ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives
Abstract
:1. Introduction
2. ADP-Ribosyltransferase Activity during Evolution in the Three Domains of Life
3. Human Proteins in ADPr Biology
3.1. ADP-Ribosyltransferases
3.2. ADP-Ribosylhydrolases
3.3. PAR-Binding Proteins
3.4. ADPr of Macromolecules
4. The Role of ADPr in RNA Biology
4.1. ADPr-Mediated Regulation of Nuclear RNA Processing
4.1.1. RNA Regulation at the Nucleus
4.1.2. rRNA at the Nucleolus
4.2. ADPr-Mediated Regulation of RNA Processing at the Cytoplasm
5. Targeting ADPr Signaling for Therapeutic Treatment
5.1. Chemical Inhibition of ARTDs
5.2. New Avenues: The Post-Transcriptional Regulation of ARTDs mRNA
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pope, S.D.; Medzhitov, R. Emerging Principles of Gene Expression Programs and Their Regulation. Mol. Cell 2018, 71, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef]
- Ramazi, S.; Zahiri, J. Posttranslational modifications in proteins: Resources, tools and prediction methods. Database 2021, 2021, baab012. [Google Scholar] [CrossRef]
- Vu, L.D.; Gevaert, K.; De Smet, I. Protein Language: Post-Translational Modifications Talking to Each Other. Trends Plant Sci. 2018, 23, 1068–1080. [Google Scholar] [CrossRef]
- van der Laarse, S.A.M.; Leney, A.C.; Heck, A.J.R. Crosstalk between phosphorylation and O-GlcNAcylation: Friend or foe. FEBS J. 2018, 285, 3152–3167. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Brickner, J.R.; Majid, M.C.; Mosammaparast, N. Crosstalk between ubiquitin and other post-translational modifications on chromatin during double-strand break repair. Trends Cell Biol. 2014, 24, 426–434. [Google Scholar] [CrossRef] [Green Version]
- Kishore, C. Epigenetic Regulation and Promising Therapies in Colorectal Cancer. Curr. Mol. Pharmacol. 2021, 14, 838–852. [Google Scholar] [CrossRef]
- He, W.; Wei, L.; Zou, Q. Research progress in protein posttranslational modification site prediction. Brief. Funct. Genom. 2018, 18, 220–229. [Google Scholar] [CrossRef]
- Ueda, K.; Hayaishi, O. ADP-ribosylation. Annu. Rev. Biochem. 1985, 54, 73–100. [Google Scholar] [CrossRef]
- Corda, D.; Di Girolamo, M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003, 22, 1953–1958. [Google Scholar] [CrossRef] [Green Version]
- Aravind, L.; Zhang, D.; de Souza, R.F.; Anand, S.; Iyer, L.M. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 2015, 384, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.S.; Chang, P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat. Chem. Biol. 2018, 14, 236–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munnur, D.; Ahel, I. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017, 284, 4002–4016. [Google Scholar] [CrossRef] [Green Version]
- Otto, H.; Reche, P.A.; Bazan, F.; Dittmar, K.; Haag, F.; Koch-Nolte, F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genom. 2005, 6, 139. [Google Scholar] [CrossRef] [Green Version]
- Teloni, F.; Altmeyer, M. Readers of poly(ADP-ribose): Designed to be fit for purpose. Nucleic Acids Res. 2016, 44, 993–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassa, P.O.; Haenni, S.S.; Elser, M.; Hottiger, M.O. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006, 70, 789–829. [Google Scholar] [CrossRef] [Green Version]
- Hawse, W.F.; Wolberger, C. Structure-based mechanism of ADP-ribosylation by sirtuins. J. Biol. Chem. 2009, 284, 33654–33661. [Google Scholar] [CrossRef] [Green Version]
- Iyer, L.M.; Zhang, D.; Rogozin, I.B.; Aravind, L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011, 39, 9473–9497. [Google Scholar] [CrossRef] [Green Version]
- Perina, D.; Mikoč, A.; Ahel, J.; Ćetković, H.; Žaja, R.; Ahel, I. Distribution of protein poly(ADP-ribosyl)ation systems across all domains of life. DNA Repair 2014, 23, 4–16. [Google Scholar] [CrossRef] [Green Version]
- Palazzo, L.; Mikoč, A.; Ahel, I. ADP-ribosylation: New facets of an ancient modification. FEBS J. 2017, 284, 2932–2946. [Google Scholar] [CrossRef]
- Faraone-Mennella, M.R.; Gambacorta, A.; Nicolaus, B.; Farina, B. Purification and biochemical characterization of a poly(ADP-ribose) polymerase-like enzyme from the thermophilic archaeon Sulfolobus solfataricus. Biochem. J. 1998, 335 Pt 2, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, G.; Corda, D.; Catara, G. From toxins to mammalian enzymes: The diversity of mono-ADP-ribosylation. Front. Biosci. 2015, 20, 389–404. [Google Scholar] [CrossRef]
- Mikolčević, P.; Hloušek-Kasun, A.; Ahel, I.; Mikoč, A. ADP-ribosylation systems in bacteria and viruses. Comput. Struct. Biotechnol. J. 2021, 19, 2366–2383. [Google Scholar] [CrossRef] [PubMed]
- Catara, G.; Corteggio, A.; Valente, C.; Grimaldi, G.; Palazzo, L. Targeting ADP-ribosylation as an antimicrobial strategy. Biochem. Pharmacol. 2019, 167, 13–26. [Google Scholar] [CrossRef]
- Koch-Nolte, F. Endogenous ADP-Ribosylation. Endog. Adp-Ribosylation 2015, 384, 1–213. [Google Scholar] [CrossRef]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef] [Green Version]
- Ludden, P.W. Reversible ADP-ribosylation as a mechanism of enzyme regulation in procaryotes. Mol. Cell. Biochem. 1994, 138, 123–129. [Google Scholar] [CrossRef]
- Ma, Y.; Ludden, P.W. Role of the dinitrogenase reductase arginine 101 residue in dinitrogenase reductase ADP-ribosyltransferase binding, NAD binding, and cleavage. J. Bacteriol. 2001, 183, 250–256. [Google Scholar] [CrossRef] [Green Version]
- Simon, N.C.; Aktories, K.; Barbieri, J.T. Novel bacterial ADP-ribosylating toxins: Structure and function. Nat. Rev. Microbiol. 2014, 12, 599–611. [Google Scholar] [CrossRef] [Green Version]
- Leung, A.K.L.; McPherson, R.L.; Griffin, D.E. Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog. 2018, 14, e1006864. [Google Scholar] [CrossRef]
- Krueger, K.M.; Barbieri, J.T. The family of bacterial ADP-ribosylating exotoxins. Clin. Microbiol. Rev. 1995, 8, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Debing, Y.; Jankevicius, G.; Neyts, J.; Ahel, I.; Coutard, B.; Canard, B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J. Virol. 2016, 90, 8478–8486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katada, T.; Ui, M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc. Natl. Acad. Sci. USA 1982, 79, 3129–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Souza, R.F.; Aravind, L. Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions. Mol. Biosyst. 2012, 8, 1661–1677. [Google Scholar] [CrossRef] [PubMed]
