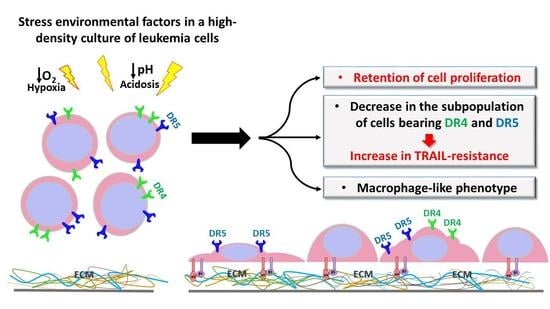
Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Protocol of izTRAIL Preparation
2.3. Cell Cultures
2.4. Cell Proliferation and Cell Viability Assays
2.5. Short Tandem Repeat (STR) Profiling
2.6. Microarray-Based Comparative Genomic Hybridization (aCGH) Assay
2.7. Cell Viability Assay
2.8. Cell Morphology Assay
2.9. Cell Adhesion Assay
2.10. Cell Immunophenotyping
2.11. Phagocytosis Assay
2.12. Intracellular ROS Assay
2.13. Intracellular NO Assay
2.14. Multiplex Analysis of Cytokine Production
2.15. Analysis of TRAIL Receptor Expression
2.16. Statistical Analysis
3. Results
3.1. Derivation and Characteristics of THP-1ad Cells
3.2. Adhesion of THP-1ad Cells to Extracellular Matrix Is Mediated by Integrin aVβ5
3.3. THP-1ad Cells Have a Similar Immunophenotype to Macrophages
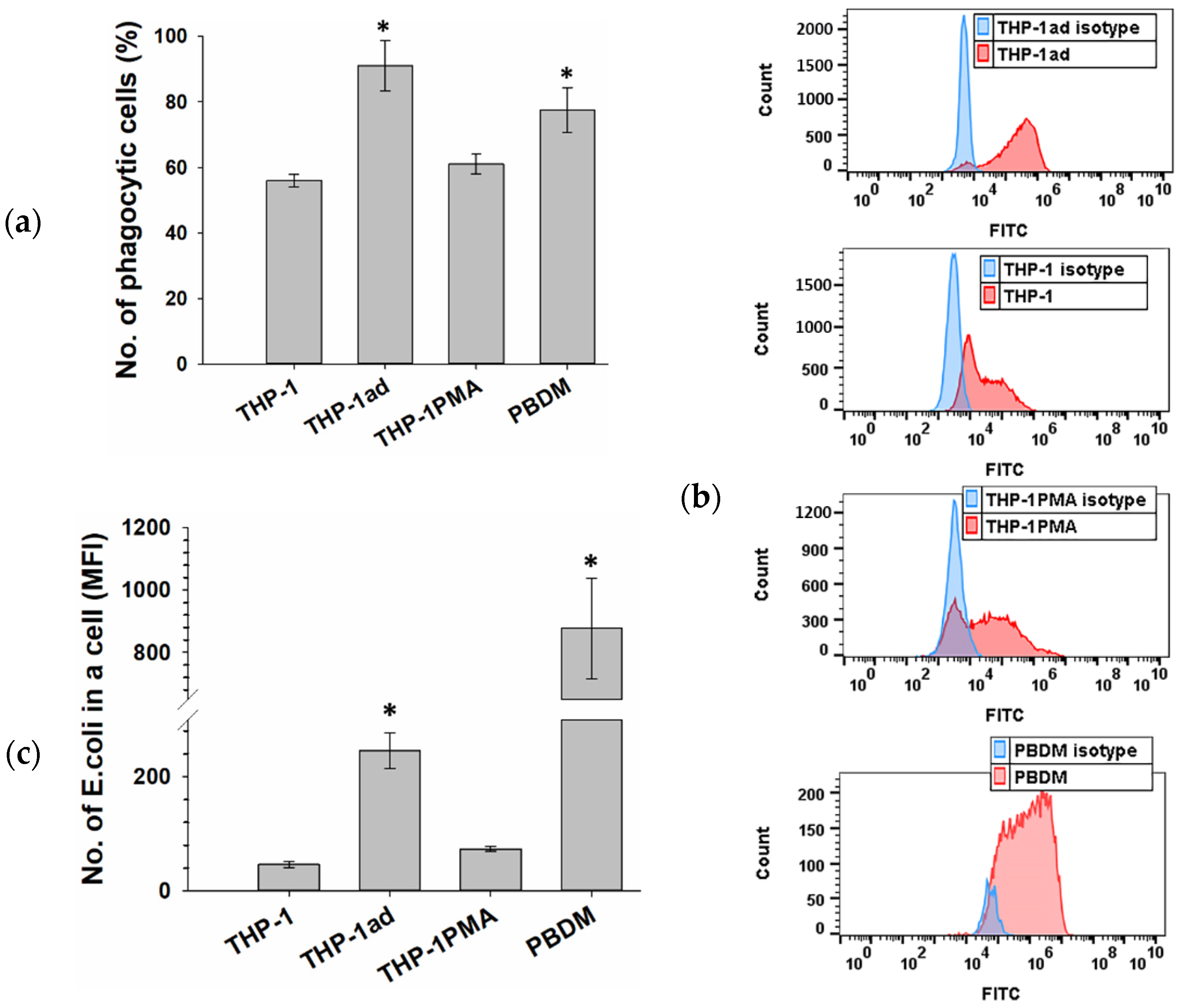
3.4. THP-1ad Cells Are Capable of High Phagocytic Activity of pHrodo Green E. coli
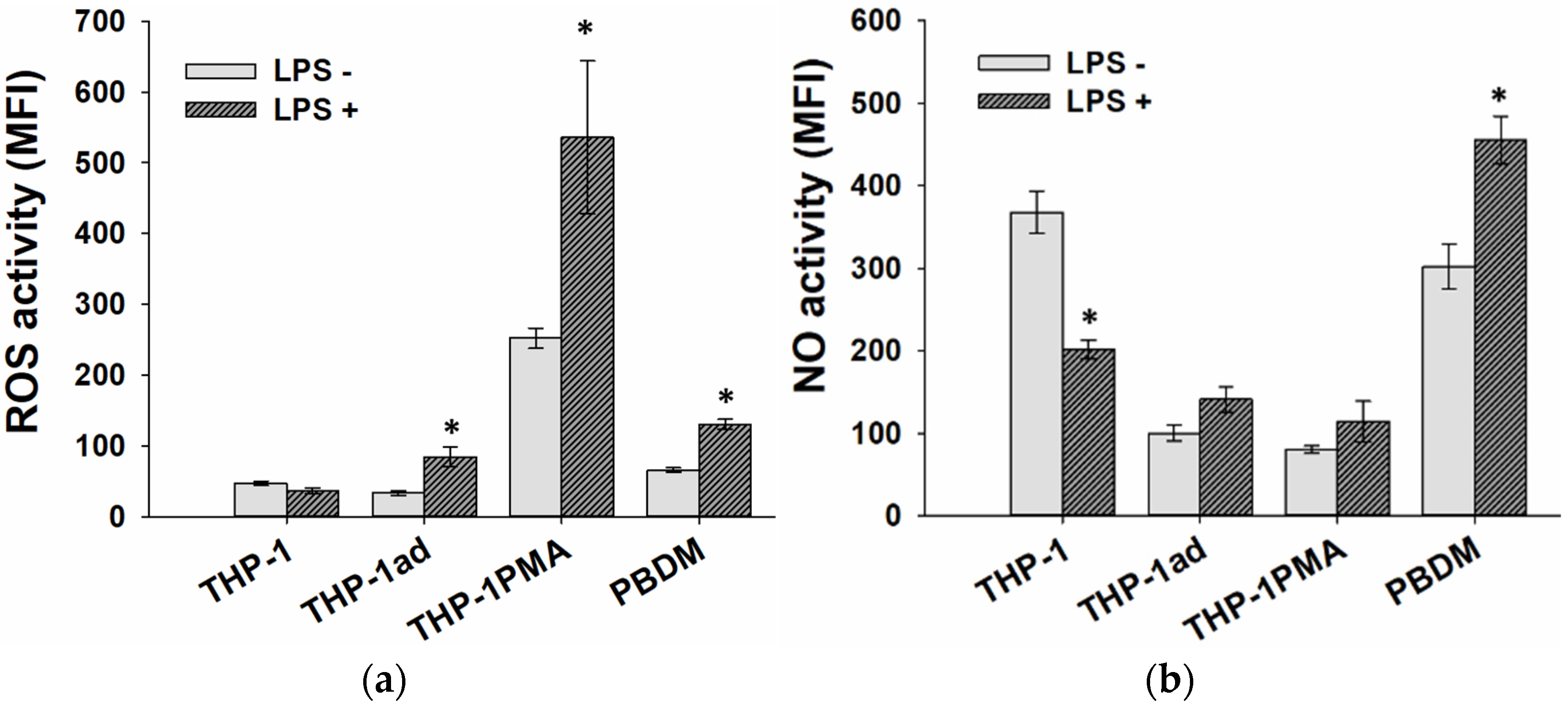
3.5. ROS and NO Production Is Activated in THP-1ad Cells as Well as Macrophage Cells
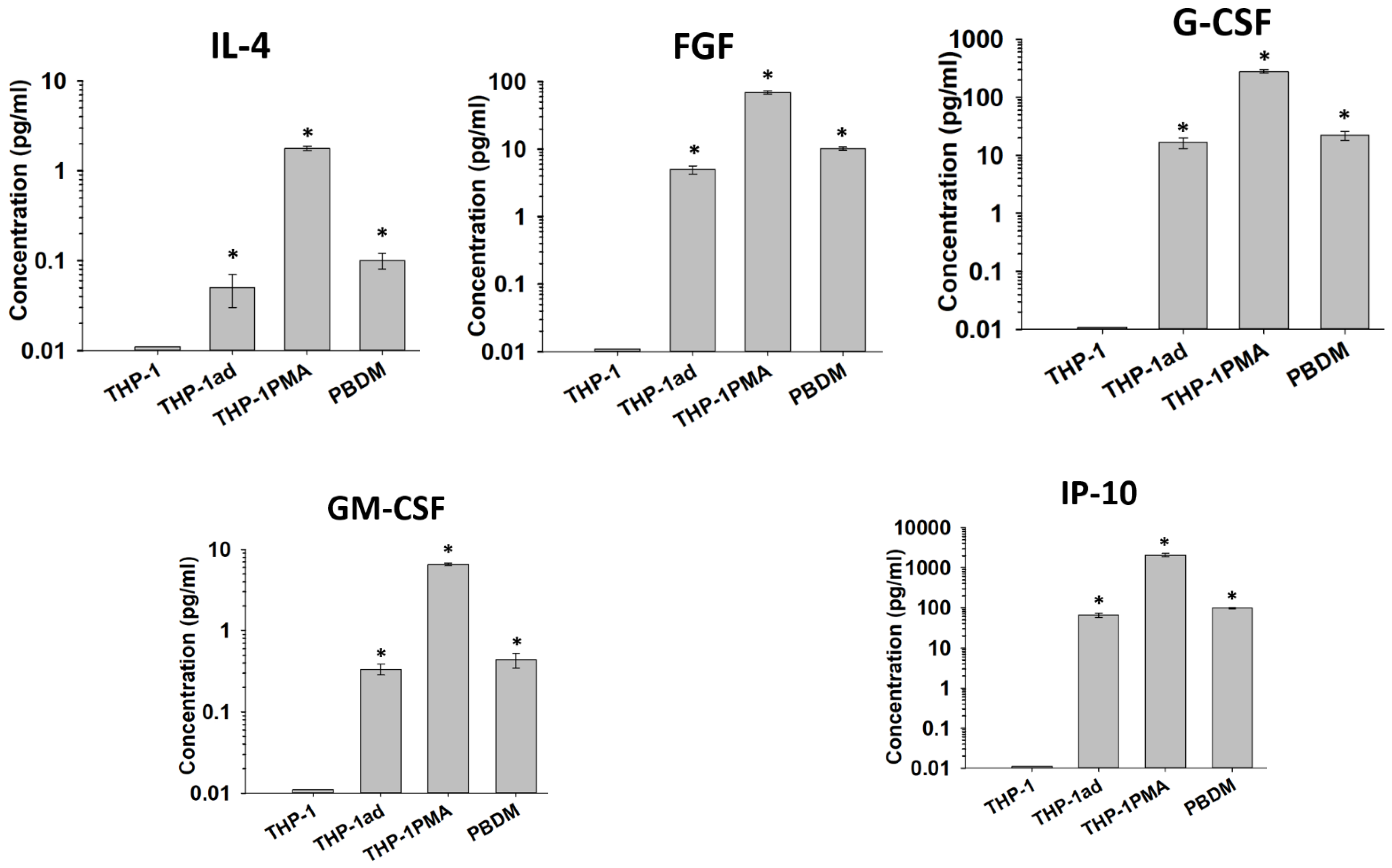
3.6. Cytokine Secretion by THP-1ad Cells Is Similar to Macrophages
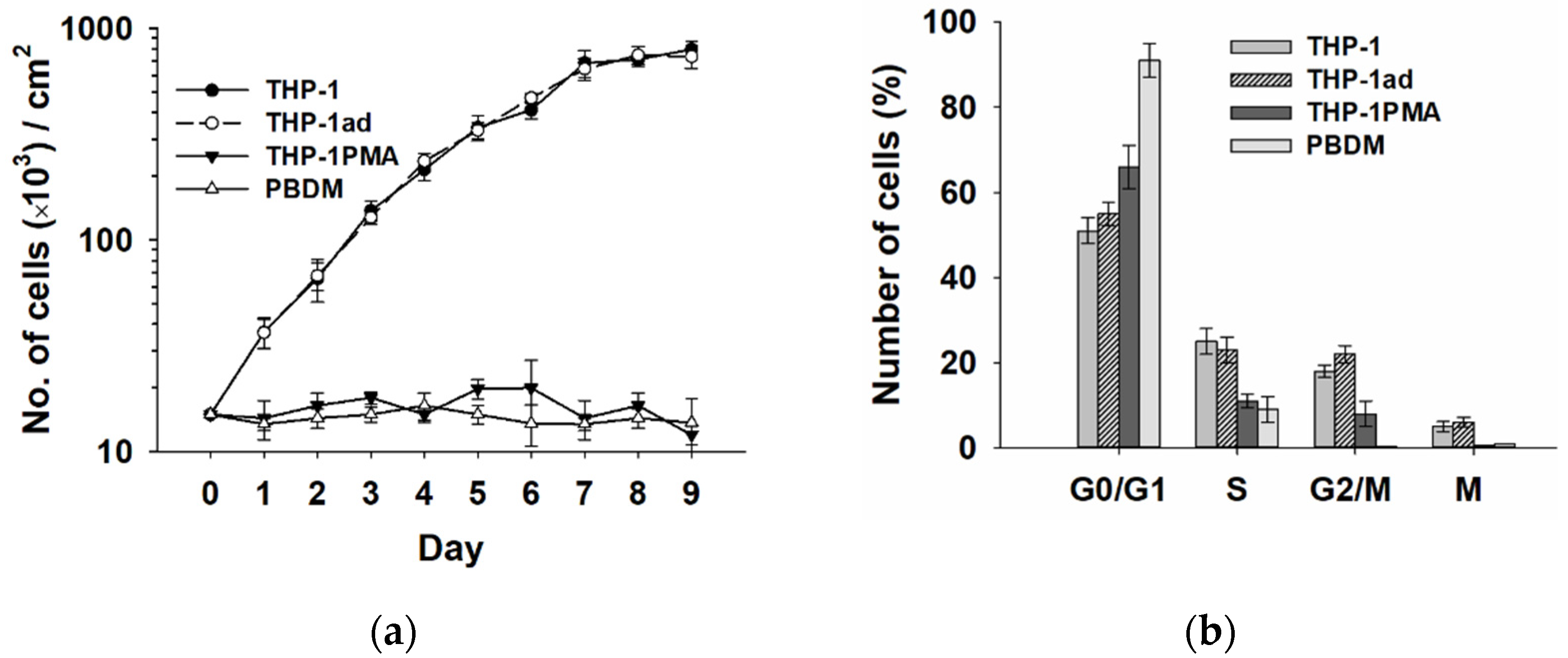
3.7. The Proliferative Activity of Macrophage-like THP-1ad Cells Does Not Differ from Parental Cells
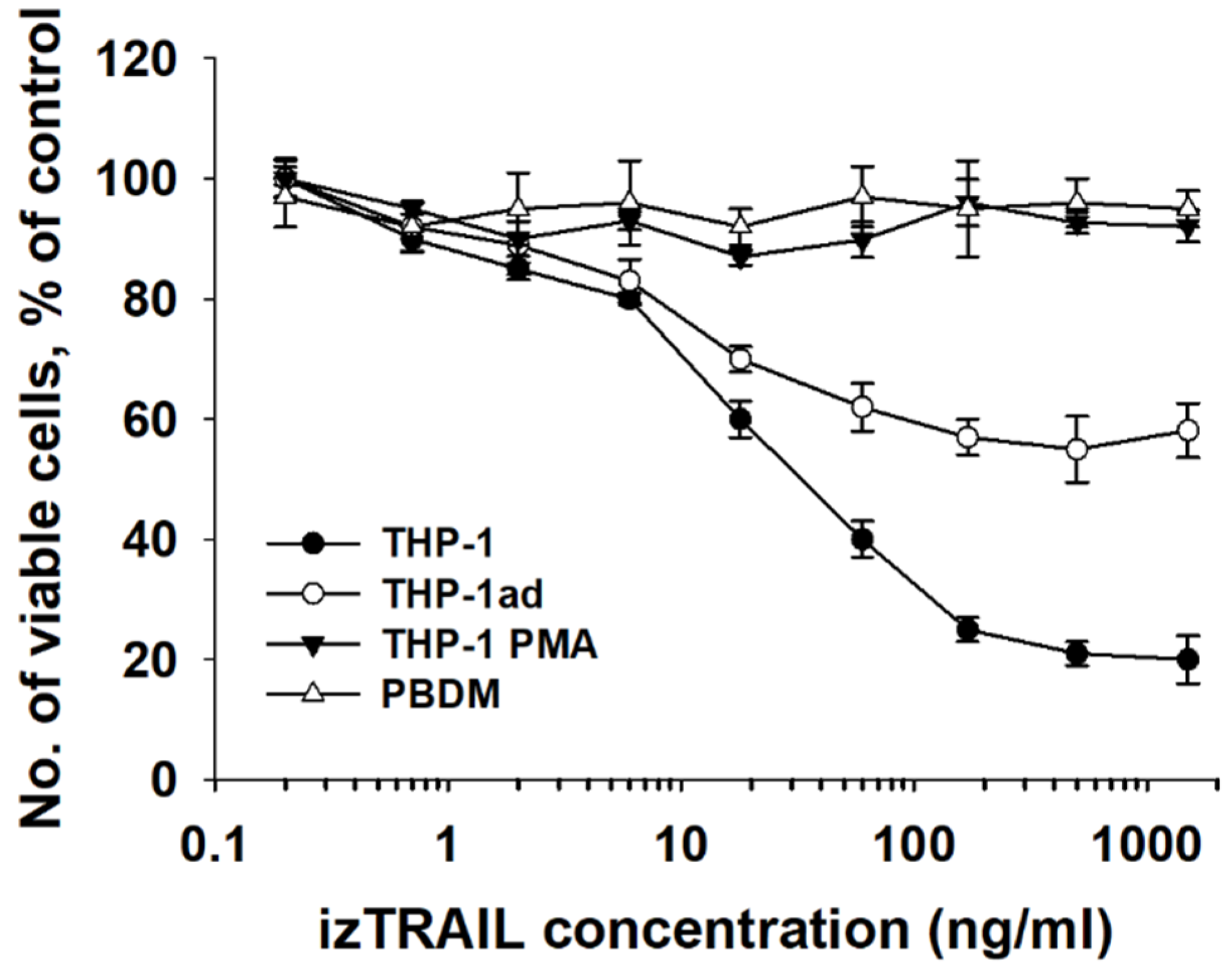
3.8. Macrophage-like THP-1ad Cells Acquire Resistance to TRAIL Induced Cell Death
3.9. Resistance of THP-1ad Cells to TRAIL Is Associated with a Decrease in Expression of Death Receptors DR4 and DR5
