, B.; Zemann, A.; Madej, M.J.; Schuelke, M.; Techritz, S.; Ruf, S.; Bock, R.; Hüttenhofer, A. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res. 2006, 34, 3842–3852. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef]
- Gregory, R.I.; Chendrimada, T.P.; Cooch, N.; Shiekhattar, R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005, 123, 631–640. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Finn, K.J.; Ji, X.; Baillat, D.; Gregory, R.I.; Liebhaber, S.A.; Pasquinelli, A.E.; Shiekhattar, R. MicroRNA silencing through RISC recruitment of eIF. Nature 2007, 447, 823–828. [Google Scholar] [CrossRef]
- Chen, H.W.; Koehler, C.M.; Teitell, M.A. Human polynucleotide phosphorylase: Location matters. Trends Cell Biol. 2007, 17, 600–608. [Google Scholar] [CrossRef]
- Wang, G.; Chen, H.W.; Oktay, Y.; Zhang, J.; Allen, E.L.; Smith, G.M.; Fan, K.C.; Hong, J.S.; French, S.W.; McCaffery, J.M.; et al. PNPASE regulates RNA import into mitochondria. Cell 2010, 142, 456–467. [Google Scholar] [CrossRef]
- Shepherd, D.L.; Hathaway, Q.A.; Pinti, M.V.; Nichols, C.E.; Durr, A.J.; Sreekumar, S.; Hughes, K.M.; Stine, S.M.; Martinez, I.; Hollander, J.M. Exploring the mitochondrial microRNA import pathway through Polynucleotide Phosphorylase (PNPase). J. Mol. Cell Cardiol. 2017, 110, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Shu, Z.; Lieser, S.A.; Chen, P.L.; Lee, W.H. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J. Biol. Chem. 2009, 284, 20812–20821. [Google Scholar] [CrossRef] [PubMed]
- Erturk, E.; Enes Onur, O.; Akgun, O.; Tuna, G.; Yildiz, Y.; Ari, F. Mitochondrial miRNAs (MitomiRs): Their potential roles in breast and other cancers. Mitochondrion 2022, 66, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, X.; Wang, F.; Zhou, L.; Yin, Z.; Fan, J.; Nie, X.; Wang, P.; Fu, X.D.; Chen, C.; et al. MicroRNA-21 Lowers Blood Pressure in Spontaneous Hypertensive Rats by Upregulating Mitochondrial Translation. Circulation 2016, 134, 734–751. [Google Scholar] [CrossRef]
- Harma, P.; Sharma, V.; Ahluwalia, T.S.; Dogra, N.; Kumar, S.; Singh, S. Let-7a induces metabolic reprogramming in breast cancer cells via targeting mitochondrial encoded ND. Cancer Cell Int. 2021, 21, 629. [Google Scholar] [CrossRef]
- Das, S.; Bedja, D.; Campbell, N.; Dunkerly, B.; Chenna, V.; Maitra, A.; Steenbergen, C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. PLoS ONE 2014, 9, e96820. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial miRNA Determines Chemoresistance by Reprogramming Metabolism and Regulating Mitochondrial Transcription. Cancer Res. 2019, 79, 1069–1084. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.-H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Li, K.; Smagula, C.S.; Parsons, W.J.; Richardson, J.A.; Gonzalez, M.; Hagler, H.K.; Williams, R.S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 1994, 124, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, S.; Zhou, L.; Li, X.; Meng, Y.; Li, Y.; Li, L.; Jiao, B.; Bai, L.; Yu, Y.; et al. Aberrant shuttling of long noncoding RNAs during the mitochondria-nuclear crosstalk in hepatocellular carcinoma cells. Am. J. Cancer Res. 2019, 9, 999–1008. [Google Scholar] [PubMed]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, Y.; Fujiu, K.; Yuki, R.; Oishi, Y.; Morioka, M.S.; Isagawa, T.; Matsuda, J.; Oshima, T.; Matsubara, T.; Sugita, J.; et al. A long noncoding RNA regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation. Proc. Natl. Acad. Sci. USA 2020, 117, 14365–14375. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010, 39, 925–938. [Google Scholar] [CrossRef]
- Mohammad, G.; Kowluru, R.A. Nuclear Genome-Encoded Long Noncoding RNAs and Mitochondrial Damage in Diabetic Retinopathy. Cells 2021, 10, 3271. [Google Scholar] [CrossRef]
- Villegas, J.; Zárraga, A.M.; Muller, I.; Montecinos, L.; Werner, E.; Brito, M.; Meneses, A.M.; Burzio, L.O. A novel chimeric mitochondrial RNA localized in the nucleus of mouse sperm. DNA Cell Biol. 2000, 19, 579–588. [Google Scholar] [CrossRef]