- Fieldhouse, R.J.; Turgeon, Z.; White, D.; Merrill, A.R. Cholera- and anthrax-like toxins are among several new ADP-ribosyltransferases. PLoS Comput. Biol. 2010, 6, e1001029. [Google Scholar] [CrossRef]
- Hottiger, M.O.; Hassa, P.O.; Lüscher, B.; Schüler, H.; Koch-Nolte, F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010, 35, 208–219. [Google Scholar] [CrossRef]
- Castellano, S.; Farina, B.; Faraone-Mennella, M.R. The ADP-ribosylation of Sulfolobus solfataricus Sso7 modulates protein/DNA interactions in vitro. FEBS Lett. 2009, 583, 1154–1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Maro, A.; De Maio, A.; Castellano, S.; Parente, A.; Farina, B.; Faraone-Mennella, M.R. The ADP-ribosylating thermozyme from Sulfolobus solfataricus is a DING protein. Biol. Chem. 2009, 390, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Porzio, E.; Bianchi, A.R.; Baccigalupi, L.; Isticato, R.; Mennella, M.R.F. The DINGGG thermoprotein is membrane bound in the Crenarchaeon Sulfolobus solfataricus. Chem. Biol. Technol. Agric. 2016, 3, 8. [Google Scholar] [CrossRef] [Green Version]
- Porzio, E.; De Maio, A.; Ricciardi, T.; Mistretta, C.; Manco, G.; Faraone-Mennella, M.R. Comparison of the DING protein from the archaeon Sulfolobus solfataricus with human phosphate-binding protein and Pseudomonas fluorescence DING counterparts. Extremophiles 2018, 22, 177–188. [Google Scholar] [CrossRef] [PubMed]
- De Maio, A.; Porzio, E.; Rotondo, S.; Bianchi, A.R.; Faraone-Mennella, M.R. In Sulfolobus solfataricus, the Poly(ADP-Ribose) Polymerase-Like Thermoprotein Is a Multifunctional Enzyme. Microorganisms 2020, 8, 1523. [Google Scholar] [CrossRef] [PubMed]
- Porzio, E.; Faraone Mennella, M.R.; Manco, G. DING Proteins Extend to the Extremophilic World. Int. J. Mol. Sci. 2021, 22, 2035. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Buckle, A.M.; Cordell, S.C.; Löwe, J.; Bycroft, M. The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non-histone domain of macroH2A. J. Mol. Biol. 2003, 330, 503–511. [Google Scholar] [CrossRef]
- Karras, G.I.; Kustatscher, G.; Buhecha, H.R.; Allen, M.D.; Pugieux, C.; Sait, F.; Bycroft, M.; Ladurner, A.G. The macro domain is an ADP-ribose binding module. EMBO J. 2005, 24, 1911–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosenthal, F.; Feijs, K.L.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O.; et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yamauchi, Y.; Kamakura, M.; Murayama, T.; Goshima, F.; Kimura, H.; Nishiyama, Y. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J. Virol. 2012, 86, 492–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattiussi, S.; Tempera, I.; Matusali, G.; Mearini, G.; Lenti, L.; Fratarcangeli, S.; Mosca, L.; D’Erme, M.; Mattia, E. Inhibition of Poly(ADP-ribose)polymerase impairs Epstein Barr Virus lytic cycle progression. Infect. Agent Cancer 2007, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Saikatendu, K.S.; Joseph, J.S.; Subramanian, V.; Clayton, T.; Griffith, M.; Moy, K.; Velasquez, J.; Neuman, B.W.; Buchmeier, M.J.; Stevens, R.C.; et al. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1’’-phosphate dephosphorylation by a conserved domain of nsP3. Structure 2005, 13, 1665–1675. [Google Scholar] [CrossRef] [Green Version]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020, 11, 4940. [Google Scholar] [CrossRef]
- Tchasovnikarova, I.A.; Timms, R.T.; Matheson, N.J.; Wals, K.; Antrobus, R.; Göttgens, B.; Dougan, G.; Dawson, M.A.; Lehner, P.J. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 2015, 348, 1481–1485. [Google Scholar] [CrossRef] [Green Version]
- Wyżewski, Z.; Gradowski, M.; Krysińska, M.; Dudkiewicz, M.; Pawłowski, K. A novel predicted ADP-ribosyltransferase-like family conserved in eukaryotic evolution. PeerJ 2021, 9, e11051. [Google Scholar] [CrossRef]
- Haigis, M.C.; Sinclair, D.A. Mammalian sirtuins: Biological insights and disease relevance. Annu. Rev. Pathol. 2010, 5, 253–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, H.C.; Guarente, L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol. Metab. 2014, 25, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Challa, S.; Stokes, M.S.; Kraus, W.L. MARTs and MARylation in the Cytosol: Biological Functions, Mechanisms of Action, and Therapeutic Potential. Cells 2021, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Lüscher, B.; Ahel, I.; Altmeyer, M.; Ashworth, A.; Bai, P.; Chang, P.; Cohen, M.; Corda, D.; Dantzer, F.; Daugherty, M.D.; et al. ADP-ribosyltransferases, an update on function and nomenclature. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.S.; Challa, S.; Jones, A.; Kraus, W.L. PARPs and ADP-ribosylation in RNA biology: From RNA expression and processing to protein translation and proteostasis. Genes Dev. 2020, 34, 302–320. [Google Scholar] [CrossRef]
- Kim, C.; Wang, X.D.; Yu, Y. PARP1 inhibitors trigger innate immunity via PARP1 trapping-induced DNA damage response. Elife 2020, 9, e01969. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Kassab, M.A.; Dantzer, F.; Yu, X. PARP2 mediates branched poly ADP-ribosylation in response to DNA damage. Nat. Commun. 2018, 9, 3233. [Google Scholar] [CrossRef]
- Rouleau, M.; McDonald, D.; Gagné, P.; Ouellet, M.E.; Droit, A.; Hunter, J.M.; Dutertre, S.; Prigent, C.; Hendzel, M.J.; Poirier, G.G. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 2007, 100, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Boehler, C.; Gauthier, L.R.; Mortusewicz, O.; Biard, D.S.; Saliou, J.M.; Bresson, A.; Sanglier-Cianferani, S.; Smith, S.; Schreiber, V.; Boussin, F.; et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc. Natl. Acad. Sci. USA 2011, 108, 2783–2788. [Google Scholar] [CrossRef] [Green Version]
- Kickhoefer, V.A.; Siva, A.C.; Kedersha, N.L.; Inman, E.M.; Ruland, C.; Streuli, M.; Rome, L.H. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 1999, 146, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.; Coughlin, M.; Mitchison, T.J. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat. Cell Biol. 2005, 7, 1133–1139. [Google Scholar] [CrossRef] [PubMed]
