Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity
Abstract
1. Introduction
2. Typical Workflow of Single-Particle Cryo-EM
3. Sources of the Cryo-EM Image Data Heterogeneity
4. Conventional Approaches to Study Cryo-EM Data Heterogeneity
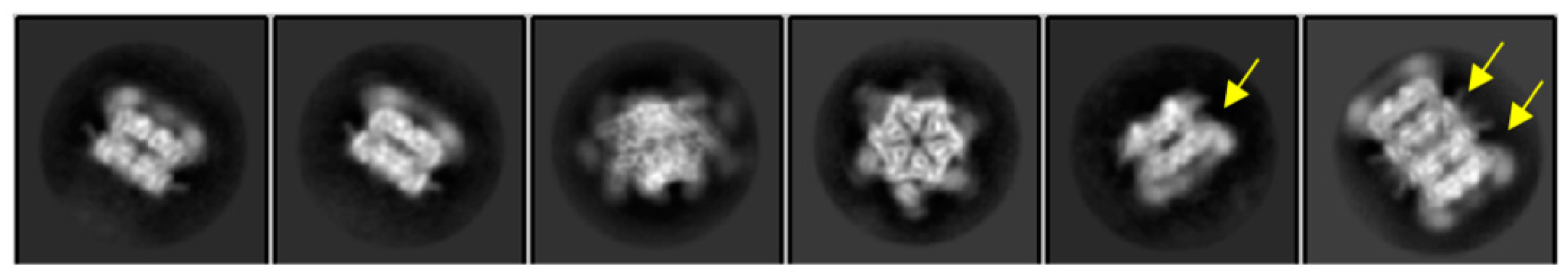
4.1. Conventional Multivariate Statistical Analysis (MSA)
4.2. Regularized Likelihood Approach
5. Continuous Structural Heterogeneity Derived from Cryo-EM Data
5.1. Covariance Matrix Estimation
5.2. Hyper-Molecules
5.3. 3DVA (3D Variability Analysis) Approach
5.4. CryoDRGN
6. Mapping Energy Landscape from Cryo-EM Data
7. Hybrid Approaches with Molecular Dynamic Simulations
7.1. Detecting Structural Variability Based on the Resolution Anisotropy
7.2. Molecular Dynamics Flexible Fitting (MDFF)
8. Time-Resolved Cryo-EM Studies
9. Interpretation of the Extracted Information for Biomolecular Perturbation
10. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Callaway, E. Revolutionary Cryo-EM Is Taking over Structural Biology. Nature 2020, 578, 201. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.-C.; McMullan, G.; Scheres, S.H.W. How Cryo-EM Is Revolutionizing Structural Biology. Trends Biochem. Sci. 2015, 40, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, N.; Turchetta, R.; Van Hoften, G.; Henderson, R.; McMullan, G.; Faruqi, A.R. A High Frame Rate, 16 Million Pixels, Radiation Hard CMOS Sensor. J. Instrum. 2011, 6, C03003. [Google Scholar] [CrossRef]
- Mooney, P.; Contarato, D.; Denes, P.; Gubbens, A.; Lee, B.; Lent, M.; Agard, D. A High-Speed Electron-Counting Direct Detection Camera for TEM. Microsc. Microanal. 2011, 17, 1004–1005. [Google Scholar] [CrossRef]
- Milazzo, A.-C.; Moldovan, G.; Lanman, J.; Jin, L.; Bouwer, J.C.; Klienfelder, S.; Peltier, S.T.; Ellisman, M.H.; Kirkland, A.I.; Xuong, N.-H. Characterization of a Direct Detection Device Imaging Camera for Transmission Electron Microscopy. Ultramicroscopy 2010, 110, 744–747. [Google Scholar] [CrossRef] [PubMed]
- Frank, J. Three-Dimensional Electron. Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in Their Native State; Oxford University Press: New York, NY, USA, 2006; ISBN 9780198034384. [Google Scholar]
- Van Heel, M. Angular Reconstitution: A Posteriori Assignment of Projection Directions for 3D Reconstruction. Ultramicroscopy 1987, 21, 111–123. [Google Scholar] [CrossRef]
- Glaeseral, R.M. Radiation Damage and High Resolution Biological Electron Microscopy. Proc. Annu. Meet. Electron. Microsc. Soc. Am. 1973, 31, 226–227. [Google Scholar] [CrossRef]
- Isaacson, M.; Johnson, D.; Crewe, A.V. Electron Beam Excitation and Damage of Biological Molecules; Its Implications for Specimen Damage in Electron Microscopy. Radiat. Res. 1973, 55, 205–224. [Google Scholar] [CrossRef]
- Glaeser, R.M.; Taylor, K.A. Radiation Damage Relative to Transmission Electron Microscopy of Biological Specimens at Low Temperature: A Review. J. Microsc. 1978, 112, 127–138. [Google Scholar] [CrossRef]
- Hayward, S.B.; Glaeser, R.M. Radiation Damage of Purple Membrane at Low Temperature. Ultramicroscopy 1979, 4, 201–210. [Google Scholar] [CrossRef]