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Austin, R.; Smyth, M.J.; Lane, S.W. Harnessing the immune system in acute myeloid leukaemia. Crit. Rev. Oncol. Hematol. 2016, 103, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Sag, D.; Ayyildiz, Z.O.; Gunalp, S.; Wingender, G. The Role of TRAIL/DRs in the Modulation of Immune Cells and Responses. Cancers 2019, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fang, B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005, 12, 228–237. [Google Scholar] [CrossRef]
- Chamuleau, M.E.; Ossenkoppele, G.J.; van Rhenen, A.; van Dreunen, L.; Jirka, S.M.; Zevenbergen, A.; Schuurhuis, G.J.; van de Loosdrecht, A.A. High TRAIL-R3 expression on leukemic blasts is associated with poor outcome and induces apoptosis-resistance which can be overcome by targeting TRAIL-R2. Leuk. Res. 2011, 35, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Hylander, B.L.; Baer, M.R.; Chen, X.; Repasky, E.A. Multiple mechanisms underlie resistance of leukemia cells to Apo2 Ligand/TRAIL. Mol. Cancer Ther. 2006, 5, 1844–1853. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Riccioni, R.; Pasquini, L.; Mariani, G.; Saulle, E.; Rossini, A.; Diverio, D.; Pelosi, E.; Vitale, A.; Chierichini, A.; Cedrone, M.; et al. TRAIL decoy receptors mediate resistance of acute myeloid leukemia cells to TRAIL. Haematologica 2005, 90, 612–624. [Google Scholar] [PubMed]
- Buchsbaum, D.J.; Zhou, T.; Lobuglio, A.F. TRAIL receptor-targeted therapy. Future Oncol. 2006, 2, 493–508. [Google Scholar] [CrossRef] [PubMed]
- Mirandola, P.; Gobbi, G.; Ponti, C.; Sponzilli, I.; Cocco, L.; Vitale, M. PKCepsilon controls protection against TRAIL in erythroid progenitors. Blood 2006, 107, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Secchiero, P.; Gonelli, A.; Mirandola, P.; Melloni, E.; Zamai, L.; Celeghini, C.; Milani, D.; Zauli, G. Tumor necrosis factor-related apoptosis-inducing ligand induces monocytic maturation of leukemic and normal myeloid precursors through a caspase-dependent pathway. Blood 2002, 100, 2421–2429. [Google Scholar] [CrossRef] [PubMed]