- Azarm, K.; Smith, S. Nuclear PARPs and genome integrity. Genes Dev. 2020, 34, 285–301. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.M.; Mishina, Y.M.; Liu, S.; Cheung, A.; Stegmeier, F.; Michaud, G.A.; Charlat, O.; Wiellette, E.; Zhang, Y.; Wiessner, S.; et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 2009, 461, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Damale, M.G.; Pathan, S.K.; Shinde, D.B.; Patil, R.H.; Arote, R.B.; Sangshetti, J.N. Insights of tankyrases: A novel target for drug discovery. Eur. J. Med. Chem. 2020, 207, 112712. [Google Scholar] [CrossRef]
- Huang, J.Y.; Wang, K.; Vermehren-Schmaedick, A.; Adelman, J.P.; Cohen, M.S. PARP6 is a Regulator of Hippocampal Dendritic Morphogenesis. Sci. Rep. 2016, 6, 18512. [Google Scholar] [CrossRef] [Green Version]
- Atasheva, S.; Frolova, E.I.; Frolov, I. Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J. Virol. 2014, 88, 2116–2130. [Google Scholar] [CrossRef] [Green Version]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Mao, D.; Roswit, W.T.; Jin, X.; Patel, A.C.; Patel, D.A.; Agapov, E.; Wang, Z.; Tidwell, R.M.; Atkinson, J.J.; et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 2015, 16, 1215–1227. [Google Scholar] [CrossRef]
- Chou, H.Y.; Chou, H.T.; Lee, S.C. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J. Biol. Chem. 2006, 281, 15201–15207. [Google Scholar] [CrossRef] [Green Version]
- Venkannagari, H.; Verheugd, P.; Koivunen, J.; Haikarainen, T.; Obaji, E.; Ashok, Y.; Narwal, M.; Pihlajaniemi, T.; Lüscher, B.; Lehtiö, L. Small-Molecule Chemical Probe Rescues Cells from Mono-ADP-Ribosyltransferase ARTD10/PARP10-Induced Apoptosis and Sensitizes Cancer Cells to DNA Damage. Cell Chem. Biol. 2016, 23, 1251–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer-Ficca, M.L.; Ihara, M.; Bader, J.J.; Leu, N.A.; Beneke, S.; Meyer, R.G. Spermatid head elongation with normal nuclear shaping requires ADP-ribosyltransferase PARP11 (ARTD11) in mice. Biol. Reprod. 2015, 92, 80. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Shi, Y.; Li, S.; Liu, J.; Zu, S.; Xu, X.; Gao, M.; Sun, N.; Pan, C.; Peng, L.; et al. ADP-ribosyltransferase PARP11 suppresses Zika virus in synergy with PARP12. Cell Biosci. 2021, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Vyas, S.; Rood, J.E.; Bhutkar, A.; Sharp, P.A.; Chang, P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 2011, 42, 489–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catara, G.; Grimaldi, G.; Schembri, L.; Spano, D.; Turacchio, G.; Lo Monte, M.; Beccari, A.R.; Valente, C.; Corda, D. PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Sci. Rep. 2017, 7, 14035. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, G.; Filograna, A.; Schembri, L.; Lo Monte, M.; Di Martino, R.; Pirozzi, M.; Spano, D.; Beccari, A.R.; Parashuraman, S.; Luini, A.; et al. PKD-dependent PARP12-catalyzed mono-ADP-ribosylation of Golgin-97 is required for E-cadherin transport from Golgi to plasma membrane. Proc. Natl. Acad. Sci. USA 2022, 119, e2026494119. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Todorova, T.; Bock, F.J.; Chang, P. PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat. Commun. 2014, 5, 5362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunewald, M.E.; Chen, Y.; Kuny, C.; Maejima, T.; Lease, R.; Ferraris, D.; Aikawa, M.; Sullivan, C.S.; Perlman, S.; Fehr, A.R. The coronavirus macrodomain is required to prevent PARP-mediated inhibition of virus replication and enhancement of IFN expression. PLoS Pathog. 2019, 15, e1007756. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.B.; Johns, M.; Cao, J.; Liu, Y.; Yu, S.C.; Hyde, G.D.; Laffan, M.A.; Marchese, F.P.; Cho, S.H.; Clark, A.R.; et al. PARP-14 combines with tristetraprolin in the selective posttranscriptional control of macrophage tissue factor expression. Blood 2014, 124, 3646–3655. [Google Scholar] [CrossRef] [Green Version]
- Jwa, M.; Chang, P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat. Cell Biol. 2012, 14, 1223–1230. [Google Scholar] [CrossRef] [Green Version]
- Demény, M.A.; Virág, L. The PARP Enzyme Family and the Hallmarks of Cancer Part 1. Cell Intrinsic Hallmarks. Cancers 2021, 13, 2042. [Google Scholar] [CrossRef]
- Karlberg, T.; Klepsch, M.; Thorsell, A.G.; Andersson, C.D.; Linusson, A.; Schüler, H. Structural basis for lack of ADP-ribosyltransferase activity in poly(ADP-ribose) polymerase-13/zinc finger antiviral protein. J. Biol. Chem. 2015, 290, 7336–7344. [Google Scholar] [CrossRef] [Green Version]
- Xue, G.; Braczyk, K.; Gonçalves-Carneiro, D.; Dawidziak, D.M.; Sanchez, K.; Ong, H.; Wan, Y.; Zadrozny, K.K.; Ganser-Pornillos, B.K.; Bieniasz, P.D.; et al. Poly(ADP-ribose) potentiates ZAP antiviral activity. PLoS Pathog. 2022, 18, e1009202. [Google Scholar] [CrossRef]
- Menzel, S.; Adriouch, S.; Bannas, P.; Haag, F.; Koch-Nolte, F. Monitoring Expression and Enzyme Activity of Ecto-ARTCs. Methods Mol. Biol. 2018, 1813, 167–186. [Google Scholar] [CrossRef]
- Di Girolamo, M.; Fabrizio, G. Overview of the mammalian ADP-ribosyl-transferases clostridia toxin-like (ARTCs) family. Biochem. Pharmacol. 2019, 167, 86–96. [Google Scholar] [CrossRef]
- O’Sullivan, J.; Tedim Ferreira, M.; Gagné, J.P.; Sharma, A.K.; Hendzel, M.J.; Masson, J.Y.; Poirier, G.G. Emerging roles of eraser enzymes in the dynamic control of protein ADP-ribosylation. Nat. Commun. 2019, 10, 1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashimo, M.; Kato, J.; Moss, J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc. Natl. Acad. Sci. USA 2013, 110, 18964–18969. [Google Scholar] [CrossRef] [Green Version]
- Fontana, P.; Bonfiglio, J.J.; Palazzo, L.; Bartlett, E.; Matic, I.; Ahel, I. Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife 2017, 6, e28533. [Google Scholar] [CrossRef] [PubMed]
- Barkauskaite, E.; Jankevicius, G.; Ahel, I. Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation. Mol. Cell 2015, 58, 935–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alhammad, Y.M.O.; Kashipathy, M.M.; Roy, A.; Gagné, J.P.; McDonald, P.; Gao, P.; Nonfoux, L.; Battaile, K.P.; Johnson, D.K.; Holmstrom, E.D.; et al. The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase. J. Virol. 2021, 95, e01969-20. [Google Scholar] [CrossRef] [PubMed]