- Cheng, A.; Eng, E.T.; Alink, L.; Rice, W.J.; Jordan, K.D.; Kim, L.Y.; Potter, C.S.; Carragher, B. High Resolution Single Particle Cryo-Electron Microscopy Using Beam-Image Shift. J. Struct. Biol. 2018, 204, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Nakane, T.; Kimanius, D.; Lindahl, E.; Scheres, S.H. Characterisation of Molecular Motions in Cryo-EM Single-Particle Data by Multi-Body Refinement in RELION. Elife 2018, 7, e36861. [Google Scholar] [CrossRef] [PubMed]
- Mitra, K.; Frank, J. Ribosome Dynamics: Insights from Atomic Structure Modeling into Cryo-Electron Microscopy Maps. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Tama, F.; Valle, M.; Frank, J.; Brooks, C.L., 3rd. Dynamic Reorganization of the Functionally Active Ribosome Explored by Normal Mode Analysis and Cryo-Electron Microscopy. Proc. Natl. Acad. Sci. USA 2003, 100, 9319–9323. [Google Scholar] [CrossRef] [PubMed]
- Zhong, E.D.; Bepler, T.; Berger, B.; Davis, J.H. CryoDRGN: Reconstruction of Heterogeneous Cryo-EM Structures Using Neural Networks. Nat. Methods 2021, 18, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Fica, S.M.; Nagai, K. Cryo-Electron Microscopy Snapshots of the Spliceosome: Structural Insights into a Dynamic Ribonucleoprotein Machine. Nat. Struct. Mol. Biol. 2017, 24, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Haselbach, D.; Komarov, I.; Agafonov, D.E.; Hartmuth, K.; Graf, B.; Dybkov, O.; Urlaub, H.; Kastner, B.; Lührmann, R.; Stark, H. Structure and Conformational Dynamics of the Human Spliceosomal Bact Complex. Cell 2018, 172, 454–464. [Google Scholar] [CrossRef]
- Marino, J.; Schertler, G.F.X. A Set of Common Movements within GPCR-G-Protein Complexes from Variability Analysis of Cryo-EM Datasets. J. Struct. Biol. 2021, 213, 107699. [Google Scholar] [CrossRef]
- Dong, M.; Deganutti, G.; Piper, S.J.; Liang, Y.-L.; Khoshouei, M.; Belousoff, M.J.; Harikumar, K.G.; Reynolds, C.A.; Glukhova, A.; Furness, S.G.B.; et al. Structure and Dynamics of the Active Gs-Coupled Human Secretin Receptor. Nat. Commun. 2020, 11, 4137. [Google Scholar] [CrossRef]
- Josephs, T.M.; Belousoff, M.J.; Liang, Y.-L.; Piper, S.J.; Cao, J.; Garama, D.J.; Leach, K.; Gregory, K.J.; Christopoulos, A.; Hay, D.L.; et al. Structure and Dynamics of the CGRP Receptor in Apo and Peptide-Bound Forms. Science 2021, 372, eabf7258. [Google Scholar] [CrossRef]
- Hilger, D. The Role of Structural Dynamics in GPCR-Mediated Signaling. FEBS J. 2021, 288, 2461–2489. [Google Scholar] [CrossRef] [PubMed]
- Chiu, P.-L.; Kelly, D.F.; Walz, T. The Use of Trehalose in the Preparation of Specimens for Molecular Electron Microscopy. Micron 2011, 42, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Dubochet, J.; McDowall, A.W. Vitrification of Pure Water for Electron Microscopy. J. Microsc. 1981, 124, 3–4. [Google Scholar] [CrossRef]
- Adrian, M.; Dubochet, J.; Lepault, J.; McDowall, A.W. Cryo-Electron Microscopy of Viruses. Nature 1984, 308, 32–36. [Google Scholar] [CrossRef]