- Shiiki, K.; Yoshikawa, H.; Kinoshita, H.; Takeda, M.; Ueno, A.; Nakajima, Y.; Tasaka, K. Potential mechanisms of resistance to TRAIL/Apo2L-induced apoptosis in human promyelocytic leukemia HL-60 cells during granulocytic differentiation. Cell Death Differ. 2000, 7, 939–946. [Google Scholar] [CrossRef]
- Evstratova, Y.V.; Kobyakova, M.I.; Novikova, V.V.; Senotov, A.S.S.; Akatov, V.S.; Fadeev, R.S. Monocyte-Macrophage Differentiation Suppresses the Expression of Proapoptotic Receptors to Apo2L/TRAIL and Increases Resistance to TRAIL-Induced Apoptosis. Biophysics 2019, 64, 729–731. [Google Scholar] [CrossRef]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, T.; Suzuki, M.; Saeki, H.; Matsuno, S.; Tachibana, T.; Kudo, T. Establishment of an activated macrophage cell line, A-THP-1, and its properties. Tohoku J. Exp. Med. 1998, 186, 99–119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sadofsky, L.R.; Hayman, Y.A.; Vance, J.; Cervantes, J.L.; Fraser, S.D.; Wilkinson, H.N.; Williamson, J.D.; Hart, S.P.; Morice, A.H. Characterisation of a New Human Alveolar Macrophage-Like Cell Line (Daisy). Lung 2019, 197, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Agarwal, M.; Mukherjee, R.; Sen, P.; Sinha, D.K. 3D micro-environment regulates NF-kappabeta dependent adhesion to induce monocyte differentiation. Cell Death Dis. 2018, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, R.; Chekanov, A.; Solovieva, M.; Bezborodova, O.; Nemtsova, E.; Dolgikh, N.; Fadeeva, I.; Senotov, A.; Kobyakova, M.; Evstratova, Y.; et al. Improved Anticancer Effect of Recombinant Protein izTRAIL Combined with Sorafenib and Peptide iRGD. Int. J. Mol. Sci. 2019, 20, 525. [Google Scholar] [CrossRef] [PubMed]