- Slade, D.; Dunstan, M.S.; Barkauskaite, E.; Weston, R.; Lafite, P.; Dixon, N.; Ahel, M.; Leys, D.; Ahel, I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature 2011, 477, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Dunstan, M.S.; Barkauskaite, E.; Lafite, P.; Knezevic, C.E.; Brassington, A.; Ahel, M.; Hergenrother, P.J.; Leys, D.; Ahel, I. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat. Commun. 2012, 3, 878. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, R.; Morra, R.; Appel, C.D.; Tallis, M.; Chioza, B.; Jankevicius, G.; Simpson, M.A.; Matic, I.; Ozkan, E.; Golia, B.; et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [Green Version]
- Jankevicius, G.; Hassler, M.; Golia, B.; Rybin, V.; Zacharias, M.; Timinszky, G.; Ladurner, A.G. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat. Struct. Mol. Biol. 2013, 20, 508–514. [Google Scholar] [CrossRef]
- Rack, J.G.M.; Zorzini, V.; Zhu, Z.; Schuller, M.; Ahel, D.; Ahel, I. Viral macrodomains: A structural and evolutionary assessment of the pharmacological potential. Open Biol. 2020, 10, 200237. [Google Scholar] [CrossRef]
- Palazzo, L.; Thomas, B.; Jemth, A.S.; Colby, T.; Leidecker, O.; Feijs, K.L.; Zaja, R.; Loseva, O.; Puigvert, J.C.; Matic, I.; et al. Processing of protein ADP-ribosylation by Nudix hydrolases. Biochem. J. 2015, 468, 293–301. [Google Scholar] [CrossRef]
- Palazzo, L.; Daniels, C.M.; Nettleship, J.E.; Rahman, N.; McPherson, R.L.; Ong, S.E.; Kato, K.; Nureki, O.; Leung, A.K.; Ahel, I. ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 2016, 283, 3371–3388. [Google Scholar] [CrossRef]
- Krietsch, J.; Rouleau, M.; Pic, É.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.Y.; Poirier, G.G.; Gagné, J.P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Asp. Med. 2013, 34, 1066–1087. [Google Scholar] [CrossRef]
- Sowa, S.T.; Galera-Prat, A.; Wazir, S.; Alanen, H.I.; Maksimainen, M.M.; Lehtiö, L. A molecular toolbox for ADP-ribosyl binding proteins. Cell Rep. Methods 2021, 1, 100121. [Google Scholar] [CrossRef] [PubMed]
- Althaus, F.R.; Kleczkowska, H.E.; Malanga, M.; Müntener, C.R.; Pleschke, J.M.; Ebner, M.; Auer, B. Poly ADP-ribosylation: A DNA break signal mechanism. Mol. Cell Biochem. 1999, 193, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yu, X. Functions of PARylation in DNA Damage Repair Pathways. Genom. Proteom. Bioinform. 2016, 14, 131–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Popp, O.; Veith, S.; Fahrer, J.; Bohr, V.A.; Bürkle, A.; Mangerich, A. Site-specific noncovalent interaction of the biopolymer poly(ADP-ribose) with the Werner syndrome protein regulates protein functions. ACS Chem. Biol. 2013, 8, 179–188. [Google Scholar] [CrossRef]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, M.; Takamura-Enya, T.; Kanazawa, T.; Totsuka, Y.; Matsushima-Hibiya, Y.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Mono(ADP-ribosyl)ation of DNA by apoptosis-inducing protein, pierisin. Nucleic Acids Res. Suppl. 2002, 2, 243–244. [Google Scholar] [CrossRef] [Green Version]
- Carpusca, I.; Jank, T.; Aktories, K. Bacillus sphaericus mosquitocidal toxin (MTX) and pierisin: The enigmatic offspring from the family of ADP-ribosyltransferases. Mol. Microbiol. 2006, 62, 621–630. [Google Scholar] [CrossRef]
- Munnur, D.; Bartlett, E.; Mikolčević, P.; Kirby, I.T.; Rack, J.G.M.; Mikoč, A.; Cohen, M.S.; Ahel, I. Reversible ADP-ribosylation of RNA. Nucleic Acids Res. 2019, 47, 5658–5669. [Google Scholar] [CrossRef] [Green Version]
- Weixler, L.; Schäringer, K.; Momoh, J.; Lüscher, B.; Feijs, K.L.H.; Žaja, R. ADP-ribosylation of RNA and DNA: From in vitro characterization to in vivo function. Nucleic Acids Res. 2021, 49, 3634–3650. [Google Scholar] [CrossRef]
- Palazzo, L.; Leidecker, O.; Prokhorova, E.; Dauben, H.; Matic, I.; Ahel, I. Serine is the major residue for ADP-ribosylation upon DNA damage. Elife 2018, 7, e34334. [Google Scholar] [CrossRef] [PubMed]
- Larsen, S.C.; Hendriks, I.A.; Lyon, D.; Jensen, L.J.; Nielsen, M.L. Systems-wide Analysis of Serine ADP-Ribosylation Reveals Widespread Occurrence and Site-Specific Overlap with Phosphorylation. Cell Rep. 2018, 24, 2493–2505. [Google Scholar] [CrossRef] [Green Version]
- Leslie Pedrioli, D.M.; Leutert, M.; Bilan, V.; Nowak, K.; Gunasekera, K.; Ferrari, E.; Imhof, R.; Malmström, L.; Hottiger, M.O. Comprehensive ADP-ribosylome analysis identifies tyrosine as an ADP-ribose acceptor site. EMBO Rep. 2018, 19, e45310. [Google Scholar] [CrossRef] [PubMed]
- Zarkovic, G.; Belousova, E.A.; Talhaoui, I.; Saint-Pierre, C.; Kutuzov, M.M.; Matkarimov, B.T.; Biard, D.; Gasparutto, D.; Lavrik, O.I.; Ishchenko, A.A. Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: New insights into DNA ADP-ribosylation. Nucleic Acids Res. 2018, 46, 2417–2431. [Google Scholar] [CrossRef]
- Tao, Z.; Gao, P.; Liu, H.W. Identification of the ADP-ribosylation sites in the PARP-1 automodification domain: Analysis and implications. J. Am. Chem. Soc. 2009, 131, 14258–14260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Ding, M.; Yu, Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 2013, 10, 981–984. [Google Scholar] [CrossRef]
- Carusone, T.M.; Cardiero, G.; Cerreta, M.; Mandrich, L.; Moran, O.; Porzio, E.; Catara, G.; Lacerra, G.; Manco, G. WTAP and BIRC3 are involved in the posttranscriptional mechanisms that impact on the expression and activity of the human lactonase PON2. Cell Death Dis. 2020, 11, 324. [Google Scholar] [CrossRef]
- Yang, C.S.; Jividen, K.; Spencer, A.; Dworak, N.; Ni, L.; Oostdyk, L.T.; Chatterjee, M.; Kuśmider, B.; Reon, B.; Parlak, M.; et al. Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516. [Google Scholar] [CrossRef] [Green Version]
- Gomez, A.; Bindesbøll, C.; Satheesh, S.V.; Grimaldi, G.; Hutin, D.; MacPherson, L.; Ahmed, S.; Tamblyn, L.; Cho, T.; Nebb, H.I.; et al. Characterization of TCDD-inducible poly-ADP-ribose polymerase (TIPARP/ARTD14) catalytic activity. Biochem. J. 2018, 475, 3827–3846. [Google Scholar] [CrossRef] [Green Version]
- Kamata, T.; Yang, C.S.; Paschal, B.M. PARP7 mono-ADP-ribosylates the agonist conformation of the androgen receptor in the nucleus. Biochem. J. 2021, 478, 2999–3014. [Google Scholar] [CrossRef]
