Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative
Abstract
:1. Introduction
2. Materials and Methods
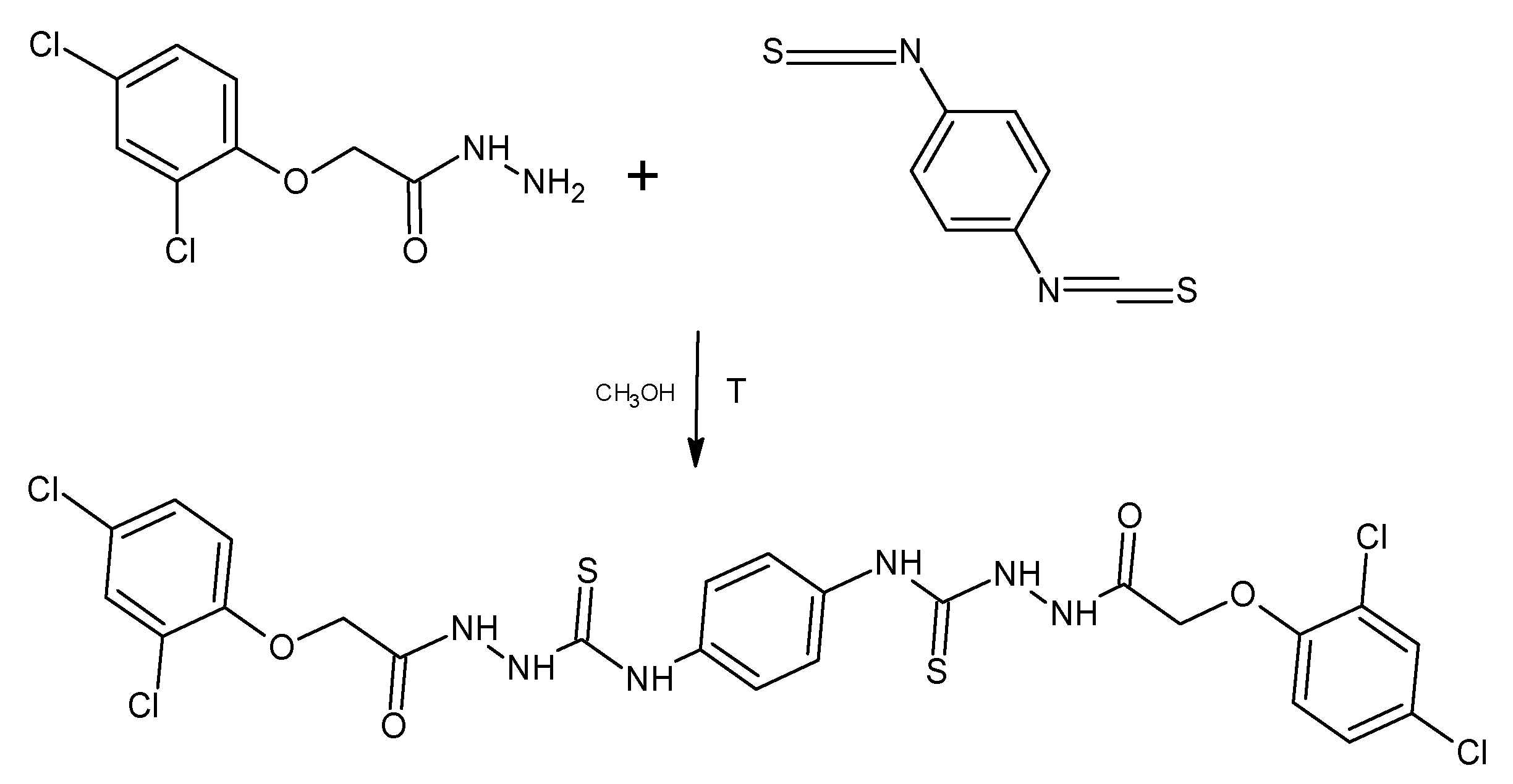
2.1. General Procedure for the Synthesis of Thiosemicarbazide Derivatives (PK1-PK13)