- Rasul, A.; Bao, R.; Malhi, M.; Zhao, B.; Tsuji, I.; Li, J.; Li, X. Induction of apoptosis by costunolide in bladder cancer cells is mediated through ROS generation and mitochondrial dysfunction. Molecules 2013, 18, 1418–1433. [Google Scholar] [CrossRef] [PubMed]
- Nunez, R. DNA measurement and cell cycle analysis by flow cytometry. Curr. Issues Mol. Biol. 2001, 3, 67–70. [Google Scholar] [PubMed]
- Avril, P.; Vidal, L.; Barille-Nion, S.; Le Nail, L.R.; Redini, F.; Layrolle, P.; Pinault, M.; Chevalier, S.; Perrot, P.; Trichet, V. Epinephrine Infiltration of Adipose Tissue Impacts MCF7 Breast Cancer Cells and Total Lipid Content. Int. J. Mol. Sci. 2019, 20, 5626. [Google Scholar] [CrossRef] [PubMed]
- Mihaila, M.; Hotnog, C.M.; Bostan, M.; Munteanu, A.C.; Vacaroiu, I.A.; Brasoveanu, L.I.; Uivarosi, V. Anticancer Activity of Some Ruthenium(III) Complexes with Quinolone Antibiotics: In Vitro Cytotoxicity, Cell Cycle Modulation, and Apoptosis-Inducing Properties in LoVo Colon Cancer Cell Line. Appl. Sci. 2021, 11, 8594. [Google Scholar] [CrossRef]
- Solovieva, M.E.; Soloviev, V.V.; Faskhutdinova, A.A.; Kudryavtsev, A.A.; Akatov, V.S. Prooxidant and cytotoxic action of N-acetylcysteine and glutathione in combinations with vitamin B12b. Cell Tissue Biol. 2007, 1, 40–49. [Google Scholar] [CrossRef]
- Ragazzo, M.; Carboni, S.; Caputo, V.; Buttini, C.; Manzo, L.; Errichiello, V.; Puleri, G.; Giardina, E. Interpreting Mixture Profiles: Comparison between Precision ID GlobalFiler NGS STR Panel v2 and Traditional Methods. Genes 2020, 11, 591. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; Ohe, J.V.; Luo, T.; Hass, R. Spontaneous Fusion of MSC with Breast Cancer Cells Can Generate Tumor Dormancy. Int. J. Mol. Sci. 2021, 22, 5930. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, B.S.; Kessler, H.; Kossatz, S.; Reuning, U. RGD-Binding Integrins Revisited: How Recently Discovered Functions and Novel Synthetic Ligands (Re-)Shape an Ever-Evolving Field. Cancers 2021, 13, 1711. [Google Scholar] [CrossRef] [PubMed]