- Cowley, J.M. Image Contrast in a Transmission Scanning Electron Microscope. Appl. Phys. Lett. 1969, 15, 58–59. [Google Scholar] [CrossRef]
- Spence, J.C.H. High-Resolution Electron Microscopy; OUP Oxford University Press: Oxford, UK, 2013; ISBN 9780191508400. [Google Scholar]
- Erickson, H.; Klug, A. Measurement and Compensation of Defocusing and Aberrations by Fourier Processing of Electron Micrographs. Philos. Trans. R. Soc. Lond. 1971, 261, 105–118. [Google Scholar] [CrossRef]
- Wade, R.H. A Brief Look at Imaging and Contrast Transfer. Ultramicroscopy 1992, 46, 145–156. [Google Scholar] [CrossRef]
- Wu, S.; Armache, J.-P.; Cheng, Y. Single-Particle Cryo-EM Data Acquisition by Using Direct Electron Detection Camera. Microscopy 2016, 65, 35–41. [Google Scholar] [CrossRef]
- Chiu, P.-L.; Li, X.; Li, Z.; Beckett, B.; Brilot, A.F.; Grigorieff, N.; Agard, D.A.; Cheng, Y.; Walz, T. Evaluation of Super-Resolution Performance of the K2 Electron-Counting Camera Using 2D Crystals of Aquaporin-0. J. Struct. Biol. 2015, 192, 163–173. [Google Scholar] [CrossRef]
- Li, X.; Mooney, P.; Zheng, S.; Booth, C.R.; Braunfeld, M.B.; Gubbens, S.; Agard, D.A.; Cheng, Y. Electron Counting and Beam-Induced Motion Correction Enable near-Atomic-Resolution Single-Particle Cryo-EM. Nat. Methods 2013, 10, 584–590. [Google Scholar] [CrossRef] [PubMed]
- D’Imprima, E.; Kühlbrandt, W. Current Limitations to High-Resolution Structure Determination by Single-Particle CryoEM. Q. Rev. Biophys. 2021, 54, e4. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.R.; Henderson, R. Single Particle Electron Cryomicroscopy: Trends, Issues and Future Perspective. Q. Rev. Biophys. 2016, 49, e13. [Google Scholar] [CrossRef]
- Russo, C.J.; Henderson, R. Charge Accumulation in Electron Cryomicroscopy. Ultramicroscopy 2018, 187, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.J.; Henderson, R. Microscopic Charge Fluctuations Cause Minimal Contrast Loss in CryoEM. Ultramicroscopy 2018, 187, 56–63. [Google Scholar] [CrossRef]
- Glaeser, R.M.; Han, B.-G.; Csencsits, R.; Killilea, A.; Pulk, A.; Cate, J.H.D. Factors That Influence the Formation and Stability of Thin, Cryo-EM Specimens. Biophys. J. 2016, 110, 749–755. [Google Scholar] [CrossRef]
- Grant, T.; Grigorieff, N. Measuring the Optimal Exposure for Single Particle Cryo-EM Using a 2.6 Å Reconstruction of Rotavirus VP6. Elife 2015, 4, e06980. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Palovcak, E.; Armache, J.-P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic Correction of Beam-Induced Motion for Improved Cryo-Electron Microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef]
- Sigworth, F.J.; Doerschuk, P.C.; Carazo, J.-M.; Scheres, S.H.W. An Introduction to Maximum-Likelihood Methods in Cryo-EM. Methods Enzymol. 2010, 482, 263–294. [Google Scholar] [CrossRef] [PubMed]