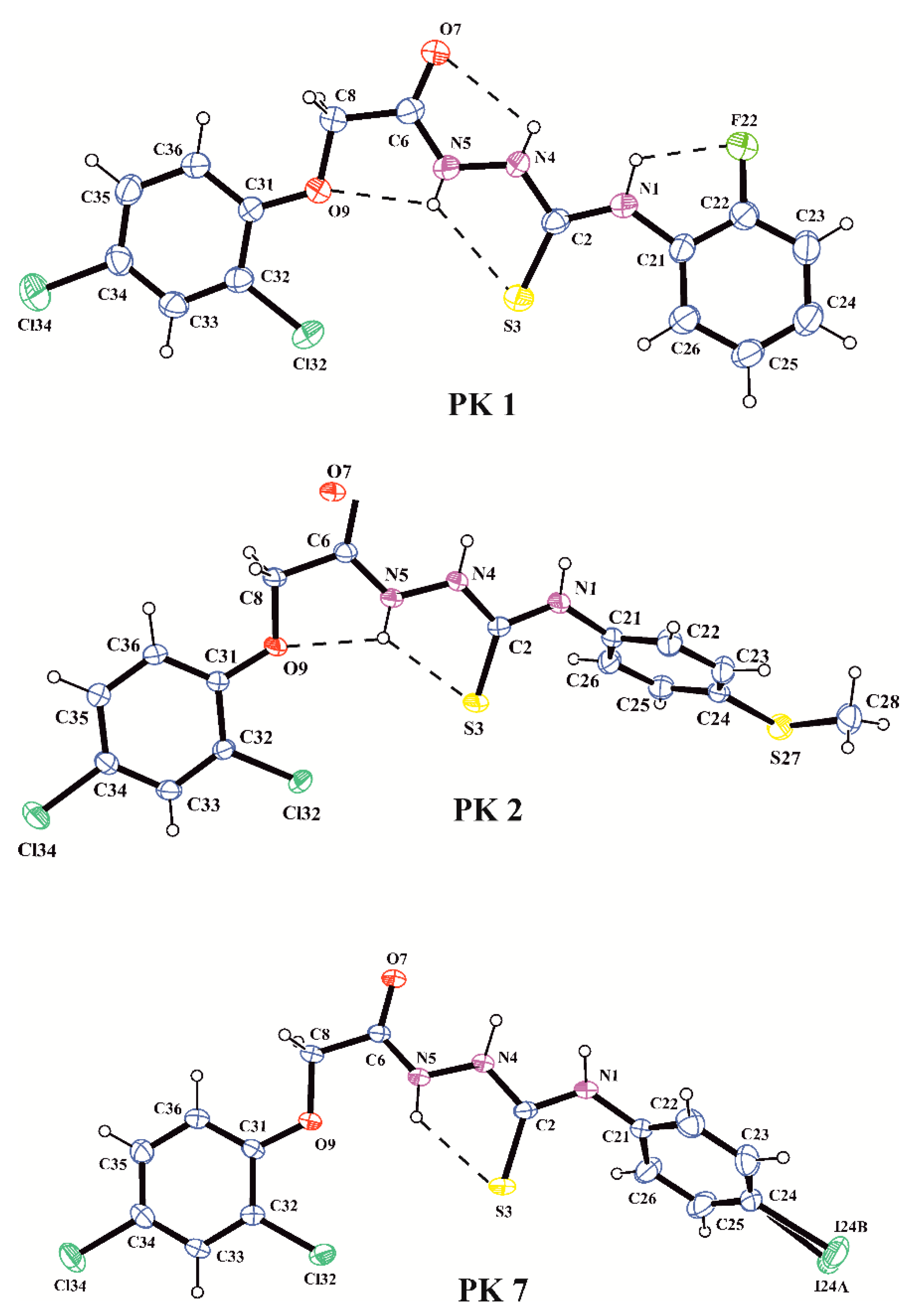
2.2. X-Ray Structure Determination
2.3. HPLC Procedure
2.4. TLC Procedure
2.5. Biological Assays
2.5.1. Cell Culture
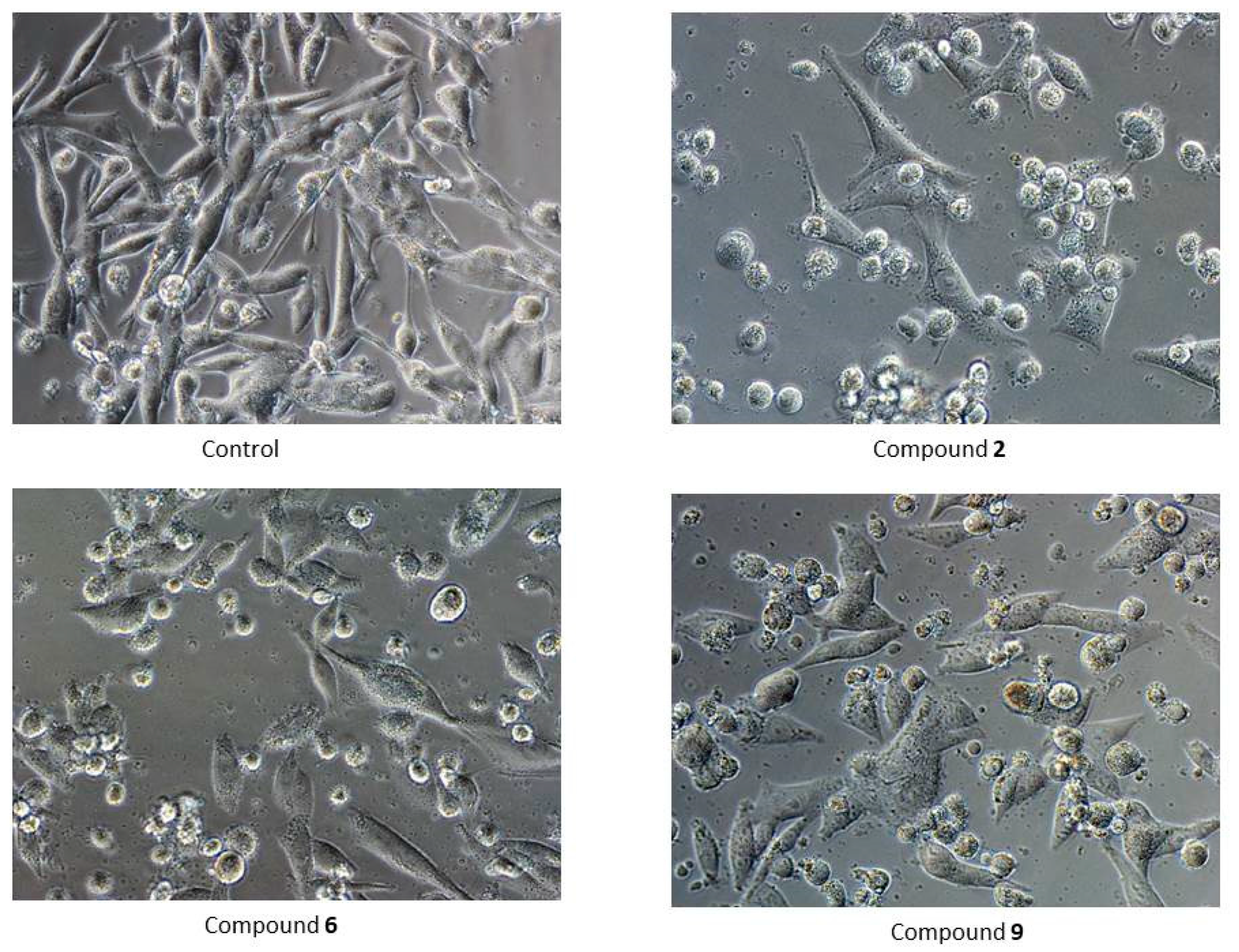
