The Exoproteome of Staphylococcus pasteuri Isolated from Cervical Mucus during the Estrus Phase in Water Buffalo (Bubalus bubalis)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Protein Isolation
2.2. Protein Precipitation and Estimation
2.3. SDS-PAGE Analysis
2.4. Trypsin Digestion
2.5. Liquid Chromatography-Tandem Mass Spectrometry
2.6. Comparative Gene Ontology
2.7. Protein Selection for Modelling
2.8. Physicochemical Properties and Secondary Structure Prediction of Lpp
2.9. Multiple Sequence Alignment and Phylogenetic Tree Construction
2.10. Homology Modelling, Structure Validation and Binding Site Analysis
2.11. Molecular Docking Studies
2.12. Protein Optimization and Grid Construction
2.13. Ligand Collection and Optimization
3. Results
3.1. Total Exoproteome of S. pasteuri
3.2. Mass Spectrometry Analysis
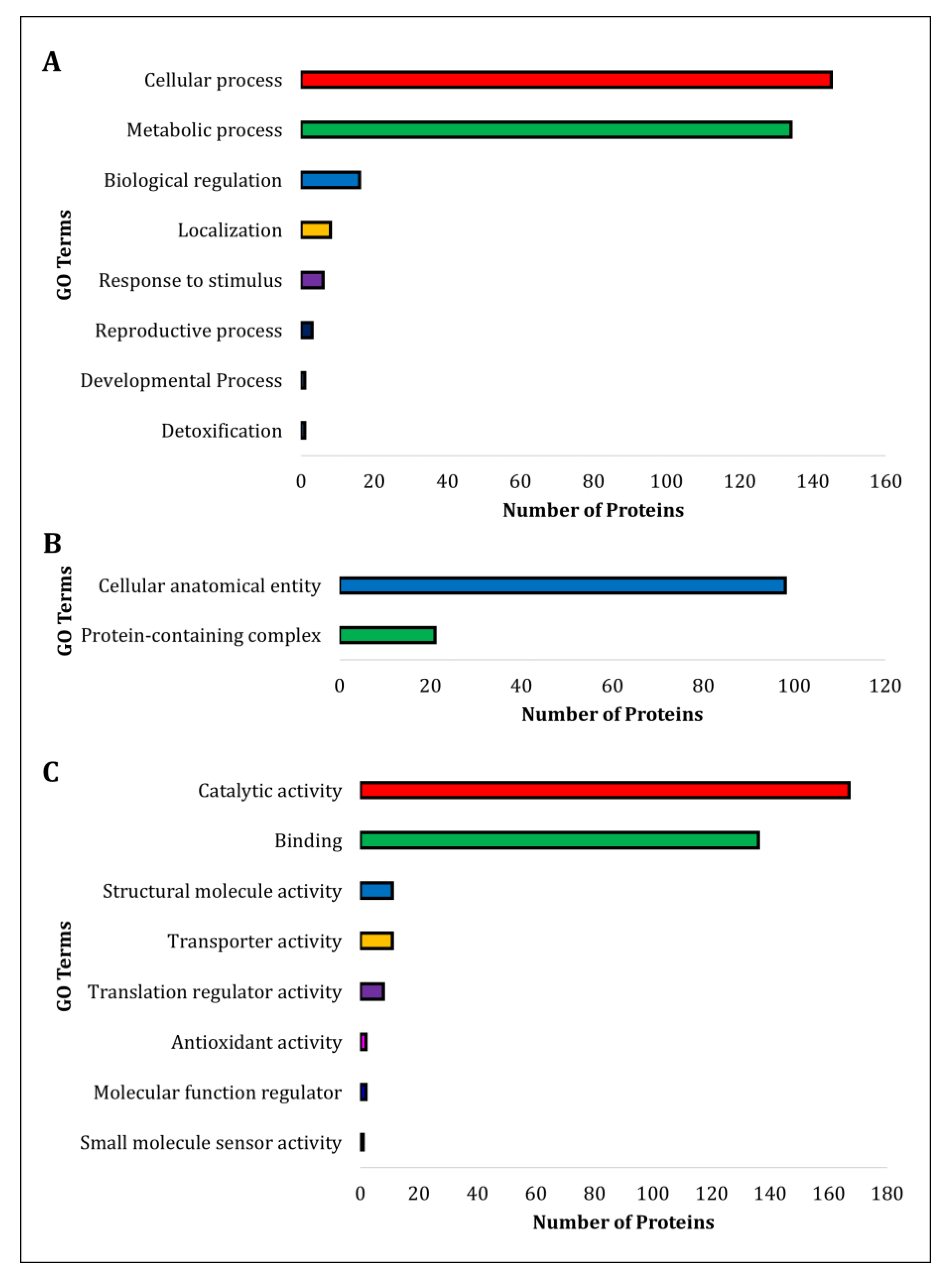
3.3. Comparative Gene Ontology
3.4. Properties of Lipoprotein
3.5. Secondary Structure Prediction
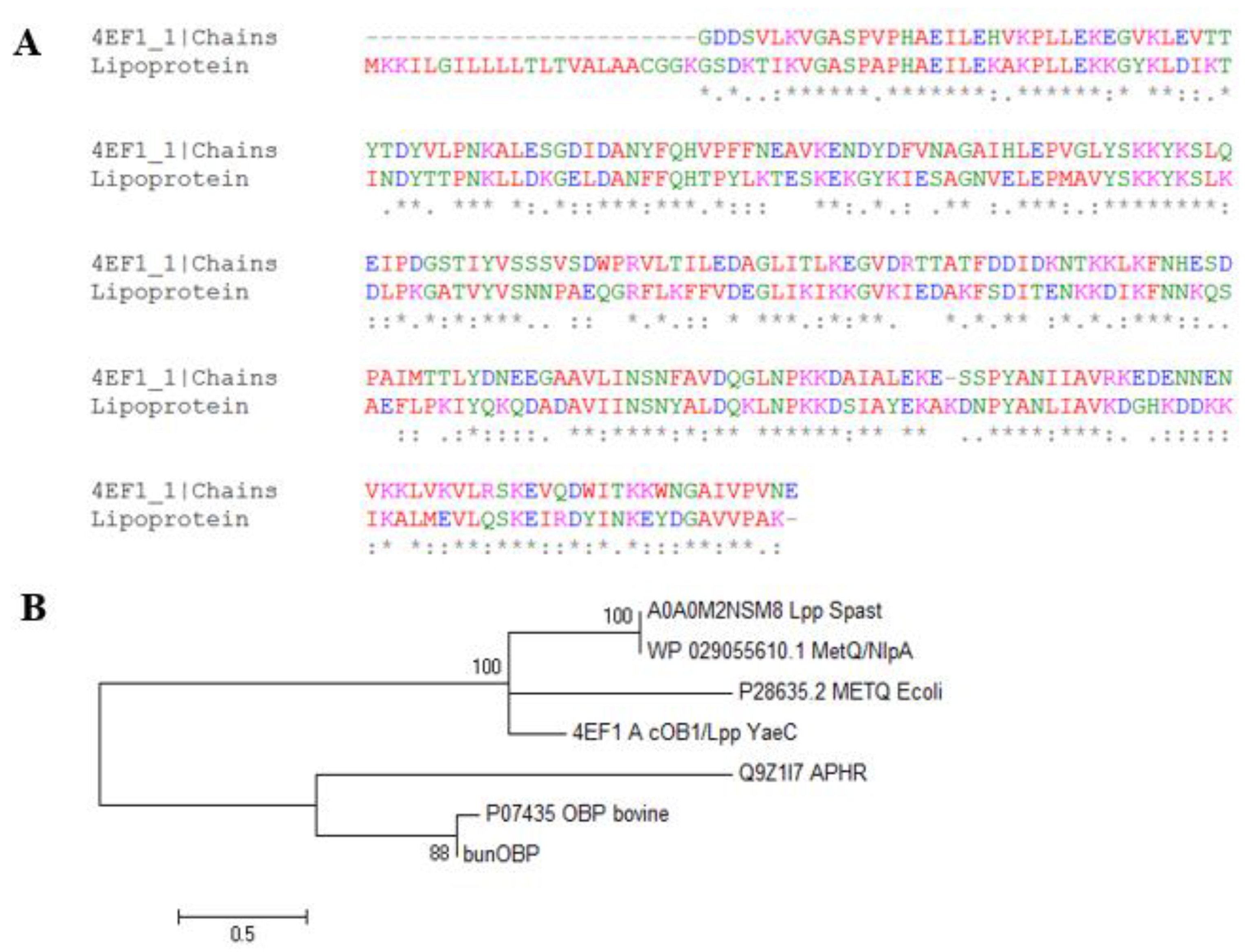
3.6. Multiple Sequence Alignment
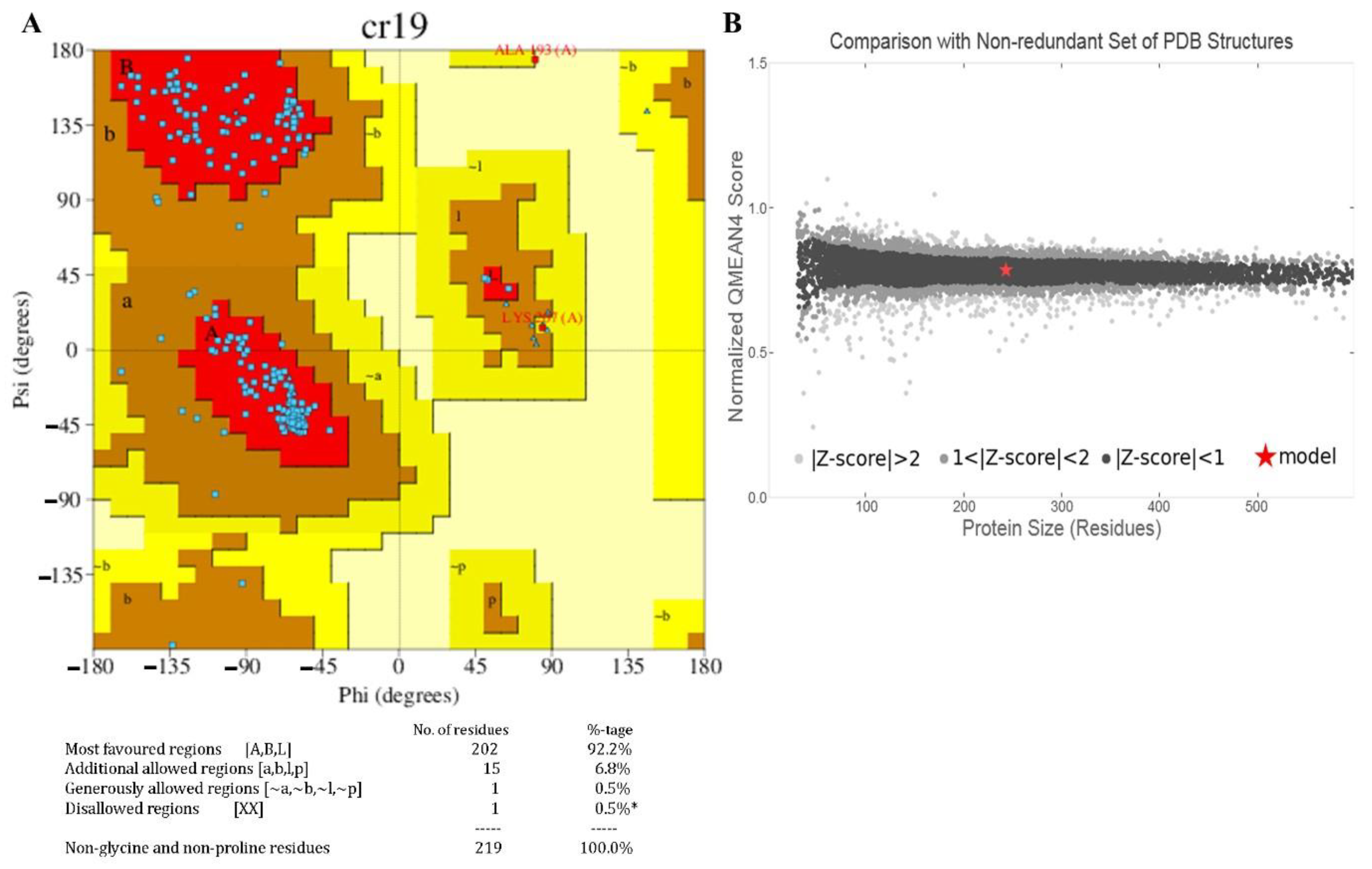
3.7. Homology Modelling and Validation
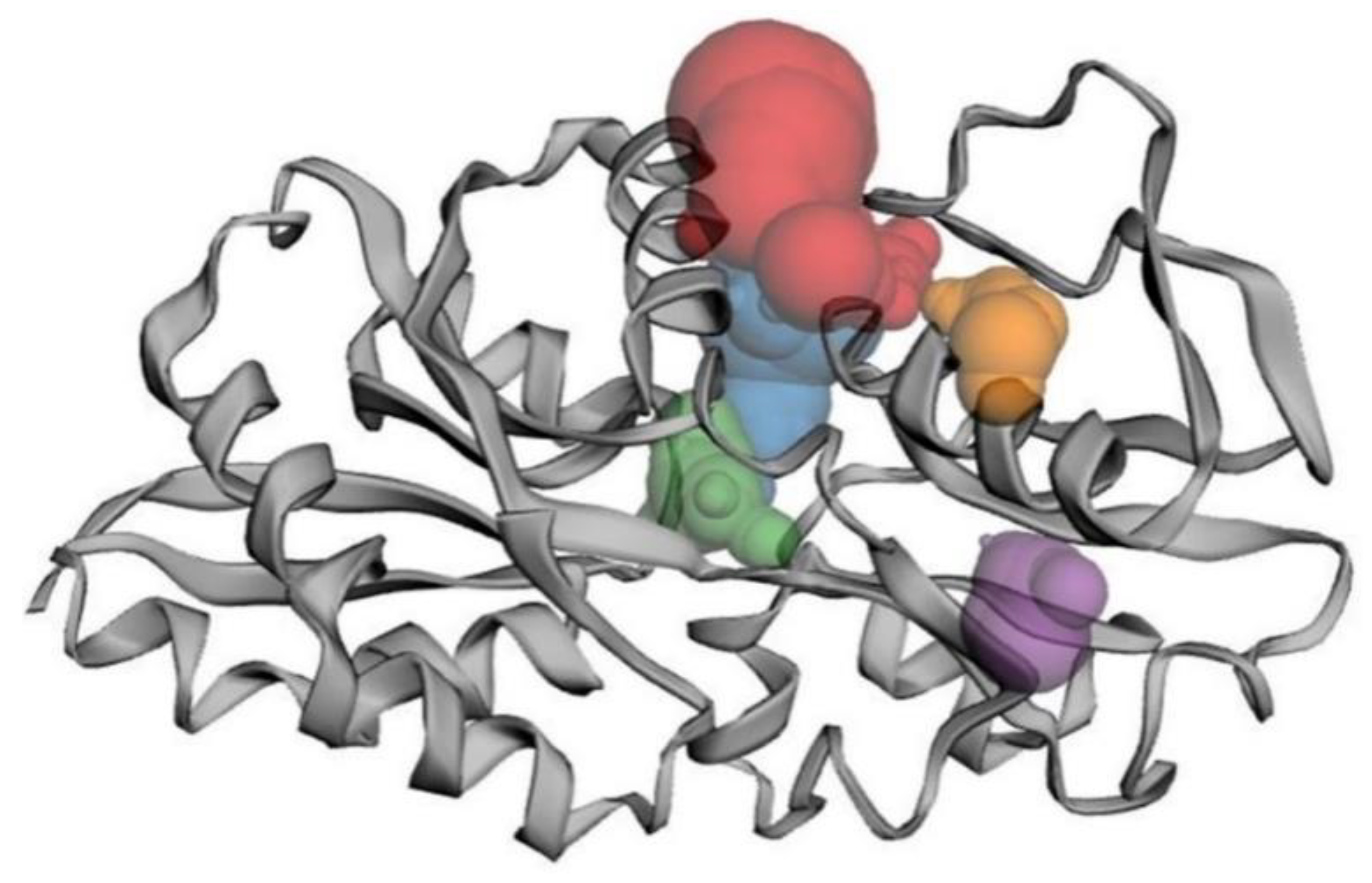
3.8. Binding Site Analysis
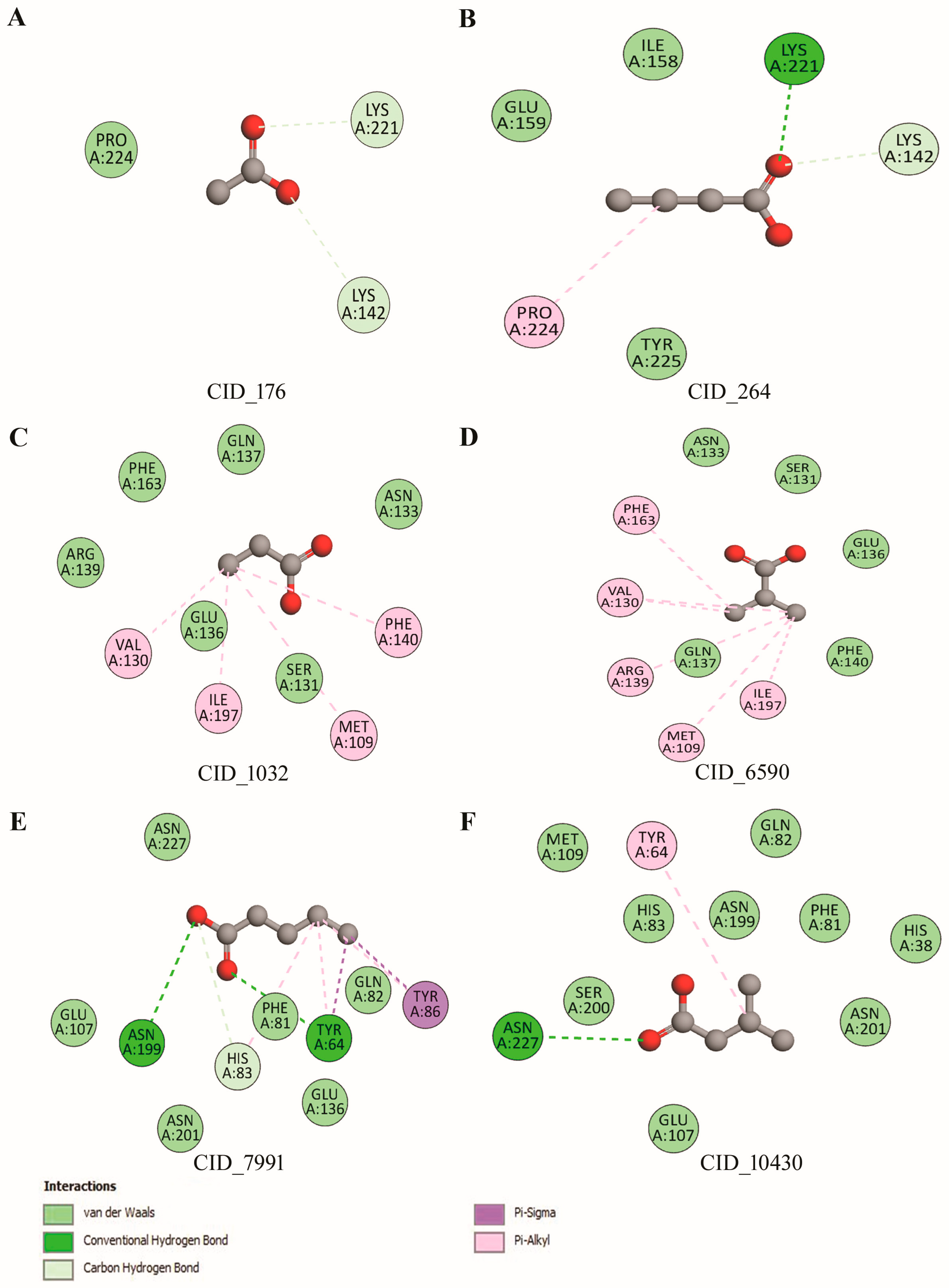
3.9. Molecular Interaction of Lpp
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tjalsma, H.; Antelmann, H.; Jongbloed, J.D.H.; Braun, P.G.; Darmon, E.; Dorenbos, R.; Dubois, J.-Y.F.; Westers, H.; Zanen, G.; Quax, W.J.; et al. Proteomics of protein secretion by Bacillus subtilis: Separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 2004, 68, 207–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, E.R.; Mecsas, J. Bacterial Secretion Systems: An Overview. Microbiol. Spectr. 2016, 4, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallin, E.; Heijne, G.V. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998, 7, 1029–1038. [Google Scholar] [CrossRef]