- Calle, P.; Jativa, S.; Torrico, S.; Munoz, A.; Garcia, M.; Sola, A.; Serra, D.; Mera, P.; Herrero, L.; Hotter, G. Infusion of Phagocytic Macrophages Overexpressing CPT1a Ameliorates Kidney Fibrosis in the UUO Model. Cells 2021, 10, 1650. [Google Scholar] [CrossRef] [PubMed]
- Kobyakova, M.I.; Evstratova, Y.V.; Senotov, A.S.S.; Lomovsky, A.I.; Novikova, V.V.; Krasnov, K.S.; Fadeeva, I.S.; Akatov, V.S.; Fadeev, R.S. A Study of the Macrophage Differentiation of Acute Myeloid Leukemia Cells in Multicellular Aggregates. Biophysics 2020, 65, 277–282. [Google Scholar] [CrossRef]
- Zollbrecht, C.; Persson, A.E.; Lundberg, J.O.; Weitzberg, E.; Carlstrom, M. Nitrite-mediated reduction of macrophage NADPH oxidase activity is dependent on xanthine oxidoreductase-derived nitric oxide but independent of S-nitrosation. Redox Biol. 2016, 10, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Jang, H.D. Carnosic Acid Attenuates an Early Increase in ROS Levels during Adipocyte Differentiation by Suppressing Translation of Nox4 and Inducing Translation of Antioxidant Enzymes. Int. J. Mol. Sci. 2021, 22, 6096. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.A.; Jiang, Y.; Yang, H.W.; Hwang, J.; Jeon, Y.J.; Ryu, B. Diphlorethohydroxycarmalol Isolated from Ishige okamurae Exerts Vasodilatory Effects via Calcium Signaling and PI3K/Akt/eNOS Pathway. Int. J. Mol. Sci. 2021, 22, 1610. [Google Scholar] [CrossRef]
- Klimczak, A.; Zimna, A.; Malcher, A.; Kozlowska, U.; Futoma, K.; Czarnota, J.; Kemnitz, P.; Bryl, A.; Kurpisz, M. Co-Transplantation of Bone Marrow-MSCs and Myogenic Stem/Progenitor Cells from Adult Donors Improves Muscle Function of Patients with Duchenne Muscular Dystrophy. Cells 2020, 9, 1119. [Google Scholar] [CrossRef]