2.5.2. Cytotoxicity Analysis
2.5.3. Cell Cycle Assay
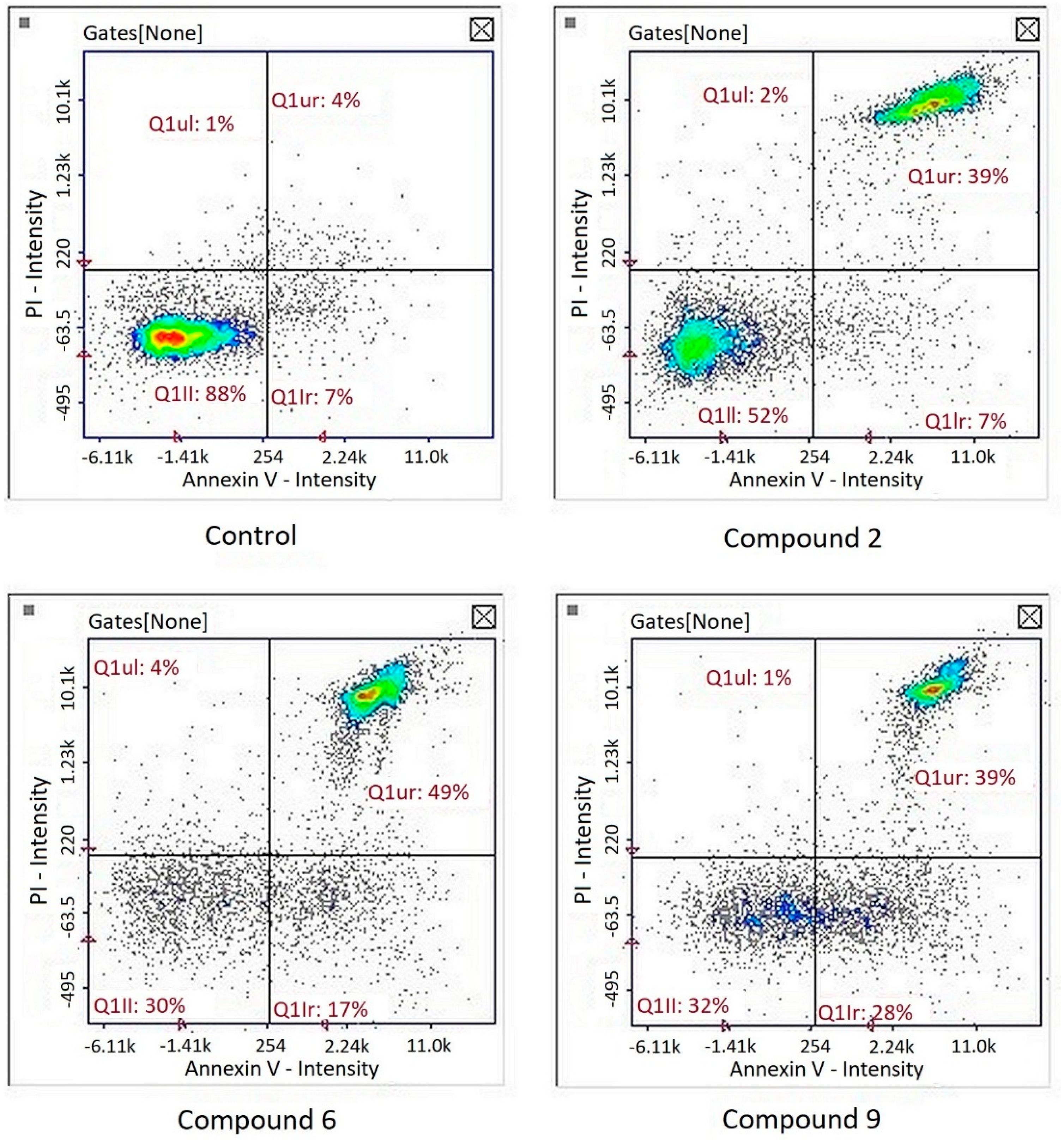
2.5.4. Cell Apoptosis Assay
2.5.5. Analysis of Intracellular Thiol Levels
2.5.6. Quantitative Real-Time PCR Analysis (qRT-PCR)
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Siegel, R.L.; Ward, E.M.; Jemal, A. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiol. Biomark. Prev. 2016, 25, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 8 September 2021).