- Sigworth, F.J. A Maximum-Likelihood Approach to Single-Particle Image Refinement. J. Struct. Biol. 1998, 122, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W. Classification of Structural Heterogeneity by Maximum-Likelihood Methods. Methods Enzymol. 2010, 482, 295–320. [Google Scholar] [CrossRef] [PubMed]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Soc. 1977, 39, 1–22. [Google Scholar] [CrossRef]
- Radermacher, M. Weighted Back-Projection Methods. In Electron Tomography; Springer: New York, NY, USA, 2008; pp. 245–273. ISBN 9780387312347. [Google Scholar]
- Nogales, E.; Scheres, S.H.W. Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol. Cell 2015, 58, 677–689. [Google Scholar] [CrossRef]
- Henderson, R. Avoiding the Pitfalls of Single Particle Cryo-Electron Microscopy: Einstein from Noise. Proc. Natl. Acad. Sci. USA 2013, 110, 18037–18041. [Google Scholar] [CrossRef]
- Scheres, S.H.W.; Chen, S. Prevention of Overfitting in Cryo-EM Structure Determination. Nat. Methods 2012, 9, 853–854. [Google Scholar] [CrossRef] [PubMed]
- Penczek, P.A. Fundamentals of Three-Dimensional Reconstruction from Projections. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2010; pp. 1–33. [Google Scholar]
- Allen, T.W.; Andersen, O.S.; Roux, B. On the Importance of Atomic Fluctuations, Protein Flexibility, and Solvent in Ion Permeation. J. Gen. Physiol. 2004, 124, 679–690. [Google Scholar] [CrossRef]
- Skjaerven, L.; Reuter, N.; Martinez, A. Dynamics, Flexibility and Ligand-Induced Conformational Changes in Biological Macromolecules: A Computational Approach. Future Med. Chem. 2011, 3, 2079–2100. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.V.; Grubmüller, H. Effects of Cryo-EM Cooling on Structural Ensembles. Nat. Commun. 2022, 13, 1709. [Google Scholar] [CrossRef]
- Swint-Kruse, L.; Brown, C.S. Resmap: Automated Representation of Macromolecular Interfaces as Two-Dimensional Networks. Bioinformatics 2005, 21, 3327–3328. [Google Scholar] [CrossRef] [PubMed]
- Aiyer, S.; Zhang, C.; Baldwin, P.R.; Lyumkis, D. Evaluating Local and Directional Resolution of Cryo-EM Density Maps. Methods Mol. Biol. 2021, 2215, 161–187. [Google Scholar] [CrossRef]
- Local Resolution of Cryo-EM Maps with MonoRes. Nat. Methods 2018, 15, 246. [CrossRef]
- Nandi, P.; Li, S.; Columbres, R.C.A.; Wang, F.; Williams, D.R.; Poh, Y.-P.; Chou, T.-F.; Chiu, P.-L. Structural and Functional Analysis of Disease-Linked P97 ATPase Mutant Complexes. Int. J. Mol. Sci. 2021, 22, 8079. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Rubinstein, J.L. Cryo-EM of ATP Synthases. Curr. Opin. Struct. Biol. 2018, 52, 71–79. [Google Scholar] [CrossRef]
- Yang, J.-H.; Williams, D.; Kandiah, E.; Fromme, P.; Chiu, P.-L. Structural Basis of Redox Modulation on Chloroplast ATP Synthase. Commun. Biol. 2020, 3, 482. [Google Scholar] [CrossRef] [PubMed]
- Hisabori, T.; Sunamura, E.-I.; Kim, Y.; Konno, H. The Chloroplast ATP Synthase Features the Characteristic Redox Regulation Machinery. Antioxid. Redox Signal. 2013, 19, 1846–1854. [Google Scholar] [CrossRef] [PubMed]
- van Heel, M.; Frank, J. Use of Multivariate Statistics in Analysing the Images of Biological Macromolecules. Ultramicroscopy 1981, 6, 187–194. [Google Scholar] [CrossRef]