- Villegas, J.; Burzio, V.; Villota, C.; Landerer, E.; Martinez, R.; Santander, M.; Martinez, R.; Pinto, R.; Vera, M.I.; Boccardo, E.; et al. Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res. 2007, 35, 7336–7347. [Google Scholar] [CrossRef]
- Landerer, E.; Villegas, J.; Burzio, V.A.; Oliveira, L.; Villota, C.; Lopez, C.; Restovic, F.; Martinez, R.; Castillo, O.; Burzio, L.O. Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell. Oncol. 2011, 34, 297–305. [Google Scholar] [CrossRef]
- Rackham, O.; Shearwood, A.M.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef]
- Gao, S.; Tian, X.; Chang, H.; Sun, Y.; Wu, Z.; Cheng, Z.; Dong, P.; Zhao, Q.; Ruan, J.; Bu, W. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion 2018, 38, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Bauters, C.; Volkmann, I.; Maury, F.; Fetisch, J.; Holzmann, A.; Lemesle, G.; de Groote, P.; Pinet, F.; Thum, T. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 2014, 114, 1569–1575. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ramat, A.; Simonelig, M.; Liu, M.F. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat. Rev. Mol. Cell Biol. 2022. [Google Scholar] [CrossRef]
- Honda, S.; Kirino, Y.; Maragkakis, M.; Alexiou, P.; Ohtaki, A.; Murali, R.; Mourelatos, Z.; Kirino, Y. Mitochondrial protein BmPAPI modulates the length of mature piRNAs. RNA 2013, 19, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Izumi, N.; Shoji, K.; Sakaguchi, Y.; Honda, S.; Kirino, Y.; Suzuki, T.; Katsuma, S.; Tomari, Y. Identification and Functional Analysis of the Pre-piRNA 3′ Trimmer in Silkworms. Cell 2016, 164, 962–973. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications [published correction appears in Cell 2022, 185, 2390]. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Zheng, H.; Huang, S.; Wei, G.; Sun, Y.; Li, C.; Si, X.; Chen, Y.; Tang, Z.; Li, X.; Chen, Y.; et al. CircRNA Samd4 induces cardiac repair after myocardial infarction by blocking mitochondria-derived ROS output. Mol. Ther. 2022, 30, 3477–3498. [Google Scholar] [CrossRef]
- Gong, W.; Xu, J.; Wang, Y.; Min, Q.; Chen, X.; Zhang, W.; Chen, J.; Zhan, Q. Nuclear genome-derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 2022, 7, 40. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Wu, Z.; Sun, H.; Wang, C.; Liu, W.; Liu, M.; Zhu, Y.; Xu, W.; Jin, H.; Li, J. Mitochondrial Genome-Derived circRNA mc-COX2 Functions as an Oncogene in Chronic Lymphocytic Leukemia. Mol. Ther. Nucleic Acids 2020, 20, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Liu, J.; Deng, H.; Ma, R.; Liao, J.-Y.; Liang, H.; Hu, J.; Li, J.; Guo, Z.; Cai, J.; et al. Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 2020, 183, 76–93.e22. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial complex III Rieske Fe-S protein processing and assembly. Cell Cycle 2018, 17, 681–687. [Google Scholar] [CrossRef]
- Alavian, K.N.; Beutner, G.; Lazrove, E.; Sacchetti, S.; Park, H.-A.; Licznerski, P.; Li, H.; Nabili, P.; Hockensmith, K.; Graham, M.; et al. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. USA 2014, 111, 10580–10585. [Google Scholar] [CrossRef]
- Kwong, J.Q.; Molkentin, J.D. Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab. 2015, 21, 206–214. [Google Scholar] [CrossRef]
- Elrod, J.W.; Molkentin, J.D. Physiologic functions of cyclophilin D and the mitochondrial permeability transition pore. Circ. J. 2013, 77, 1111–1122. [Google Scholar] [CrossRef]
- Giorgio, V.; von Stockum, S.; Antoniel, M.; Fabbro, A.; Fogolari, F.; Forte, M.; Glick, G.D.; Petronilli, V.; Zoratti, M.; Szabó, I.; et al. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. USA 2013, 110, 5887–5892. [Google Scholar] [CrossRef]
- Giorgio, V.; Burchell, V.; Schiavone, M.; Bassot, C.; Minervini, G.; Petronilli, V.; Argenton, F.; Forte, M.; Tosatto, S.; Lippe, G.; et al. Ca2+ binding to F-ATP synthase β subunit triggers the mitochondrial permeability transition. EMBO Rep. 2017, 18, 1065–1076. [Google Scholar] [CrossRef]