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Kalinowski, D.S.; Quach, P.; Richardson, D.R. Thiosemicarbazones: The new wave in cancer treatment. Future Med. Chem. 2009, 1, 1143–1151. [Google Scholar] [CrossRef]
- Shakya, B.; Yadav, P.N. Thiosemicarbazones as Potent Anticancer Agents and their Modes of Action. Mini Rev. Med. Chem. 2020, 20, 638–661. [Google Scholar] [CrossRef]
- Pitucha, M.; Korga-Plewko, A.; Czylkowska, A.; Rogalewicz, B.; Dozd, M.; Iwan, M.; Kubik, J.; Humeniuk, E.; Adamczuk, G.; Karczmarzyk, Z.; et al. Influence of Complexation of Thiosemicarbazone Derivatives with Cu (II) Ions on Their Antitumor Activity against Melanoma Cells. Int. J. Mol. Sci. 2021, 22, 3104. [Google Scholar] [CrossRef] [PubMed]
- Padma, V.V. An Overview of Targeted Cancer Therapy. Biomedicine 2015, 5, 19. [Google Scholar] [CrossRef]
- Pitucha, M.; Rzymowska, J.; Olender, A.; Grzybowska-Szatkowska, L. Synthesis of 1,6-Bis(Semicarbazide)Hexanes and 1,6-Bis(1,2,4-Triazol-5-One)Hexanes and Their Antiproliferative and Antimicrobial Activity. J. Serb. Chem. Soc. 2012, 77, 1–8. [Google Scholar] [CrossRef]
- Kaczor, A.A.; Pitucha, M.; Karczmarzyk, Z.; Wysocki, W.; Rzymowska, J.; Matosiuk, D. Structural and Molecular Docking Studies of 4-Benzyl-3-[(1-Methylpyrrol- 2-Yl)Methyl]-4,5-Dihydro-1H-1,2,4-Triazol-5-One with Anticancer Activity. Med. Chem. 2013, 9, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Pitucha, M.; Woś, M.; Miazga-Karska, M.; Klimek, K.; Mirosław, B.; Pachuta-Stec, A.; Gładysz, A.; Ginalska, G. Synthesis, Antibacterial and Antiproliferative Potential of Some New 1-Pyridinecarbonyl-4-Substituted Thiosemicarbazide Derivatives. Med. Chem. Res. 2016, 25, 1666–1677. [Google Scholar] [CrossRef] [Green Version]
- Wos, M.; Miazga-Karska, M.; Kaczor, A.A.; Klimek, K.; Karczmarzyk, Z.; Kowalczuk, D.; Wysocki, W.; Ginalska, G.; Urbanczyk-Lipkowska, Z.; Morawiak, M.; et al. Novel Thiosemicarbazide Derivatives with 4-Nitrophenyl Group as Multi-Target Drugs: α-Glucosidase Inhibitors with Antibacterial and Antiproliferative Activity. Biomed. Pharmacother. 2017, 93, 1269–1276. [Google Scholar] [CrossRef]
- Singhal, S.; Arora, S.; Agarwal, S.; Sharma, R.; Singhal, N. A Review on potential biological activity of thiosemicarbazides. World J. Pharm. Pharm. Sci. 2013, 2, 4661–4681. [Google Scholar]
- Zhang, X.; Lei, P.; Sun, T.; Jin, X.; Yang, X.; Ling, Y. Design, Synthesis, and Fungicidal Activity of Novel Thiosemicarbazide Derivatives Containing Piperidine Fragments. Molecules 2017, 22, 2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arora, S.; Agarwal, S.; Singhal, S. Anticancer activities of thiosemicarbazides/thiosemicarbazones: A review. Int. J. Pharm. Pharm. Sci. 2014, 6, 34–41. [Google Scholar]
- Chen, R.; Huo, L.; Jaiswal, Y.; Huang, J.; Zhong, Z.; Zhong, J.; Williams, L.; Xia, X.; Liang, Y.; Yan, Z. Design, Synthesis, Antimicrobial, and Anticancer Activities of Acridine Thiosemicarbazides Derivatives. Molecules 2019, 24, 2065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Wang, X.; Zhao, X.; Liang, Y.; He, H.; Fu, L. Synthesis and Antitumor Activity of Novel Quinazoline Derivatives Containing Thiosemicarbazide Moiety. Eur. J. Med. Chem. 2012, 54, 925–930. [Google Scholar] [CrossRef]