- Kudva, R.; Denks, K.; Kuhn, P.; Vogt, A.; Müller, M.; Koch, H.-G. Protein translocation across the inner membrane of Gram-negative bacteria: The Sec and Tat dependent protein transport pathways. Res. Microbiol. 2013, 164, 505–534. [Google Scholar] [CrossRef] [PubMed]
- Zaneveld, J.; Turnbaugh, P.J.; Lozupone, C.; Ley, R.E.; Hamady, M.; Gordon, J.I.; Knight, R. Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 2008, 12, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vindigni, S.M.; Zisman, T.L.; Suskind, D.L.; Damman, C.J. The intestinal microbiome, barrier function, and immune system in inflammatory bowel disease: A tripartite pathophysiological circuit with implications for new therapeutic directions. Therap. Adv. Gastroenterol. 2016, 9, 606–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quereda, J.J.; Barba, M.; Moce, M.L.; Gomis, J.; Jimenez-Trigos, E.; Garcia-Munoz, A.; Gomez-Martin, A.; Gonzalez-Torres, P.; Carbonetto, B.; Garcia-Rosello, E. Vaginal microbiota changes during estrous cycle in dairy heifers. Front. Vet. Sci. 2020, 7, 371. [Google Scholar] [CrossRef]
- Srinivasan, M.; Dhanasekaran, D.; Archunan, G. Vaginal microbiome analysis of buffalo (Bubalus bubalis) during estrous cycle using high-throughput amplicon sequence of 16S rRNA gene. Symbiosis 2019, 78, 97–106. [Google Scholar] [CrossRef]