- van Heel, M. Multivariate Statistical Classification of Noisy Images (Randomly Oriented Biological Macromolecules). Ultramicroscopy 1984, 13, 165–183. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Postma, E. Dimensionality Reduction: A Comparative Review. J. Mach. Learn. Res. 2009, 10, 66–71. [Google Scholar]
- Van Heel, M.F.J. Classification of Particles in Noisy Electron. Micrographs Using Correspondence Analysis. Pattern Recognition in Practice; North-Holland Publishing: Amsterdam, The Netherlands, 1980. [Google Scholar]
- Frank, J.; van Heel, M. Correspondence Analysis of Aligned Images of Biological Particles. J. Mol. Biol. 1982, 161, 134–137. [Google Scholar] [CrossRef]
- Frank, J. Differential Averaging of Single Molecule Images Using Multivariate Statistical Classification. Proc. Annu. Meet. Electron. Microsc. Soc. Am. 1982, 40, 706–709. [Google Scholar] [CrossRef]
- van Heel, M.; Portugal, R.V.; Schatz, M. Multivariate Statistical Analysis of Large Datasets: Single Particle Electron Microscopy. Open J. Stat. 2016, 06, 701–739. [Google Scholar] [CrossRef][Green Version]
- Harauz, G.; Welsh, E.A. Multivariate Statistical Analysis of Electron Micrographs of a Mammalian Transcription Initiation Complex. J. Electron. Microsc. 1992, 41, 264–266. [Google Scholar] [CrossRef]
- Likas, A.; Vlassis, N.J.; Verbeek, J. The Global K-Means Clustering Algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef]
- Yang, Z.; Fang, J.; Chittuluru, J.; Asturias, F.J.; Penczek, P.A. Iterative Stable Alignment and Clustering of 2D Transmission Electron Microscope Images. Structure 2012, 20, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Sorzano, C.O.S.; Bilbao-Castro, J.R.; Shkolnisky, Y.; Alcorlo, M.; Melero, R.; Caffarena-Fernández, G.; Li, M.; Xu, G.; Marabini, R.; Carazo, J.M. A Clustering Approach to Multireference Alignment of Single-Particle Projections in Electron Microscopy. J. Struct. Biol. 2010, 171, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Ma, Y.-B.; Congdon, C.; Brett, B.; Chen, S.; Xu, Y.; Ouyang, Q.; Mao, Y. Massively Parallel Unsupervised Single-Particle Cryo-EM Data Clustering via Statistical Manifold Learning. PLoS ONE 2017, 12, e0182130. [Google Scholar] [CrossRef]
- Chen, T.-L.; Hsieh, D.-N.; Hung, H.; Tu, I.-P.; Wu, P.-S.; Wu, Y.-M.; Chang, W.-H.; Huang, S.-Y. γ-SUP: A Clustering Algorithm for Cryo-Electron Microscopy Images of Asymmetric Particles. Ann. Appl. Stat. 2014, 8, 259–285. [Google Scholar] [CrossRef]
- Rao, R.; Moscovich, A.; Singer, A. Wasserstein K-Means for Clustering Tomographic Projections. arXiv 2020, arXiv:2010.09989. [Google Scholar] [CrossRef]
- Ludtke, S.J.; Baldwin, P.R.; Chiu, W. EMAN: Semiautomated Software for High-Resolution Single-Particle Reconstructions. J. Struct. Biol. 1999, 128, 82–97. [Google Scholar] [CrossRef]
- Frank, J.; Radermacher, M.; Penczek, P.; Zhu, J.; Li, Y.; Ladjadj, M.; Leith, A. SPIDER and WEB: Processing and Visualization of Images in 3D Electron Microscopy and Related Fields. J. Struct. Biol. 1996, 116, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, P.B.; Henderson, R. Optimal Determination of Particle Orientation, Absolute Hand, and Contrast Loss in Single-Particle Electron Cryomicroscopy. J. Mol. Biol. 2003, 333, 721–745. [Google Scholar] [CrossRef]