- Curtis, A.S.S.; Forrester, J.V.; McInnes, C.; Lawrie, F. Adhesion of cells to polystyrene surfaces. J. Cell Biol. 1983, 97, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Horbett, T. The role of adsorbed proteins in animal cell adhesion. Colloids Surf. B Biointerfaces 1994, 2, 225–240. [Google Scholar] [CrossRef]
- Bharadwaj, M.; Strohmeyer, N.; Colo, G.P.; Helenius, J.; Beerenwinkel, N.; Schiller, H.B.; Fassler, R.; Muller, D.J. alphaV-class integrins exert dual roles on alpha5beta1 integrins to strengthen adhesion to fibronectin. Nat. Commun. 2017, 8, 14348. [Google Scholar] [CrossRef] [PubMed]
- Mas-Moruno, C.; Rechenmacher, F.; Kessler, H. Cilengitide: The first anti-angiogenic small molecule drug candidate design, synthesis and clinical evaluation. Anticancer Agents Med. Chem. 2010, 10, 753–768. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, Z.; Xu, Z.; Chen, L.; Wang, Y. 2′,7′-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: Forty years of application and controversy. Free Radic. Res. 2010, 44, 587–604. [Google Scholar] [CrossRef]
- Traore, K.; Trush, M.A.; George, M., Jr.; Spannhake, E.W.; Anderson, W.; Asseffa, A. Signal transduction of phorbol 12-myristate 13-acetate (PMA)-induced growth inhibition of human monocytic leukemia THP-1 cells is reactive oxygen dependent. Leuk. Res. 2005, 29, 863–879. [Google Scholar] [CrossRef] [PubMed]
- Akatov, V.S.; Lezhnev, E.I.; Vexler, A.M.; Kublik, L.N. Low pH value of pericellular medium as a factor limiting cell proliferation in dense cultures. Exp. Cell Res. 1985, 160, 412–418. [Google Scholar] [CrossRef]