- Pitucha, M.; Korga-Plewko, A.; Kozyra, P.; Iwan, M.; Kaczor, A.A. 2,4-Dichlorophenoxyacetic Thiosemicarbazides as a New Class of Compounds against Stomach Cancer Potentially Intercalating with DNA. Biomolecules 2020, 10, 296. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sheikh, M.S. Melanoma: Molecular Pathogenesis and Therapeutic Management. Mol. Cell Pharmacol. 2014, 6, 228. [Google Scholar]
- Domingues, B.; Lopes, J.M.; Soares, P.; Pópulo, H. Melanoma Treatment in Review. Immunotargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [Green Version]
- CrysAlisPro, Agilent Technologies. Version 1.171.37.35h, 2015 (release 09-02-2015 CrysAlis171.NET), (Compiled Feb 9 2015, 16: 28: 20); Agilent Technologies: Oxfordshire, UK, 2015. [Google Scholar]
- Sheldrick, G.M. A Short History of SHELX. Acta Crystallogr. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrugia, L.J. WinGX and ORTEP for Windows: An Update. J. Appl. Cryst. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Allen, F.H.; Kennard, O.; Watson, D.G.; Brammer, L.; Orpen, A.G.; Taylor, R. Tables of Bond Lengths Determined by X-ray and Neutron Diffraction. Part 1. Bond Lengths in Organic Compounds. J. Chem. Soc. Perkin Trans. 1987, 13, S1–S19. [Google Scholar] [CrossRef]
- Pitucha, M.; Karczmarzyk, Z.; Swatko-Ossor, M.; Wysocki, W.; Wos, M.; Chudzik, K.; Ginalska, G.; Fruzinski, A. Synthesis, In Vitro Screening and Docking Studies of New Thiosemicarbazide Derivatives as Antitubercular Agents. Molecules 2019, 24, 251. [Google Scholar] [CrossRef] [Green Version]
- Andrić, F.; Héberger, K. Chromatographic and Computational Assessment of Lipophilicity Using Sum of Ranking Differences and Generalized Pair-Correlation. J. Chromatogr. A 2015, 1380, 130–138. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Lian, H. Recent Advances in Lipophilicity Measurement by Reversed-Phase High-Performance Liquid Chromatography. TrAC Trends Anal. Chem. 2015, 68, 28–36. [Google Scholar] [CrossRef]
- Dolan, J.W.; Gant, J.R.; Snyder, L.R. Gradient Elution in High-Performance Liquid Chromatography: II. Practical Application to Reversed-Phase Systems. J. Chromatogr. A 1979, 165, 31–58. [Google Scholar] [CrossRef]
- Bate-Smith, E.C.; Westall, R.G. Chromatographic Behaviour and Chemical Structure I. Some Naturally Occuring Phenolic Substances. Biochim. Biophys. Acta 1950, 4, 427–440. [Google Scholar] [CrossRef]
- Soczewiński, E.; Wachtmeister, C.A. The Relation between the Composition of Certain Ternary Two-Phase Solvent Systems and RM Values. J. Chromatogr. A 1962, 7, 311–320. [Google Scholar] [CrossRef]
- Jaffé, H.H. A Reëxamination of the Hammett Equation. Chem. Rev. 1953, 53, 191–261. [Google Scholar] [CrossRef]
- Biagi, G.L.; Barbaro, A.M.; Sapone, A.; Recanatini, M. Determination of Lipophilicity by Means of Reversed-Phase Thin-Layer Chromatography: I. Basic Aspects and Relationship between Slope and Intercept of TLC Equations. J. Chromatogr. A 1994, 662, 341–361. [Google Scholar] [CrossRef]
- Pastore, A.; Federici, G.; Bertini, E.; Piemonte, F. Analysis of Glutathione: Implication in Redox and Detoxification. Clin. Chim. Acta 2003, 333, 19–39. [Google Scholar] [CrossRef]