- Pannu, N.S.; Read, R.J. Improved Structure Refinement through Maximum Likelihood. Acta Crystallogr. A 1996, 52, 659–668. [Google Scholar] [CrossRef]
- Provencher, S.W.; Vogel, R.H. Three-Dimensional Reconstruction from Electron Micrographs of Disordered Specimens. I. Method. Ultramicroscopy 1988, 25, 209–221. [Google Scholar] [CrossRef]
- Scheres, S.H.W.; Gao, H.; Valle, M.; Herman, G.T.; Eggermont, P.P.B.; Frank, J.; Carazo, J.-M. Disentangling Conformational States of Macromolecules in 3D-EM through Likelihood Optimization. Nat. Methods 2007, 4, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.W. RELION: Implementation of a Bayesian Approach to Cryo-EM Structure Determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef]
- Rawson, S.; Iadanza, M.G.; Ranson, N.A.; Muench, S.P. Methods to Account for Movement and Flexibility in Cryo-EM Data Processing. Methods 2016, 100, 35–41. [Google Scholar] [CrossRef]
- Bai, X.-C.; Rajendra, E.; Yang, G.; Shi, Y.; Scheres, S.H.W. Sampling the Conformational Space of the Catalytic Subunit of Human γ-Secretase. eLife 2015, 4, e11182. [Google Scholar] [CrossRef]
- Cossio, P.; Hummer, G. Likelihood-Based Structural Analysis of Electron Microscopy Images. Curr. Opin. Struct. Biol. 2018, 49, 162–168. [Google Scholar] [CrossRef]
- Katsevich, G.; Katsevich, A.; Singer, A. Covariance Matrix Estimation for the Cryo-EM Heterogeneity Problem. arXiv 2013, arXiv:1309.1737. [Google Scholar] [CrossRef]
- Lederman, R. Hyper-Molecules: High Dimensional Maps of Molecular Conformations. Acta Crystallogr. A Found. Adv. 2020, 76, a61. [Google Scholar] [CrossRef]
- Lederman, R.R.; Andén, J.; Singer, A. Hyper-Molecules: On the Representation and Recovery of Dynamical Structures for Applications in Flexible Macro-Molecules in Cryo-EM. Inverse Probl. 2020, 36, 044005. [Google Scholar] [CrossRef]
- Tagare, H.D.; Kucukelbir, A.; Sigworth, F.J.; Wang, H.; Rao, M. Directly Reconstructing Principal Components of Heterogeneous Particles from Cryo-EM Images. J. Struct. Biol. 2015, 191, 245–262. [Google Scholar] [CrossRef]
- Neal, R.M.; Hinton, G.E. A View of the Em Algorithm That Justifies Incremental, Sparse, and Other Variants. In Learning in Graphical Models; Springer: Dordrecht, The Netherlands, 1998; pp. 355–368. ISBN 9789401061049. [Google Scholar]
- Tipping, M.E.; Bishop, C.M. Probabilistic Principal Component Analysis. J. R. Stat. Soc. Ser. B Stat. Methodol. 1999, 61, 611–622. [Google Scholar] [CrossRef]
- Roweis, S. EM Algorithms for PCA and SPCA. In Advances in Neural Information Processing Systems; Jordan, M., Kearns, M., Solla, S., Eds.; MIT Press: Cambridge, MA, USA, 1997; Volume 10. [Google Scholar]
- Punjani, A.; Fleet, D.J. 3D Variability Analysis: Resolving Continuous Flexibility and Discrete Heterogeneity from Single Particle Cryo-EM. J. Struct. Biol. 2021, 213, 107702. [Google Scholar] [CrossRef]