- Werrlein, R.J.; Glinos, A.D. Oxygen microenvironment and respiratory oscillations in cultured mammalian cells. Nature 1974, 251, 317–319. [Google Scholar] [CrossRef]
- Pivoriunas, A.; Savickiene, J.; Treigyte, G.; Tunaitis, V.; Navakauskiene, R.; Magnusson, K.E. PI 3-K signaling pathway suppresses PMA-induced expression of p21WAF1/Cip1 in human leukemia cells. Mol. Cell Biochem. 2007, 302, 9–18. [Google Scholar] [CrossRef]
- van der Sloot, A.M.; Tur, V.; Szegezdi, E.; Mullally, M.M.; Cool, R.H.; Samali, A.; Serrano, L.; Quax, W.J. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 8634–8639. [Google Scholar] [CrossRef] [PubMed]
- Tur, V.; van der Sloot, A.M.; Reis, C.R.; Szegezdi, E.; Cool, R.H.; Samali, A.; Serrano, L.; Quax, W.J. DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J. Biol. Chem. 2008, 283, 20560–20568. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtina, Y.; Pavet, V.; Gronemeyer, H. Dual role of DR5 in death and survival signaling leads to TRAIL resistance in cancer cells. Cell Death Dis. 2017, 8, e3025. [Google Scholar] [CrossRef] [PubMed]



| Loci | Genotype of THP-1ad Cells | Closest Match (THP-1 ATCC) |
|---|---|---|
| D5S818 | 11,12 | 11,12 |
| D13S317 | 13,14 | 13,13 |
| D7S820 | 10,10 | 10,10 |
| D16S539 | 11,12 | 11,12 |
| vWA | 16,16 | 16,16 |
| TH01 | 8,9,3 | 8,9,3 |
| AMEL | X,X | X,Y |
| TPOX | 8,11 | 8,11 |
| CSF1PO | 11,13 | 11,13 |
| CD (Clusters of Differentiation) | THP-1 | THP-1ad | THP-1 PMA | Macrophages (PBDM) |
|---|---|---|---|---|
| Integrin αL, (CD11a) | 85 ± 9% | 86 ± 1% | 91 ± 2% | 98 ± 2% |
| Integrin αM, (CD11b) | – | 18 ± 2% | – | 90 ± 3% |
| Integrin αX, (CD11c) | – | 56 ± 3% | 90 ± 1% | 66 ± 1% |
| Co-receptor for LPS (CD14) | – | 19 ± 1% | 95 ± 1% | 47 ± 2% |
| Siglec-3 (CD33) | 99 ± 1% | 99 ± 1% | 99 ± 1% | 99 ± 1% |
| SCARB3 (CD36) | – | – | 20 ± 1% | 97 ± 1% |