- Srinivasan, M.; Rengarajan, R.L.; Dhanasekaran, D.; Akbarsha, M.A.; Archunan, G. Staphylococcus pasteuri (BCVME2) Resident in buffalo cervical vaginal mucus: Apotential source of estrus-specific sex pheromone(s). J. Chem. Ecol. 2021, 48, 7–15. [Google Scholar] [CrossRef]
- Archunan, G. Reproductive enhancement in buffalo: Looking at urinary pheromones and hormones. Iran. J. Vet. Res. 2020, 21, 163–171. [Google Scholar] [PubMed]
- Archunan, G.; Rajanarayanan, S.; Karthikeyan, K. 16 Cattle pheromones. In Neurobiology of Chemical Communication, 1st ed.; Mucignat-Caretta, C., Ed.; CRC Press: New York, NY, USA, 2014; pp. 461–479. [Google Scholar]
- Tsiligianni, T.; Amiridis, G.S.; Dovolou, E.; Menegatos, I.; Chadio, S.; Rizos, D.; Gutierrez-Adan, A. Association between physical properties of cervical mucus and ovulation rate in superovulated cows. Can. J. Vet. Res. 2011, 75, 248–253. [Google Scholar]
- Nguyen, M.-T.; Matsuo, M.; Niemann, S.; Herrmann, M.; Götz, F. Lipoproteins in gram-positive bacteria: Abundance, function, fitness. Front. Microbiol. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Campanacci, V.; Bishop, R.E.; Blangy, S.; Tegoni, M.; Cambillau, C. The membrane bound bacterial lipocalin Blc is a functional dimer with binding preference for lysophospholipids. FEBS Lett. 2006, 580, 4877–4883. [Google Scholar] [CrossRef] [Green Version]
- Flower, D.R.; North, A.C.T.; Sansom, C.E. The lipocalin protein family: Structural and sequence overview. Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzymol. 2000, 1482, 9–24. [Google Scholar] [CrossRef]
- Bishop, R.E.; Penfold, S.S.; Frost, L.S.; Höltje, J.-V.; Weiner, J.H. Stationary phase expression of a novel Escherichia coli outer membrane lipoprotein and its relationship with mammalian apolipoprotein D. J. Biol. Chem. 1995, 270, 23097–23103. [Google Scholar] [CrossRef] [Green Version]