- Hambright, H.G.; Meng, P.; Kumar, A.P.; Ghosh, R. Inhibition of PI3K/AKT/mTOR axis disrupts oxidative stress-mediated survival of melanoma cells. Oncotarget 2015, 6, 7195–7208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csekes, E.; Vágvölgyi, M.; Hunyadi, A.; Račková, L. Protoflavones in melanoma therapy: Prooxidant and pro-senescence effect of protoapigenone and its synthetic alkyl derivative in A375 cells. Life Sci. 2020, 260, 118419. [Google Scholar] [CrossRef]
- Heo, J.R.; Kim, S.M.; Hwang, K.A.; Kang, J.H.; Choi, K.C. Resveratrol induced reactive oxygen species and endoplasmic reticulum stress mediated apoptosis, and cell cycle arrest in the A375SM malignant melanoma cell line. Int. J. Mol. Med. 2018, 42, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Cullen, J.J.; Weydert, C.; Hinkhouse, M.M.; Ritchie, J.; Domann, F.E.; Spitz, D.; Oberley, L.W. The Role of Manganese Superoxide Dismutase in the Growth of Pancreatic Adenocarcinoma. Cancer Res. 2003, 63, 1297–1303. [Google Scholar]
- Liu, L.; Dong, Z.; Lei, Q.; Yang, J.; Hu, H.; Li, Q.; Ji, Y.; Guo, L.; Zhang, Y.; Liu, Y.; et al. Inactivation/deficiency of DHODH induces cell cycle arrest and programed cell death in melanoma. Oncotarget 2017, 8, 112354–112370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorasamy, M.S.; Choudhary, B.; Nellore, K.; Subramanya, H.; Wong, P.F. Dihydroorotate dehydrogenase Inhibitors Target c-Myc and Arrest Melanoma, Myeloma and Lymphoma cells at S-phase. J. Cancer 2017, 8, 3086–3098. [Google Scholar] [CrossRef] [Green Version]
- Dorasamy, M.S.; Ab, A.; Nellore, K.; Wong, P.F. Synergistic inhibition of melanoma xenografts by Brequinar sodium and Doxorubicin. Biomed. Pharmacother. 2019, 110, 29–36. [Google Scholar] [CrossRef]
- Madak, J.T.; Bankhead, A.; Cuthbertson, C.R.; Showalter, H.D.; Neamati, N. Revisiting the Role of Dihydroorotate Dehydrogenase as a Therapeutic Target for Cancer. Pharmacol. Ther. 2019, 195, 111–131. [Google Scholar] [CrossRef]
- Fang, J.; Uchiumi, T.; Yagi, M.; Matsumoto, S.; Amamoto, R.; Takazaki, S.; Yamaza, H.; Nonaka, K.; Kang, D. Dihydro-Orotate Dehydrogenase Is Physically Associated with the Respiratory Complex and Its Loss Leads to Mitochondrial Dysfunction. Biosci. Rep. 2013, 33, e00021. [Google Scholar] [CrossRef] [PubMed]





| Gene Symbol. | Gene Name | RefSeq | Assay ID | |
| DHODH | dihydroorotate dehydrogenase (quinone) | NM_001361.4, XM_005255827.3, XM_005255829.3, XM_017022990.1 | Hs00361406_m1 | |
| 18SRNA | Eukaryotic 18S rRNA | X03205.1 | Hs99999901_s1 | |
| BACT | actin beta | NM_001101.3 | Hs99999903_m1 | |
| Gene Symbol | Gene Name | RefSeq | Forward | Reverse |
| CAT | catalase | NM_001752 | agcttagcgttcatccgtgt | tccaatcatccgtcaaaaca |
| SOD2 | superoxide dismutase 2, mitochondrial | NM_000636, NM_001024465.2, NM_001024466.2, NM_001322814.1, NM_001322817.1, NM_001322819.1, NM_001322820.1 | cttcagggtggtatggctgt | tggccagaccttaatgttcc |
| 18SRNA | Eukaryotic 18S rRNA | X03205.1 | gaaactgcgaatggctcattaaa | cacagttatccaagtgggagagg |
| TBP | TATA-box binding protein | NM_001172085.1, NM_003194.4 | cggctgtttaacttcgcttc | ttcttggcaaaccagaaacc |
| mRNA | ||||
| Torsion Angle | PK1 | PK2 | PK7 |
|---|---|---|---|
| C22–C21–N1–C2 | 175.3(4) | −121.0(3) | −122.9(5) |
| C21–N1–C2–N4 | 172.2(4 | 179.3(2) | −175.4(4) |
| N1–C2–N4–N5 | 178.5(4) | 176.2(2) | 177.6(4) |
| C2–N4–N5–C6 | −174.4(4) | −164.8(2) | −170.0(4) |
| N4–N5–C6–C8 | 176.8(4) | 178.50(19) | 178.5(4) |