| PTPRC (CD45) | 96 ± 8% | 96 ± 1% | 99 ± 1% | 99 ± 1% |
| Fc-γ receptor 1 (CD64) | 95 ± 1% | 95 ± 1% | 25 ± 3% | 55 ± 4% |
| Macrosialin (CD68) | 51 ± 3% | 47 ± 5% | – | 95 ± 4% |
| Receptor for the hemoglobin–haptoglobin complex (CD163) | – | – | – | – |
| TLR4 (CD284) | – | 49 ± 13% | – | 9 ± 1% |
| MHC II (HLA-DR) | 21 ± 1% | 60 ± 1% | – | 59 ± 2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lomovskaya, Y.V.; Kobyakova, M.I.; Senotov, A.S.; Lomovsky, A.I.; Minaychev, V.V.; Fadeeva, I.S.; Shtatnova, D.Y.; Krasnov, K.S.; Zvyagina, A.I.; Akatov, V.S.; et al. Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5. Biomolecules 2022, 12, 150. https://doi.org/10.3390/biom12020150
Lomovskaya YV, Kobyakova MI, Senotov AS, Lomovsky AI, Minaychev VV, Fadeeva IS, Shtatnova DY, Krasnov KS, Zvyagina AI, Akatov VS, et al. Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5. Biomolecules. 2022; 12(2):150. https://doi.org/10.3390/biom12020150
Chicago/Turabian StyleLomovskaya, Yana Vladimirovna, Margarita Igorevna Kobyakova, Anatoly Sergeevich Senotov, Alexey Igorevich Lomovsky, Vladislav Valentinovich Minaychev, Irina Sergeevna Fadeeva, Daria Yuryevna Shtatnova, Kirill Sergeevich Krasnov, Alena Igorevna Zvyagina, Vladimir Semenovich Akatov, and et al. 2022. "Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5" Biomolecules 12, no. 2: 150. https://doi.org/10.3390/biom12020150
APA StyleLomovskaya, Y. V., Kobyakova, M. I., Senotov, A. S., Lomovsky, A. I., Minaychev, V. V., Fadeeva, I. S., Shtatnova, D. Y., Krasnov, K. S., Zvyagina, A. I., Akatov, V. S., & Fadeev, R. S. (2022). Macrophage-like THP-1 Cells Derived from High-Density Cell Culture Are Resistant to TRAIL-Induced Cell Death via Down-Regulation of Death-Receptors DR4 and DR5. Biomolecules, 12(2), 150. https://doi.org/10.3390/biom12020150