- Manikkaraja, C.; Bhavika, M.; Singh, R.; Nagarathnam, B.; George, G.; Gulyani, A.; Archunan, G.; Sowdhamini, R. Molecular and functional characterization of buffalo nasal epithelial odorant binding proteins and their structural insights by in silico and biochemical approaches. J. Biomol. Struct. Dyn. 2020, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Muthukumar, S.; Rajesh, D.; Selvam, R.M.; Saibaba, G.; Suvaithenamudhan, S.; Akbarsha, M.A.; Padmanabhan, P.; Gulyas, B.; Archunan, G. Buffalo nasal odorant-binding protein (bunOBP) and its structural evaluation with putative pheromones. Sci. Rep. 2018, 8, 9323. [Google Scholar] [CrossRef]
- Cassoli, L.D.; Lima, W.J.; Esguerra, J.C.; Da Silva, J.; Machado, P.F.; Mourao, G.B. Do different standard plate counting (IDF/ISSO or AOAC) methods interfere in the conversion of individual bacteria counts to colony forming units in raw milk. J. Appl. Microbiol. 2016, 121, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Flaugnatti, N.; Journet, L. Identification of effectors: Precipitation of supernatant material. In Bacterial Protein Secretion Systems: Methods and Protocols; Journet, L., Cascales, E., Eds.; Springer: New York, NY, USA, 2017; pp. 459–464. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the asembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Patterson, R.L.; Fruzangohar, M.; Ebrahimie, E.; Ogunniyi, A.D.; Mahdi, L.K.; Paton, J.C.; Adelson, D.L. Comparative GO: A web application for comparative gene ontology and gene ontology-based gene selection in bacteria. PLoS ONE 2013, 8, e58759. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Bikadi, Z.; Hazai, E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminform. 2009, 1, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Solis, F.J.; Wets, R.J.B. Minimization by random search techniques. Math. Oper. Res. 1981, 6, 19–30. [Google Scholar] [CrossRef]
- Jeffery, C. Intracellular proteins moonlighting as bacterial adhesion factors. AIMS Microbiol. 2018, 4, 362–376. [Google Scholar] [CrossRef]
- Muthukumar, S.; Rajkumar, R.; Karthikeyan, K.; Liao, C.C.; Singh, D.; Akbarsha, M.A.; Archunan, G. Buffalo cervico-vaginal fluid proteomics with special reference to estrous cycle: Heat shock protein (HSP)-70 appears to be an estrus indicator. Biol. Reprod. 2014, 90, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maguire, J.A.; Jenkins, S.; Burnell, G.M. The effects of repeated dredging and speed of tow on undersized scallops. Fish. Res. 2002, 58, 367–377. [Google Scholar] [CrossRef]
- Frisk, A.; Ison, C.A.; Lagergård, T. GroEL heat shock protein of Haemophilus ducreyi: Association with cell surface and capacity to bind to eukaryotic cells. Infect. Immun. 1998, 66, 1252–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massengo-Tiassé, R.P.; Cronan, J.E. Diversity in enoyl-acyl carrier protein reductases. Cell. Mol. Life Sci. 2009, 66, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankar, R.; Archunan, G. Gas chromatographic/mass spectrometric analysis of volatile metabolites in bovine vaginal fluid and assessment of their bioactivity. Int. J. Anal. Chem. 2011, 2011, 256106. [Google Scholar] [CrossRef] [Green Version]