- Zhong, E.D.; Lerer, A.; Davis, J.H.; Berger, B. CryoDRGN2: Ab Initio Neural Reconstruction of 3D Protein Structures from Real Cryo-EM Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), IEEE Montreal, Montreal, QC, Canada, 10–17 October 2021; pp. 4066–4075. [Google Scholar]
- Dashti, A.; Schwander, P.; Langlois, R.; Fung, R.; Li, W.; Hosseinizadeh, A.; Liao, H.Y.; Pallesen, J.; Sharma, G.; Stupina, V.A.; et al. Trajectories of the Ribosome as a Brownian Nanomachine. Proc. Natl. Acad. Sci. USA 2014, 111, 17492–17497. [Google Scholar] [CrossRef]
- Dashti, A.; Mashayekhi, G.; Shekhar, M.; Ben Hail, D.; Salah, S.; Schwander, P.; des Georges, A.; Singharoy, A.; Frank, J.; Ourmazd, A. Retrieving Functional Pathways of Biomolecules from Single-Particle Snapshots. Nat. Commun. 2020, 11, 4734. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Ourmazd, A. Continuous Changes in Structure Mapped by Manifold Embedding of Single-Particle Data in Cryo-EM. Methods 2016, 100, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, S.; Wang, W.L.; Ma, Y.; Dong, Y.; Mao, Y. Deep Manifold Learning Reveals Hidden Dynamics of Proteasome Autoregulation. arXiv 2021, arXiv:2012.12854. [Google Scholar] [CrossRef]
- Giraldo-Barreto, J.; Ortiz, S.; Thiede, E.H.; Palacio-Rodriguez, K.; Carpenter, B.; Barnett, A.H.; Cossio, P. A Bayesian Approach to Extracting Free-Energy Profiles from Cryo-Electron Microscopy Experiments. Sci. Rep. 2021, 11, 13657. [Google Scholar] [CrossRef] [PubMed]
- Karplus, M.; McCammon, J.A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Vilas, J.L.; Gómez-Blanco, J.; Conesa, P.; Melero, R.; Miguel de la Rosa-Trevín, J.; Otón, J.; Cuenca, J.; Marabini, R.; Carazo, J.M.; Vargas, J.; et al. MonoRes: Automatic and Accurate Estimation of Local Resolution for Electron Microscopy Maps. Structure 2018, 26, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Kucukelbir, A.; Sigworth, F.J.; Tagare, H.D. Quantifying the Local Resolution of Cryo-EM Density Maps. Nat. Methods 2014, 11, 63–65. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Ishida, S.; Araki, M.; Kato, T.; Terayama, K.; Okuno, Y. Extraction of Protein Dynamics Information from Cryo-EM Maps Using Deep Learning. Nat. Mach. Intell. 2021, 3, 153–160. [Google Scholar] [CrossRef]
- Wriggers, W.; Birmanns, S. Using Situs for Flexible and Rigid-Body Fitting of Multiresolution Single-Molecule Data. J. Struct. Biol. 2001, 133, 193–202. [Google Scholar] [CrossRef] [PubMed]
- McGreevy, R.; Teo, I.; Singharoy, A.; Schulten, K. Advances in the Molecular Dynamics Flexible Fitting Method for Cryo-EM Modeling. Methods 2016, 100, 50–60. [Google Scholar] [CrossRef]
- Miyashita, O.; Kobayashi, C.; Mori, T.; Sugita, Y.; Tama, F. Flexible Fitting to Cryo-EM Density Map Using Ensemble Molecular Dynamics Simulations. J. Comput. Chem. 2017, 38, 1447–1461. [Google Scholar] [CrossRef] [PubMed]
- Kulik, M.; Mori, T.; Sugita, Y. Multi-Scale Flexible Fitting of Proteins to Cryo-EM Density Maps at Medium Resolution. Front. Mol. Biosci. 2021, 8, 631854. [Google Scholar] [CrossRef] [PubMed]