- Buch-Larsen, S.C.; Hendriks, I.A.; Lodge, J.M.; Rykær, M.; Furtwängler, B.; Shishkova, E.; Westphall, M.S.; Coon, J.J.; Nielsen, M.L. Mapping Physiological ADP-Ribosylation Using Activated Ion Electron Transfer Dissociation. Cell Rep. 2020, 32, 108176. [Google Scholar] [CrossRef] [PubMed]
- De Vos, M.; Schreiber, V.; Dantzer, F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol. 2012, 84, 137–146. [Google Scholar] [CrossRef] [PubMed]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342 Pt 2, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Eisemann, T.; Riccio, A.A.; Pascal, J.M. PARP family enzymes: Regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 2018, 53, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Krishnakumar, R.; Kraus, W.L. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 2010, 39, 736–749. [Google Scholar] [CrossRef] [Green Version]
- Gibson, B.A.; Zhang, Y.; Jiang, H.; Hussey, K.M.; Shrimp, J.H.; Lin, H.; Schwede, F.; Yu, Y.; Kraus, W.L. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science 2016, 353, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, S.J.; Smith, S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 2008, 90, 83–92. [Google Scholar] [CrossRef]
- Yu, M.; Schreek, S.; Cerni, C.; Schamberger, C.; Lesniewicz, K.; Poreba, E.; Vervoorts, J.; Walsemann, G.; Grötzinger, J.; Kremmer, E.; et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 2005, 24, 1982–1993. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, S.B.; Frommel, S.C.; Camicia, R.; Winkler, H.C.; Santoro, R.; Hassa, P.O. DTX3L and ARTD9 inhibit IRF1 expression and mediate in cooperation with ARTD8 survival and proliferation of metastatic prostate cancer cells. Mol. Cancer 2014, 13, 125. [Google Scholar] [CrossRef] [Green Version]
- Schleicher, E.M.; Galvan, A.M.; Imamura-Kawasawa, Y.; Moldovan, G.L.; Nicolae, C.M. PARP10 promotes cellular proliferation and tumorigenesis by alleviating replication stress. Nucleic Acids Res. 2018, 46, 8908–8916. [Google Scholar] [CrossRef] [Green Version]
- Andrabi, S.A.; Kim, N.S.; Yu, S.W.; Wang, H.; Koh, D.W.; Sasaki, M.; Klaus, J.A.; Otsuka, T.; Zhang, Z.; Koehler, R.C.; et al. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA 2006, 103, 18308–18313. [Google Scholar] [CrossRef] [Green Version]
- Andrabi, S.A.; Kang, H.C.; Haince, J.F.; Lee, Y.I.; Zhang, J.; Chi, Z.; West, A.B.; Koehler, R.C.; Poirier, G.G.; Dawson, T.M.; et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011, 17, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virág, L.; Robaszkiewicz, A.; Rodriguez-Vargas, J.M.; Oliver, F.J. Poly(ADP-ribose) signaling in cell death. Mol. Asp. Med. 2013, 34, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Abd Elmageed, Z.Y.; Naura, A.S.; Errami, Y.; Zerfaoui, M. The poly(ADP-ribose) polymerases (PARPs): New roles in intracellular transport. Cell. Signal. 2012, 24, 1–8. [Google Scholar] [CrossRef]
- Cardamone, M.D.; Gao, Y.; Kwan, J.; Hayashi, V.; Sheeran, M.; Xu, J.; English, J.; Orofino, J.; Emili, A.; Perissi, V. Neuralized-like protein 4 (NEURL4) mediates ADP-ribosylation of mitochondrial proteins. J. Cell Biol. 2022, 221, e202101021. [Google Scholar] [CrossRef]
- Palazzo, L.; Mikolčević, P.; Mikoč, A.; Ahel, I. ADP-ribosylation signalling and human disease. Open Biol. 2019, 9, 190041. [Google Scholar] [CrossRef] [Green Version]
- Schützenhofer, K.; Rack, J.G.M.; Ahel, I. The Making and Breaking of Serine-ADP-Ribosylation in the DNA Damage Response. Front. Cell Dev. Biol. 2021, 9, 745922. [Google Scholar] [CrossRef]
- Chung, W.C.; Song, M.J. Virus-Host Interplay Between Poly (ADP-Ribose) Polymerase 1 and Oncogenic Gammaherpesviruses. Front. Microbiol. 2021, 12, 811671. [Google Scholar] [CrossRef]
- Alhammad, Y.M.O.; Fehr, A.R. The Viral Macrodomain Counters Host Antiviral ADP-Ribosylation. Viruses 2020, 12, 384. [Google Scholar] [CrossRef] [Green Version]
- Fehr, A.R.; Singh, S.A.; Kerr, C.M.; Mukai, S.; Higashi, H.; Aikawa, M. The impact of PARPs and ADP-ribosylation on inflammation and host-pathogen interactions. Genes Dev. 2020, 34, 341–359. [Google Scholar] [CrossRef]
- Richard, I.A.; Burgess, J.T.; O’Byrne, K.J.; Bolderson, E. Beyond PARP1: The Potential of Other Members of the Poly (ADP-Ribose) Polymerase Family in DNA Repair and Cancer Therapeutics. Front. Cell Dev. Biol. 2021, 9, 801200. [Google Scholar] [CrossRef]
- Brady, P.N.; Goel, A.; Johnson, M.A. Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity. Microbiol. Mol. Biol. Rev. 2019, 83, e00038-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehr, A.R.; Jankevicius, G.; Ahel, I.; Perlman, S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2018, 26, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Lüscher, B.; Bütepage, M.; Eckei, L.; Krieg, S.; Verheugd, P.; Shilton, B.H. ADP-Ribosylation, a Multifaceted Posttranslational Modification Involved in the Control of Cell Physiology in Health and Disease. Chem. Rev. 2018, 118, 1092–1136. [Google Scholar] [CrossRef]
- Kunze, F.A.; Hottiger, M.O. Regulating Immunity via ADP-Ribosylation: Therapeutic Implications and Beyond. Trends Immunol. 2019, 40, 159–173. [Google Scholar] [CrossRef] [PubMed]
- Atasheva, S.; Akhrymuk, M.; Frolova, E.I.; Frolov, I. New PARP gene with an anti-alphavirus function. J. Virol. 2012, 86, 8147–8160. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhao, H.; Liu, P.; Li, C.; Quanquin, N.; Ji, X.; Sun, N.; Du, P.; Qin, C.F.; Lu, N.; et al. suppresses Zika virus infection through PARP-dependent degradation of NS1 and NS3 viral proteins. Sci. Signal. 2018, 11, eaas9332. [Google Scholar] [CrossRef] [Green Version]
- Velázquez-Cruz, A.; Baños-Jaime, B.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation. Front. Mol. Biosci. 2021, 8, 658852. [Google Scholar] [CrossRef]
- Moore, M.J.; Proudfoot, N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009, 136, 688–700. [Google Scholar] [CrossRef] [Green Version]