- Karthikeyan, K.; Archunan, G. Gas chromatographic mass spectrometric analysis of estrus specific volatile compounds in buffalo vaginal mucus after initial sexual foreplay. J. Buffalo Sci. 2013, 2, 1–7. [Google Scholar] [CrossRef]
- Saibaba, G.; Rajesh, D.; Muthukumar, S.; Sathiyanarayanan, G.; Padmanabhan, P.; Akbarsha, M.A.; Gulyás, B.; Archunan, G. Proteomic analysis of human saliva: An approach to find the marker protein for ovulation. Reprod. Biol. 2016, 16, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Achiraman, S.; Archunan, G. Characterization of urinary volatiles in Swiss male mice (Mus musculus): Bioassay of identified compounds. J. Biosci. 2002, 27, 679–686. [Google Scholar] [CrossRef]
- Vanmuylder, N.; Werry-Huet, A.; Rooze, M.; Louryan, S. Heat shock protein HSP86 expression during mouse embryo development, especially in the germ-line. Anat. Embryol. 2002, 205, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Muthukumar, S.; Rajkumar, R.; Rajesh, D.; Saibaba, G.; Liao, C.C.; Archunan, G.; Padmanabhan, P.; Gulyas, B. Exploration of salivary proteins in buffalo: An approach to find marker proteins for estrus. FASEB J. 2014, 28, 4700–4709. [Google Scholar] [CrossRef]
- Tokuda, H.; Matsuyama, S. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 2004, 1694, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, H.; Kurokawa, K.; Lee, B.L. Lipoproteins in bacteria: Structures and biosynthetic pathways. FEBS J. 2012, 279, 4247–4268. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, A.; Matsuyama, S.-I.; Tokuda, H. Mutant of LolA, a lipoprotein-specific molecular chaperone of Escherichia coli, defective in the transfer of lipoproteins to LolB. Biochem. Biophys. Res. Commun. 