| N5–C6–C8–09 | 0.4(5) | −10.0(3) | −8.5(5) |
| C6–C8–O9–C31 | 174.5(3) | 172.41(18) | 177.2(3) |
| C8–O9–C31–C32 | −176.9(4) | 170.36(18) | −177.8(4) |
| C21–N1–C2–S3 | −8.7(7) | 0.2(4) | 7.5(7) |
| N4–N5–C6–O7 | −4.1(6) | −2.0(3) | −1.6(6) |
| D–H...A | D–H | H...A | D...A | D–H...A |
|---|---|---|---|---|
| PK1 | ||||
| N5–H5...S3 | 0.78(5) | 2.49(5) | 2.932(4) | 117(4) |
| N5–H5...O9 | 0.78(5) | 2.27(5) | 2.563(5) | 103(4) |
| N1–H1...F22 | 0.92(5) | 2.15(5) | 2.633(4) | 111(4) |
| N4–H4...O7 | 0.79(5) | 2.41(5) | 2.697(5) | 103(4) |
| N1–H1...O7i | 0.92(5) | 2.11(5) | 2.944(5) | 150(4) |
| N4–H4...O7i | 0.79(5) | 2.02(5) | 2.760(5) | 156(5) |
| (i) = 1 − x, 1 − y, 1 − z | ||||
| PK2 | ||||
| N5–H5...S3 | 0.83(3) | 2.50(2) | 2.911(2) | 112(2) |
| N5–H5...O9 | 0.83(3) | 2.19(3) | 2.552(20 | 107(2) |
| N1–H1...O7i | 0.85(3) | 2.19(3) | 2.975(2) | 154(3) |
| N4–H4...O7i | 0.84(3) | 2.12(3) | 2.902(3) | 155(2) |
| (i) = 1 − x, 1 − y, 1 − z | ||||
| PK5 | ||||
| N5–H5...S3 | 0.86(6) | 2.33(5) | 2.915(4) | 126(4) |
| N1–H1...O7i | 0.85(6) | 2.10(6) | 2.936(4) | 167(4) |
| N4–H4...O7i | 0.98(6) | 1.93(6) | 2.833(4) | 152(4) |
| (i) = 1 − x, 1 − y, −z |
| Compound | IC50 [µM] | |
|---|---|---|
| G-361 | BJ | |
| PK1 | >250 | >250 |
| PK2 | 108 ± 6 | >250 (828 ± 8) |
| PK3 | >250 | >250 |
| PK4 | >250 | >250 |
| PK5 | >250 | >250 |
| PK6 | 112 ± 4.76 | >250 (631 ± 11) |
| PK7 | 99 ± 4 | 216±76 |
| PK8 | >250 | >250 |
| PK9 | 113 ± 4 | >250 (894 ± 9) |
| PK10 | 218 ± 5.34 | 223 ± 7 |
| PK11 | 162 ± 2 | 217.46 ± 64.33 |
| PK12 | 2078 ± 4 | 189 ± 7 |
| PK13 | >250 | >250 |
| DHODH | CAT | SOD2 | Scale (RQ) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 1.000 ± 0.016 | 1.016 ± 0.206 | 1.001 ± 0.053 | >2.00 | |||||||
| PK2 | 0.463 ± 0.016 | 0.107 ± 0.013 | 1.486 ± 0.056 | 1.51–1.99 | |||||||
| PK6 | 0.824 ± 0.030 | 2.691 ± 0.127 | 1.249 ± 0.080 | 1.11–1.50 | |||||||
| PK9 | 0.375 ± 0.018 | 0.354 ± 0.022 | 1.293 ± 0.100 | 0.91–1.10 | |||||||
| 0.61–0.90 | |||||||||||
| 0.21–0.6 | |||||||||||
| <0.21 | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozyra, P.; Korga-Plewko, A.; Karczmarzyk, Z.; Hawrył, A.; Wysocki, W.; Człapski, M.; Iwan, M.; Ostrowska-Leśko, M.; Fornal, E.; Pitucha, M. Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative. Biomolecules 2022, 12, 151. https://doi.org/10.3390/biom12020151
Kozyra P, Korga-Plewko A, Karczmarzyk Z, Hawrył A, Wysocki W, Człapski M, Iwan M, Ostrowska-Leśko M, Fornal E, Pitucha M. Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative. Biomolecules. 2022; 12(2):151. https://doi.org/10.3390/biom12020151
Chicago/Turabian StyleKozyra, Paweł, Agnieszka Korga-Plewko, Zbigniew Karczmarzyk, Anna Hawrył, Waldemar Wysocki, Michał Człapski, Magdalena Iwan, Marta Ostrowska-Leśko, Emilia Fornal, and Monika Pitucha. 2022. "Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative" Biomolecules 12, no. 2: 151. https://doi.org/10.3390/biom12020151
APA StyleKozyra, P., Korga-Plewko, A., Karczmarzyk, Z., Hawrył, A., Wysocki, W., Człapski, M., Iwan, M., Ostrowska-Leśko, M., Fornal, E., & Pitucha, M. (2022). Potential Anticancer Agents against Melanoma Cells Based on an As-Synthesized Thiosemicarbazide Derivative. Biomolecules, 12(2), 151. https://doi.org/10.3390/biom12020151