- Chaudhury, A.; Chander, P.; Howe, P.H. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: Focus on hnRNP E1’s multifunctional regulatory roles. RNA 2010, 16, 1449–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gagné, J.P.; Hunter, J.M.; Labrecque, B.; Chabot, B.; Poirier, G.G. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 2003, 371, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Tulin, A.V. Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int. J. Mol. Sci. 2013, 14, 16168–16183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malanga, M.; Czubaty, A.; Girstun, A.; Staron, K.; Althaus, F.R. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008, 283, 19991–19998. [Google Scholar] [CrossRef] [Green Version]
- Isabelle, M.; Gagné, J.P.; Gallouzi, I.E.; Poirier, G.G. Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)-dependent following exposure to MNNG-induced DNA alkylation. J. Cell Sci. 2012, 125, 4555–4566. [Google Scholar] [CrossRef] [Green Version]
- Di Giammartino, D.C.; Shi, Y.; Manley, J.L. PARP1 represses PAP and inhibits polyadenylation during heat shock. Mol. Cell 2013, 49, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Han, Y.; Guo, X.; Wen, J.; Wang, K.; Jiang, X.; Tian, X.; Ba, X.; Boldogh, I.; Zeng, X. PARP1 promotes gene expression at the post-transcriptiona level by modulating the RNA-binding protein HuR. Nat. Commun. 2017, 8, 14632. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.; Lv, X.; Fu, X.; Zhang, J.; Bohio, A.A.; Zeng, X.; Hao, W.; Wang, R.; Boldogh, I.; Ba, X. Poly(ADP-ribosyl)ation enhances HuR oligomerization and contributes to pro-inflammatory gene mRNA stabilization. Cell Mol. Life Sci. 2021, 78, 1817–1835. [Google Scholar] [CrossRef]
- Kirby, I.T.; Kojic, A.; Arnold, M.R.; Thorsell, A.G.; Karlberg, T.; Vermehren-Schmaedick, A.; Sreenivasan, R.; Schultz, C.; Schüler, H.; Cohen, M.S. A Potent and Selective PARP11 Inhibitor Suggests Coupling between Cellular Localization and Catalytic Activity. Cell Chem. Biol. 2018, 25, 1547–1553. [Google Scholar] [CrossRef] [Green Version]
- Boamah, E.K.; Kotova, E.; Garabedian, M.; Jarnik, M.; Tulin, A.V. Poly(ADP-Ribose) polymerase 1 (PARP-1) regulates ribosomal biogenesis in Drosophila nucleoli. PLoS Genet. 2012, 8, e1002442. [Google Scholar] [CrossRef] [Green Version]
- Guetg, C.; Scheifele, F.; Rosenthal, F.; Hottiger, M.O.; Santoro, R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 2012, 45, 790–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, S.; Chesarone-Cataldo, M.; Todorova, T.; Huang, Y.H.; Chang, P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013, 4, 2240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Y.; Tulin, A.V. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat. Commun. 2012, 3, 760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challa, S.; Khulpateea, B.R.; Nandu, T.; Camacho, C.V.; Ryu, K.W.; Chen, H.; Peng, Y.; Lea, J.S.; Kraus, W.L. Ribosome ADP-ribosylation inhibits translation and maintains proteostasis in cancers. Cell 2021, 184, 4531–4546. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.L.; Griffin, D.E.; Bosch, J.; Fehr, A.R. The Conserved Macrodomain Is a Potential Therapeutic Target for Coronaviruses and Alphaviruses. Pathogens 2022, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Malgras, M.; Garcia, M.; Jousselin, C.; Bodet, C.; Lévêque, N. The Antiviral Activities of Poly-ADP-Ribose Polymerases. Viruses 2021, 13, 582. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Todorova, T.; Ando, Y.; Chang, P. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol. 2012, 9, 542–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, G.; Guo, X.; Goff, S.P. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 2002, 297, 1703–1706. [Google Scholar] [CrossRef]
- Zhu, Y.; Chen, G.; Lv, F.; Wang, X.; Ji, X.; Xu, Y.; Sun, J.; Wu, L.; Zheng, Y.T.; Gao, G. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl. Acad. Sci. USA 2011, 108, 15834–15839. [Google Scholar] [CrossRef] [Green Version]
- Carter-O’Connell, I.; Vermehren-Schmaedick, A.; Jin, H.; Morgan, R.K.; David, L.L.; Cohen, M.S. Combining Chemical Genetics with Proximity-Dependent Labeling Reveals Cellular Targets of Poly(ADP-ribose) Polymerase 14 (PARP14). ACS Chem. Biol. 2018, 13, 2841–2848. [Google Scholar] [CrossRef]
- Patel, M.; Nowsheen, S.; Maraboyina, S.; Xia, F. The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: A review. Cell Biosci. 2020, 10, 35. [Google Scholar] [CrossRef]
- Poltronieri, P.; Miwa, M.; Masutani, M. ADP-Ribosylation as Post-Translational Modification of Proteins: Use of Inhibitors in Cancer Control. Int. J. Mol. Sci. 2021, 22, 10829. [Google Scholar] [CrossRef] [PubMed]
- Kirby, I.T.; Cohen, M.S. Small-Molecule Inhibitors of PARPs: From Tools for Investigating ADP-Ribosylation to Therapeutics. Curr. Top. Microbiol. Immunol. 2019, 420, 211–231. [Google Scholar] [CrossRef]
- Wasyluk, W.; Zwolak, A. PARP Inhibitors: An Innovative Approach to the Treatment of Inflammation and Metabolic Disorders in Sepsis. J. Inflamm. Res. 2021, 14, 1827–1844. [Google Scholar] [CrossRef] [PubMed]
- Henning, R.J.; Bourgeois, M.; Harbison, R.D. Poly(ADP-ribose) Polymerase (PARP) and PARP Inhibitors: Mechanisms of Action and Role in Cardiovascular Disorders. Cardiovasc. Toxicol. 2018, 18, 493–506. [Google Scholar] [CrossRef]
- Pacher, P.; Szabó, C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: The therapeutic potential of PARP inhibitors. Cardiovasc. Drug Rev. 2007, 25, 235–260. [Google Scholar] [CrossRef] [Green Version]
- Fu, W.; Yao, H.; Bütepage, M.; Zhao, Q.; Lüscher, B.; Li, J. The search for inhibitors of macrodomains for targeting the readers and erasers of mono-ADP-ribosylation. Drug Discov. Today 2021, 26, 2547–2558. [Google Scholar] [CrossRef] [PubMed]
- Pollock, K.; Liu, M.; Zaleska, M.; Meniconi, M.; Pfuhl, M.; Collins, I.; Guettler, S. Fragment-based screening identifies molecules targeting the substrate-binding ankyrin repeat domains of tankyrase. Sci. Rep. 2019, 9, 19130. [Google Scholar] [CrossRef] [Green Version]
- Mullard, A. FDA approves oral version of diabetes biologic. Nat. Rev. Drug Discov. 2019, 18, 814. [Google Scholar] [CrossRef]
- Na, Z.; Peng, B.; Ng, S.; Pan, S.; Lee, J.S.; Shen, H.M.; Yao, S.Q. A small-molecule protein-protein interaction inhibitor of PARP1 that targets its BRCT domain. Angew Chem. Int. Ed. Engl. 2015, 54, 2515–2519. [Google Scholar] [CrossRef] [PubMed]