2001, 287, 1125–1128. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, D.; Muthukumar, S.; Saibaba, G.; Siva, D.; Akbarsha, M.A.; Gulyas, B.; Padmanabhan, P.; Archunan, G. Structural elucidation of estrus urinary lipocalin protein (EULP) and evaluating binding affinity with pheromones using molecular docking and fluorescence study. Sci. Rep. 2016, 6, 35900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthukumar, S.; Rajesh, D.; Saibaba, G.; Alagesan, A.; Rengarajan, R.L.; Archunan, G. Urinary lipocalin protein in a female rodent with correlation to phases in the estrous cycle: An experimental study accompanied by in silico analysis. PLoS ONE 2013, 8, e71357. [Google Scholar] [CrossRef]
- Briand, L.; Huet, J.; Perez, V.; Lenoir, G.; Nespoulous, C.; Boucher, Y.; Trotier, D.; Pernollet, J.C. Odorant and pheromone binding by aphrodisin, a hamster aphrodisiac protein. FEBS Lett. 2000, 476, 179–185. [Google Scholar] [CrossRef] [Green Version]
- Briand, L.; Trotier, D.; Pernollet, J.C. Aphrodisin, an aphrodisiac lipocalin secreted in hamster vaginal secretions. Peptides 2004, 25, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Cerna, M.; Kuntova, B.; Talacko, P.; Stopkova, R.; Stopka, P. Differential regulation of vaginal lipocalins (OBP, MUP) during the estrous cycle of the house mouse. Sci. Rep. 2017, 7, 11674. [Google Scholar] [CrossRef] [Green Version]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef]
- Flo, T.H.; Smith, K.D.; Sato, S.; Rodriguez, D.J.; Holmes, M.A.; Strong, R.K.; Akira, S.; Aderem, A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [Google Scholar] [CrossRef] [PubMed]