- Ye, Y.; Shibata, Y.; Kikkert, M.; van Voorden, S.; Wiertz, E.; Rapoport, T.A. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA 2005, 102, 14132–14138. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Peng, G.; Wang, Y.; Fang, S.; Karbowski, M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol. Biol. Cell. 2011, 22, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, Y.; Liang, H.; Meng, Y.; Liu, H.; Zhou, Y.; Huang, C.; An, B.; Mao, H.; Liao, Z. VDAC1 promotes cardiomyocyte autophagy in anoxia/reoxygenation injury via the PINK1/Parkin pathway. Cell Biol. Int. 2021, 45, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Sun, H.; Liang, X.; Lima, W.F.; Crooke, S.T. Human RNase H1 is associated with protein P32 and is involved in mitochondrial pre-rRNA processing. PLoS ONE 2013, 8, e71006. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, J. Replication protein A and more: Single-stranded DNA-binding proteins in eukaryotic cells. Acta Biochim. Biophys. Sin. 2016, 48, 665–670. [Google Scholar] [CrossRef] [PubMed]
| Methods | NcRNA Type | Sample Source | Data Size | References |
|---|---|---|---|---|
| Microarray | MiRNA | Mitochondria from adult rat liver | 15 identified miRNAs | [9] |
| Microarray | MiRNA | Mitochondria from the liver of adult male C57BL/6J mice | Patterns of miRNA expression in the mitochondria and liver tissue profiles | [10] |
| Directional deep sequencing | MiRNA and lncRNA | Mitochondria and mitoplasts from cultured human 143B cells | Full transcriptome | [11] |
| Microarray | MiRNA | Mitochondria from HeLa cells | 57 miRNAs differentially expressed in HeLa mitochondria and cytosol | [12] |
| RT-qPCR | MiRNAs | Mitochondria from human myoblasts | 160 detected miRNA | [13] |
| RNA-seq | MiRNAs | Mitochondria from HeLa and HEK293 cell | 428 and 327 mature miRNAs from HEK293 and HeLa cells | [14] |
| High-throughput microarray | MiRNAs | Mitochondria from mice heart | 78 miRNAs differentially regulated in type 1 diabetic insult and control | [15] |
| TaqMan® RT-qPCR Array | MiRNAs | Mitochondria from rat hippocampus | 285 detected miRNAs | [16] |
| RNA-seq | MiRNAs | Mitochondria from mice heart | 289 detected known miRNAs | [17] |
| DESeq2 | LncRNAs | Mitochondria from HEK293 cells | 23 mitochondrial lncRNAs | [18] |
| RNA-seq | LncRNAs | Mitochondria from HepG2 and HL7702 cells | Transcriptome | [19] |
| Bioinformatic analysis | PiRNAs | Data from Sripada et al. (PLoS ONE, 2012) and Mercer et al. (Cell, 2011) | 29 mitochondrial DNA-derived piRNAs | [20] |
| Bioinformatic analysis | MiRNAs and piRNAs | Data from Ku et al. (Natl Sci Rev., 2014) and García-López et al. (Biochim Biophys Acta. 2014) | MiRNAs and piRNAs: 56 and 3135 in primordial germ cells, 38 and 4403 in spermatogonia, 43 and 4152 in spermatozoa, 25 and 2703 in oocytes, 38 and 2502 zygotes | [21] |
| RNA-seq | PiRNAs | Embryonic gonads of mice | The genomic regions, expression level and potential roles of piRNA transcribed mitochondrial DNA | [22] |
| RNA-seq | CircRNAs | HeLa, HEK293T, RPE-1, HepG2, N2a, and NIH3T3 cells | 248 and 268 high-confidence (with ≥2 junction reads) mitochondria-encoded circRNAs in humans and mice | [23] |
| NcRNA | Involved Process | Type | Source | Target | Molecular Effect | Reference |
|---|---|---|---|---|---|---|
| miR-1 | OXPHOS | miRNA | C2C12 mice myoblasts | ND1 and COX1 mRNA | Enhancing translation of ND1 and COX1 | [34] |
| miR-21 | OXPHOS | miRNA | H9C2 rat cardiomyocytes | CYTB mRNA | Enhancing translation of CYTB | [35] |
| miR-181c | OXPHOS | miRNA | Neonatal rat ventricular myocytes | COX1 mRNA | Decreasing protein level of COX1 | [26] |
| miR-378 | OXPHOS | miRNA | HL-1 human cardiomyocyte | ATP6 mRNA | Decreasing protein level of ATP6 | [15,31] |
| let-7a | OXPHOS | miRNA | MDA-MB-231 and MCF-7 cells | ND4 mRNA | Decreasing protein level of ND4 | [36] |