- Trabuco, L.G.; Villa, E.; Schreiner, E.; Harrison, C.B.; Schulten, K. Molecular Dynamics Flexible Fitting: A Practical Guide to Combine Cryo-Electron Microscopy and X-ray Crystallography. Methods 2009, 49, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Igaev, M.; Kutzner, C.; Bock, L.V.; Vaiana, A.C.; Grubmüller, H. Automated Cryo-EM Structure Refinement Using Correlation-Driven Molecular Dynamics. eLife 2019, 8, e43542. [Google Scholar] [CrossRef]
- Orzechowski, M.; Tama, F. Flexible Fitting of High-Resolution X-ray Structures into Cryoelectron Microscopy Maps Using Biased Molecular Dynamics Simulations. Biophys. J. 2008, 95, 5692–5705. [Google Scholar] [CrossRef] [PubMed]
- Mulder, A.M.; Yoshioka, C.; Beck, A.H.; Bunner, A.E.; Milligan, R.A.; Potter, C.S.; Carragher, B.; Williamson, J.R. Visualizing Ribosome Biogenesis: Parallel Assembly Pathways for the 30S Subunit. Science 2010, 330, 673–677. [Google Scholar] [CrossRef]
- Fischer, N.; Konevega, A.L.; Wintermeyer, W.; Rodnina, M.V.; Stark, H. Ribosome Dynamics and TRNA Movement by Time-Resolved Electron Cryomicroscopy. Nature 2010, 466, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Frank, J. Time-Resolved Cryo-Electron Microscopy: Recent Progress. J. Struct. Biol. 2017, 200, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Mäeots, M.-E.; Lee, B.; Nans, A.; Jeong, S.-G.; Esfahani, M.M.N.; Ding, S.; Smith, D.J.; Lee, C.-S.; Lee, S.S.; Peter, M.; et al. Modular Microfluidics Enables Kinetic Insight from Time-Resolved Cryo-EM. Nat. Commun. 2020, 11, 3465. [Google Scholar] [CrossRef] [PubMed]
- Dandey, V.P.; Budell, W.C.; Wei, H.; Bobe, D.; Maruthi, K.; Kopylov, M.; Eng, E.T.; Kahn, P.A.; Hinshaw, J.E.; Kundu, N.; et al. Time-Resolved Cryo-EM Using Spotiton. Nat. Methods 2020, 17, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Kontziampasis, D.; Klebl, D.P.; Iadanza, M.G.; Scarff, C.A.; Kopf, F.; Sobott, F.; Monteiro, D.C.F.; Trebbin, M.; Muench, S.P.; White, H.D. A Cryo-EM Grid Preparation Device for Time-Resolved Structural Studies. IUCrJ 2019, 6, 1024–1031. [Google Scholar] [CrossRef]
- Klebl, D.P.; White, H.D.; Sobott, F.; Muench, S.P. On-Grid and in-Flow Mixing for Time-Resolved Cryo-EM. Acta Crystallogr. D Struct. Biol. 2021, 77, 1233–1240. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeVore, K.; Chiu, P.-L. Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity. Biomolecules 2022, 12, 628. https://doi.org/10.3390/biom12050628
DeVore K, Chiu P-L. Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity. Biomolecules. 2022; 12(5):628. https://doi.org/10.3390/biom12050628
Chicago/Turabian StyleDeVore, Kira, and Po-Lin Chiu. 2022. "Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity" Biomolecules 12, no. 5: 628. https://doi.org/10.3390/biom12050628
APA StyleDeVore, K., & Chiu, P.-L. (2022). Probing Structural Perturbation of Biomolecules by Extracting Cryo-EM Data Heterogeneity. Biomolecules, 12(5), 628. https://doi.org/10.3390/biom12050628