| UniProt ID | Protein Names | Function a | #Peptides | #AAs Length | MW [kDa] | Calc. pI |
|---|---|---|---|---|---|---|
| A0A0M2NQR9 | Elongation factor Tu (EF-Tu) | Translation elongation factor activity | 22 | 394 | 43.2 | 4.87 |
| A0A431ZL22 | 60 kDa chaperonin (Cpn60) | ATP binding | 14 | 541 | 57.5 | 4.67 |
| A0A269XJ15 | Enolase | Magnesium ion binding | 13 | 434 | 47.2 | 4.65 |
| A0A269XHD8 | Fructose-bisphosphate aldolase class 1 | Fructose-bisphosphate aldolase activity | 12 | 296 | 33 | 4.91 |
| A0A0M2NSM8 | Lipoprotein (Lpp) | Lipid binding protein | 2 | 269 | 30.1 | 9.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srinivasan, M.; Muthukumar, S.; Rajesh, D.; Kumar, V.; Rajakumar, R.; Akbarsha, M.A.; Gulyás, B.; Padmanabhan, P.; Archunan, G. The Exoproteome of Staphylococcus pasteuri Isolated from Cervical Mucus during the Estrus Phase in Water Buffalo (Bubalus bubalis). Biomolecules 2022, 12, 450. https://doi.org/10.3390/biom12030450
Srinivasan M, Muthukumar S, Rajesh D, Kumar V, Rajakumar R, Akbarsha MA, Gulyás B, Padmanabhan P, Archunan G. The Exoproteome of Staphylococcus pasteuri Isolated from Cervical Mucus during the Estrus Phase in Water Buffalo (Bubalus bubalis). Biomolecules. 2022; 12(3):450. https://doi.org/10.3390/biom12030450
Chicago/Turabian StyleSrinivasan, Mahalingam, Subramanian Muthukumar, Durairaj Rajesh, Vinod Kumar, Rajamanickam Rajakumar, Mohammad Abdulkader Akbarsha, Balázs Gulyás, Parasuraman Padmanabhan, and Govindaraju Archunan. 2022. "The Exoproteome of Staphylococcus pasteuri Isolated from Cervical Mucus during the Estrus Phase in Water Buffalo (Bubalus bubalis)" Biomolecules 12, no. 3: 450. https://doi.org/10.3390/biom12030450