- Brodie, S.A.; Li, G.; Harvey, D.; Khuri, F.R.; Vertino, P.M.; Brandes, J.C. Small molecule inhibition of the CHFR-PARP1 interaction as novel approach to overcome intrinsic taxane resistance in cancer. Oncotarget 2015, 6, 30773–30786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, D.; Huang, H.; Wang, J.; Zhao, Y.; Hu, X.; He, F.; Yu, L.; Wu, J. NF90 regulates PARP1 mRNA stability in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 488, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Yoon, K.; Ko, D.; Doderer, M.; Livi, C.B.; Penalva, L.O. Over-represented sequences located on 3’ UTRs are potentially involved in regulatory functions. RNA Biol. 2008, 5, 255–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, S.; Baltimore, D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009, 10, 281–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keene, J.D. RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 2007, 8, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Corso, C.; Pisapia, L.; Citro, A.; Cicatiello, V.; Barba, P.; Cigliano, L.; Abrescia, P.; Maffei, A.; Manco, G.; Del Pozzo, G. EBP1 and DRBP76/NF90 binding proteins are included in the major histocompatibility complex class II RNA operon. Nucleic Acids Res. 2011, 39, 7263–7275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberhardt, W.; Doller, A.; Akool, E.-S.; Pfeilschifter, J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol. Ther. 2007, 114, 56–73. [Google Scholar] [CrossRef]
- Harries, L.W. RNA Biology Provides New Therapeutic Targets for Human Disease. Front. Genet. 2019, 10, 205. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Z.; Dong, H.; Zhou, J.; Yuan, J.; Ni, B.; Wu, Y.; Han, C.; Tian, Y. Regulation of mRNA stability by RBPs and noncoding RNAs contributing to the pathogenicity of Th17 cells. RNA Biol. 2021, 18, 647–656. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, Y.; Richards, A.M. Effective tools for RNA-derived therapeutics: siRNA interference or miRNA mimicry. Theranostics 2021, 11, 8771–8796. [Google Scholar] [CrossRef] [PubMed]
- Chand, S.N.; Zarei, M.; Schiewer, M.J.; Kamath, A.R.; Romeo, C.; Lal, S.; Cozzitorto, J.A.; Nevler, A.; Scolaro, L.; Londin, E.; et al. Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors. Cancer Res. 2017, 77, 5011–5025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persson, H.; Kvist, A.; Rego, N.; Staaf, J.; Vallon-Christersson, J.; Luts, L.; Loman, N.; Jonsson, G.; Naya, H.; Hoglund, M.; et al. Identification of new microRNAs in paired normal and tumor breast tissue suggests a dual role for the ERBB2/Her2 gene. Cancer Res. 2011, 71, 78–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, W.J.; Cheng, S.; Campbell, C.; Wright, A.; Furneaux, H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J. Biol. Chem. 1996, 271, 8144–8151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, W.; Furuuchi, N.; Aslanukova, L.; Huang, Y.H.; Brown, S.Z.; Jiang, W.; Addya, S.; Vishwakarma, V.; Peters, E.; Brody, J.R.; et al. Elevated HuR in Pancreas Promotes a Pancreatitis-Like Inflammatory Microenvironment That Facilitates Tumor Development. Mol. Cell Biol. 2018, 38, e00427-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Srikantan, S.; Yang, X.; Lal, A.; Kim, H.H.; Kuwano, Y.; Galban, S.; Becker, K.G.; Kamara, D.; de Cabo, R.; et al. Ubiquitin-mediated proteolysis of HuR by heat shock. EMBO J. 2009, 28, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassabehji, M.; Fang, Z.M.; Hilton, E.N.; McGaughran, J.; Zhao, Z.; de Bock, C.E.; Howard, E.; Malass, M.; Donnai, D.; Diwan, A.; et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum. Mutat. 2008, 29, 1017–1027. [Google Scholar] [CrossRef]
- Lykke-Andersen, J.; Shu, M.D.; Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell 2000, 103, 1121–1131. [Google Scholar] [CrossRef] [Green Version]
- Sowa, M.E.; Bennett, E.J.; Gygi, S.P.; Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 138, 389–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jirawatnotai, S.; Hu, Y.; Michowski, W.; Elias, J.E.; Becks, L.; Bienvenu, F.; Zagozdzon, A.; Goswami, T.; Wang, Y.E.; Clark, A.B.; et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 2011, 474, 230–234. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Chimenti, M.S.; Pemble, H.; Schönichen, A.; Thompson, O.; Jacobson, M.P.; Wittmann, T. Multisite phosphorylation disrupts arginine-glutamate salt bridge networks required for binding of cytoplasmic linker-associated protein 2 (CLASP2) to end-binding protein 1 (EB1). J. Biol. Chem. 2012, 287, 17050–17064. [Google Scholar] [CrossRef] [Green Version]
- Maki, T.; Grimaldi, A.D.; Fuchigami, S.; Kaverina, I.; Hayashi, I. CLASP2 Has Two Distinct TOG Domains That Contribute Differently to Microtubule Dynamics. J. Mol. Biol. 2015, 427, 2379–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.P.; Dutra, A.; Stellrecht, C.M.; Wu, C.; Piatigorsky, J.; Saunders, G.F. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix-loop-helix transcription factor. Genomics 2002, 80, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yu, K.; Hao, Y.; Li, D.M.; Stewart, R.; Insogna, K.L.; Xu, T. LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nat. Cell Biol. 2004, 6, 609–617. [Google Scholar] [CrossRef]
- Partanen, J.; Armstrong, E.; Mäkelä, T.P.; Korhonen, J.; Sandberg, M.; Renkonen, R.; Knuutila, S.; Huebner, K.; Alitalo, K. A novel endothelial cell surface receptor tyrosine kinase with extracellular epidermal growth factor homology domains. Mol. Cell Biol. 1992, 12, 1698–1707. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Hu, M.; Wang, C.; Feng, X.; Zhao, Z.; Yang, Y.; Sahoo, N.; Gu, M.; Xiao, S.; Sah, R.; et al. LRRC8 family proteins within lysosomes regulate cellular osmoregulation and enhance cell survival to multiple physiological stresses. Proc. Natl. Acad. Sci. USA 2020, 117, 29155–29165. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, H.; Sato, N.; Yamada, M.; Mahony, D.; Seghezzi, W.; Lees, E.; Arai, K.; Masai, H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol. Cell Biol. 1999, 19, 5083–5095. [Google Scholar] [CrossRef] [Green Version]