| miR-2392 | OXPHOS | miRNA | CAL-27 and SCC-9 cells | Mitochondria DNA | Enhancing transcription of mitochondrial DNA | [38] |
| MALAT1 | OXPHOS | LncRNA | HepG2 Cells | Mitochondria DNA | Inhibiting methylation of mitochondrial DNA | [19] |
| GAS5 | TCA | LncRNA | MDA-MB-231, MDA-MB-468, HEK293T | MDH2 | Promoting the association of FH-MDH2-CS | [18] |
| LncFAO | β-oxidation | LncRNA | Mice bone marrow-derived macrophages | HADHB | Increasing of HADHB level | [44] |
| SAMMSON | Mitochondrial translation | LncRNA | Mel501 and SK-MEL-2 cells | P32 | Enhancing the mitochondrial localization of P32 | [43] |
| CircPUM1 | OXPHOS | CircRNA | Esophageal squamous cell carcinoma cells | UQCRC2 | Promoting the association of UQCRC1 and 2 | [60] |
| SCAR | MPTP opening | CircRNA | Primary liver fibroblasts | ATP5B | Inhibiting the interaction between ATP5B and CypD | [64] |
| CircSmad4 | MPTP opening | CircRNA | Mice neonatal cardiomyocytes | VCP | Enhancing the mitochondrial localization of VCP | [59] |
| MecciND1 | Mitochondrial DNA replication | CircRNA | HeLa, HEK293T, RPE-1, HepG2, N2a, and NIH3T3 cells | RPA32/70 | Enhancing the mitochondrial localization of RPA32/70 | [23] |
| NcRNA | Origination | Biogenesis | Translocation | Molecular Function |
|---|---|---|---|---|
| MiRNA | Nuclear genome | Transcribed by RNA polymerase II in nucleus. Processed by DROSHA, DGCR8 and Dicer, successively [24]. | The pre-miRNAs or mature miRNAs might be imported into mitochondria by Ago2 or PNPase [10,12,14,30,31]. | Directly bind mitochondrial mRNAs to promote or inhibit translation [15,26,34,35,36]. Directly bind mitochondrial DNA to inhibit transcription [38]. The function is Ago2-dependent. |
| Mitochondrial genome | The existence is not verified yet | |||
| LncRNA | Nuclear genome | Transcribed by RNA polymerase II and processed in nucleus [39]. | Might be translocated into mitochondria through their associated mitochondrial proteins [18,19,40]. | Directly bind nuclear-derived mitochondrial proteins to affect their translocation, stability, activity or association [18,43,44]. Interact with mitochondrial DNA to regulate DNA methylation [19]. |
| Mitochondrial genome | Transcribed in mitochondria. Might be processed by mitochondrial tRNA processing factors [50]. | Partially being translocated into nucleus [50]. The mechanism is not clear. | Might form RNA-RNA duplexes with their sense mitochondrial mRNAs to regulate stability of sense mRNAs [50]. | |
| CircRNA | Nuclear genome | Generated by back-splicing from pre-mRNAs [50,51]. | Might be translocated into mitochondria through their associated mitochondrial proteins [59]. | Interact with nuclear-derived mitochondrial protein to affect their mitochondrial localization or act as protein scaffold [59,60]. |
| Mitochondrial genome | The mechanism is not clear. Might be related to the short repeats located at the 5′ and 3′ junction sites [23]. | Some distributed in both mitochondria and the cytoplasm [23,63,64]. The mechanism is not clear. | Interact with nuclear-derived mitochondrial protein to affect their mitochondrial localization or protein association [23,64]. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Lu, Y.; Zhang, H.; Zhang, J.; Fang, X.; Wang, J.; Li, M. Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules 2022, 12, 1863. https://doi.org/10.3390/biom12121863
Sun W, Lu Y, Zhang H, Zhang J, Fang X, Wang J, Li M. Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules. 2022; 12(12):1863. https://doi.org/10.3390/biom12121863
Chicago/Turabian StyleSun, Weihan, Yijian Lu, Heng Zhang, Jun Zhang, Xinyu Fang, Jianxun Wang, and Mengyang Li. 2022. "Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis" Biomolecules 12, no. 12: 1863. https://doi.org/10.3390/biom12121863
APA StyleSun, W., Lu, Y., Zhang, H., Zhang, J., Fang, X., Wang, J., & Li, M. (2022). Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules, 12(12), 1863. https://doi.org/10.3390/biom12121863