- Tenca, P.; Brotherton, D.; Montagnoli, A.; Rainoldi, S.; Albanese, C.; Santocanale, C. Cdc7 is an active kinase in human cancer cells undergoing replication stress. J. Biol. Chem. 2007, 282, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Otterlei, M.; Haug, T.; Nagelhus, T.A.; Slupphaug, G.; Lindmo, T.; Krokan, H.E. Nuclear and mitochondrial splice forms of human uracil-DNA glycosylase contain a complex nuclear localisation signal and a strong classical mitochondrial localisation signal, respectively. Nucleic Acids Res. 1998, 26, 4611–4617. [Google Scholar] [CrossRef] [Green Version]
- Conaway, J.W.; Florens, L.; Sato, S.; Tomomori-Sato, C.; Parmely, T.J.; Yao, T.; Swanson, S.K.; Banks, C.A.; Washburn, M.P.; Conaway, R.C. The mammalian Mediator complex. FEBS Lett. 2005, 579, 904–908. [Google Scholar] [CrossRef] [Green Version]
- Devroe, E.; Erdjument-Bromage, H.; Tempst, P.; Silver, P.A. Human Mob proteins regulate the NDR1 and NDR2 serine-threonine kinases. J. Biol. Chem. 2004, 279, 24444–24451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, M. Identification and characterization of human TIPARP gene within the CCNL amplicon at human chromosome 3q25.31. Int. J. Oncol. 2003, 23, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Hill, D.A.; Ivanovich, J.; Priest, J.R.; Gurnett, C.A.; Dehner, L.P.; Desruisseau, D.; Jarzembowski, J.A.; Wikenheiser-Brokamp, K.A.; Suarez, B.K.; Whelan, A.J.; et al. DICER1 mutations in familial pleuropulmonary blastoma. Science 2009, 325, 965. [Google Scholar] [CrossRef] [Green Version]
- Türeci, O.; Sahin, U.; Koslowski, M.; Buss, B.; Bell, C.; Ballweber, P.; Zwick, C.; Eberle, T.; Zuber, M.; Villena-Heinsen, C.; et al. A novel tumour associated leucine zipper protein targeting to sites of gene transcription and splicing. Oncogene 2002, 21, 3879–3888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velayos-Baeza, A.; Vettori, A.; Copley, R.R.; Dobson-Stone, C.; Monaco, A.P. Analysis of the human VPS13 gene family. Genomics 2004, 84, 536–549. [Google Scholar] [CrossRef] [PubMed]
- Usener, D.; Schadendorf, D.; Koch, J.; Dübel, S.; Eichmüller, S. cTAGE: A cutaneous T cell lymphoma associated antigen family with tumor-specific splicing. J. Investig. Dermatol. 2003, 121, 198–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, A.; Klein, E.; Neirinckx, V.; Knudsen, A.M.; Fabian, C.; Hau, A.C.; Dieterle, M.; Oudin, A.; Nazarov, P.V.; Golebiewska, A.; et al. AN1-type zinc finger protein 3 (ZFAND3) is a transcriptional regulator that drives Glioblastoma invasion. Nat. Commun. 2020, 11, 6366. [Google Scholar] [CrossRef]
- Tauchi, H.; Matsuura, S.; Isomura, M.; Kinjo, T.; Nakamura, A.; Sakamoto, S.; Kondo, N.; Endo, S.; Komatsu, K.; Nakamura, Y. Sequence analysis of an 800-kb genomic DNA region on chromosome 8q21 that contains the Nijmegen breakage syndrome gene, NBS1. Genomics 1999, 55, 242–247. [Google Scholar] [CrossRef] [PubMed]

| ARTD | Activity | Catalytic Triad Sequence | ADP-Ribosylated Macromolecules | Key Functions | References |
|---|---|---|---|---|---|
| * PARP1 | Poly | H-Y-E | Proteins, DNA | DNA damage repair, | [56] |
| Chromatin regulation, | [57] | ||||
| Gene expression, | [58] | ||||
| Immune response | [59] | ||||
| PARP2 | Poly | H-Y-E | Proteins, DNA | DNA damage repair | [60] |
| PARP3 | Mono | H-Y-E | Proteins, DNA | DNA damage repair, | [61] |
| mitotic progression | [62] | ||||
| PARP4 | Mono | H-Y-E | Proteins | Vault particle | [63] |
| TNKS1 | Poly/Oligo | H-Y-E | Proteins | Cell division, | [64] |
| Genome integrity, | [65] | ||||
| Immune response, Protein turnover | [66] | ||||
| TNKS2 | Poly/oligo | H-Y-E | Proteins | Cell division, Genome integrity | [67] |
| PARP6 | Mono | H-Y-I | Proteins | Dendrite | [68] |
| PARP7 | Mono | H-Y-I | Proteins | Immune response | [69] |
| PARP8 | Mono | H-Y-I | Proteins | Cell Viability | [70] |
| PARP9 | Inactive/Mono | Q-Y-T | Proteins | Immune response | [71] |
| PARP10 | Mono | H-Y-I | Proteins, RNA | DNA damage repair, | [72] |
| Immune response | [73] | ||||
| PARP11 | Mono | H-Y-I | Proteins, RNA | Nuclear pore function, | [74] |
| Immune response | [75] | ||||
| PARP12 | Mono | H-Y-I | Proteins | Stress granule function | [76,77] |
| Immune Response | [69] | ||||
| Intracellular trafficking | [78] | ||||
| PARP13 | Inactive | Y-Y-T | ND | Immune response | [79,80] |
| PARP14 | Mono | H-Y-L | Proteins | Stress granule function, | [76] |
| cytoskeleton regulation | [70] | ||||
| Immune response | [81] | ||||
| Post-transcriptional gene regulation | [82] | ||||
| PARP15 | Mono | H-Y-L | Proteins, RNA | Stress granule function | [76] |
| PARP16 | Mono | H-Y-Y | Proteins | Unfolded protein response | [83] |
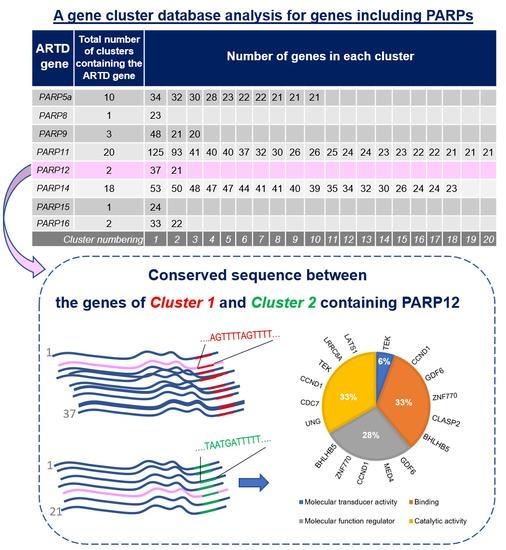
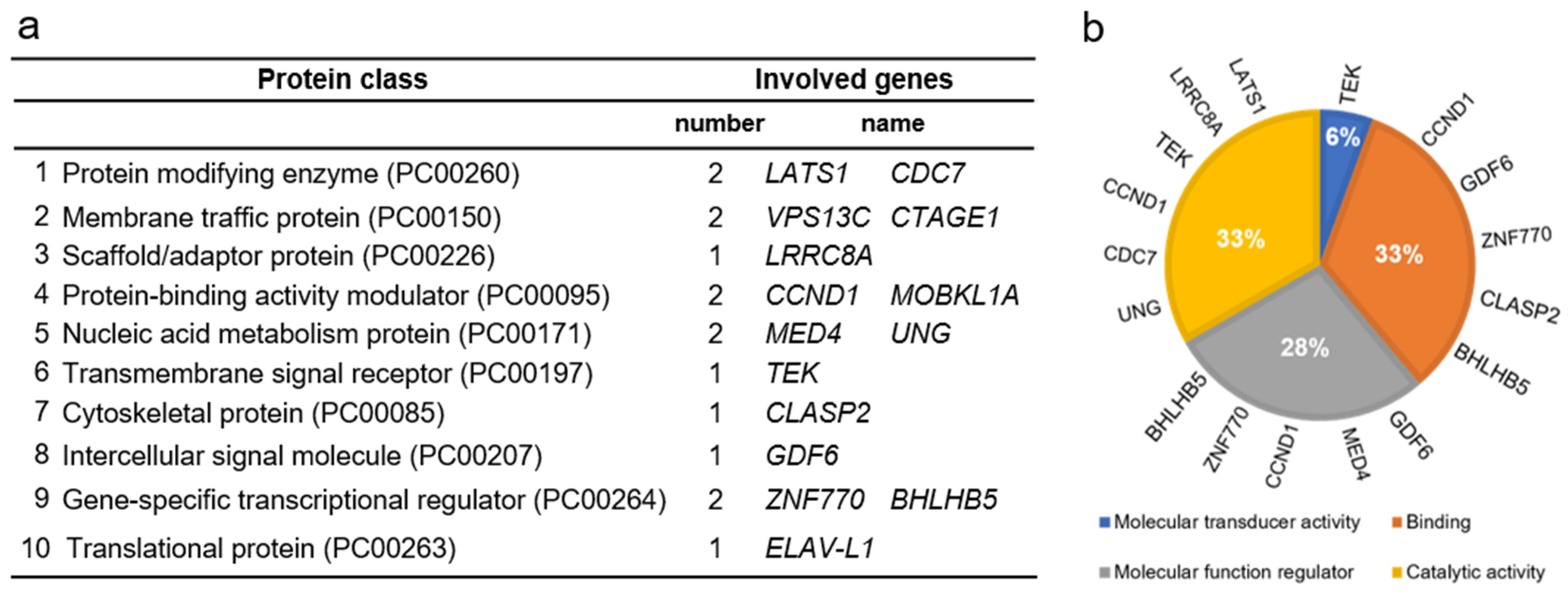
| ARTD Gene | Total Number of Clusters Containing the ARTD Gene | Number of Genes in Each Cluster | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PARP5a | 10 | 34 | 32 | 30 | 28 | 23 | 22 | 22 | 21 | 21 | 21 | ||||||||||
| PARP8 | 1 | 23 | |||||||||||||||||||
| PARP9 | 3 | 48 | 21 | 20 | |||||||||||||||||
| PARP11 | 20 | 125 | 93 | 41 | 40 | 40 | 37 | 32 | 30 | 26 | 26 | 25 | 24 | 24 | 23 | 23 | 22 | 22 | 21 | 21 | 21 |
| PARP12 | 2 | 37 | 21 | ||||||||||||||||||
| PARP14 | 18 | 53 | 50 | 48 | 47 | 47 | 44 | 41 | 41 | 40 | 39 | 35 | 34 | 32 | 30 | 26 | 24 | 24 | 23 | ||
| PARP15 | 1 | 24 | |||||||||||||||||||
| PARP16 | 2 | 33 | 22 | ||||||||||||||||||
| Cluster numbering | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manco, G.; Lacerra, G.; Porzio, E.; Catara, G. ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives. Biomolecules 2022, 12, 443. https://doi.org/10.3390/biom12030443
Manco G, Lacerra G, Porzio E, Catara G. ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives. Biomolecules. 2022; 12(3):443. https://doi.org/10.3390/biom12030443
Chicago/Turabian StyleManco, Giuseppe, Giuseppina Lacerra, Elena Porzio, and Giuliana Catara. 2022. "ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives" Biomolecules 12, no. 3: 443. https://doi.org/10.3390/biom12030443
APA StyleManco, G., Lacerra, G., Porzio, E., & Catara, G. (2022). ADP-Ribosylation Post-Translational Modification: An Overview with a Focus on RNA Biology and New Pharmacological Perspectives. Biomolecules, 12(3), 443. https://doi.org/10.3390/biom12030443