Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Recombinant Expression and Purification of Monomeric α-Synuclein
2.2. In Vitro Assembly of α-Synuclein Amyloid Fibrils
2.3. Fibril Fragmentation by Controlled Sonication
2.4. AFM Imaging and Image Analysis
3. Results
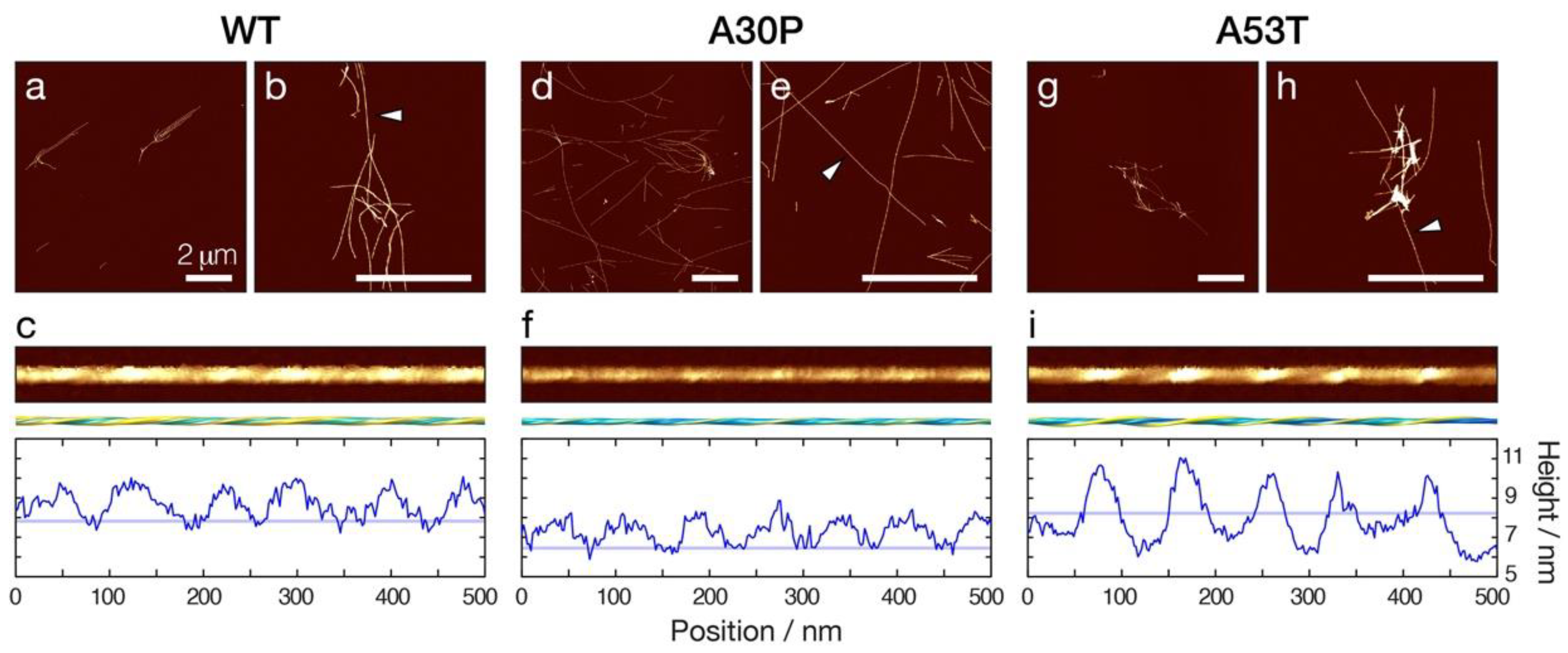
3.1. Amyloid Fibrils Formed from A30P and A53T α-Synuclein Display Distinct Polymorphic Structures Compared to WT α-Synuclein Fibrils
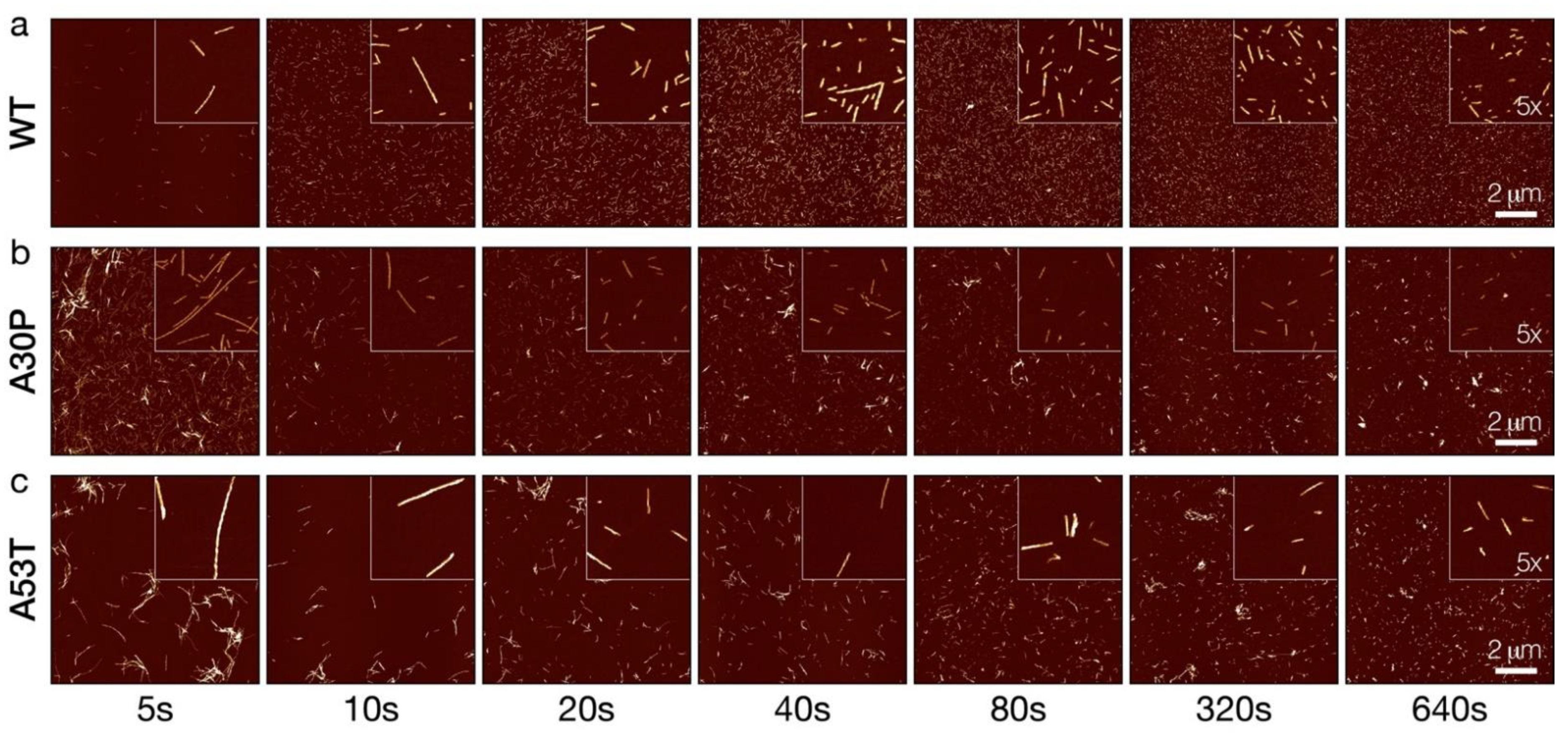
3.2. Controlled Sonication Promotes Rapid Fragmentation of α-Synuclein Amyloid Fibrils
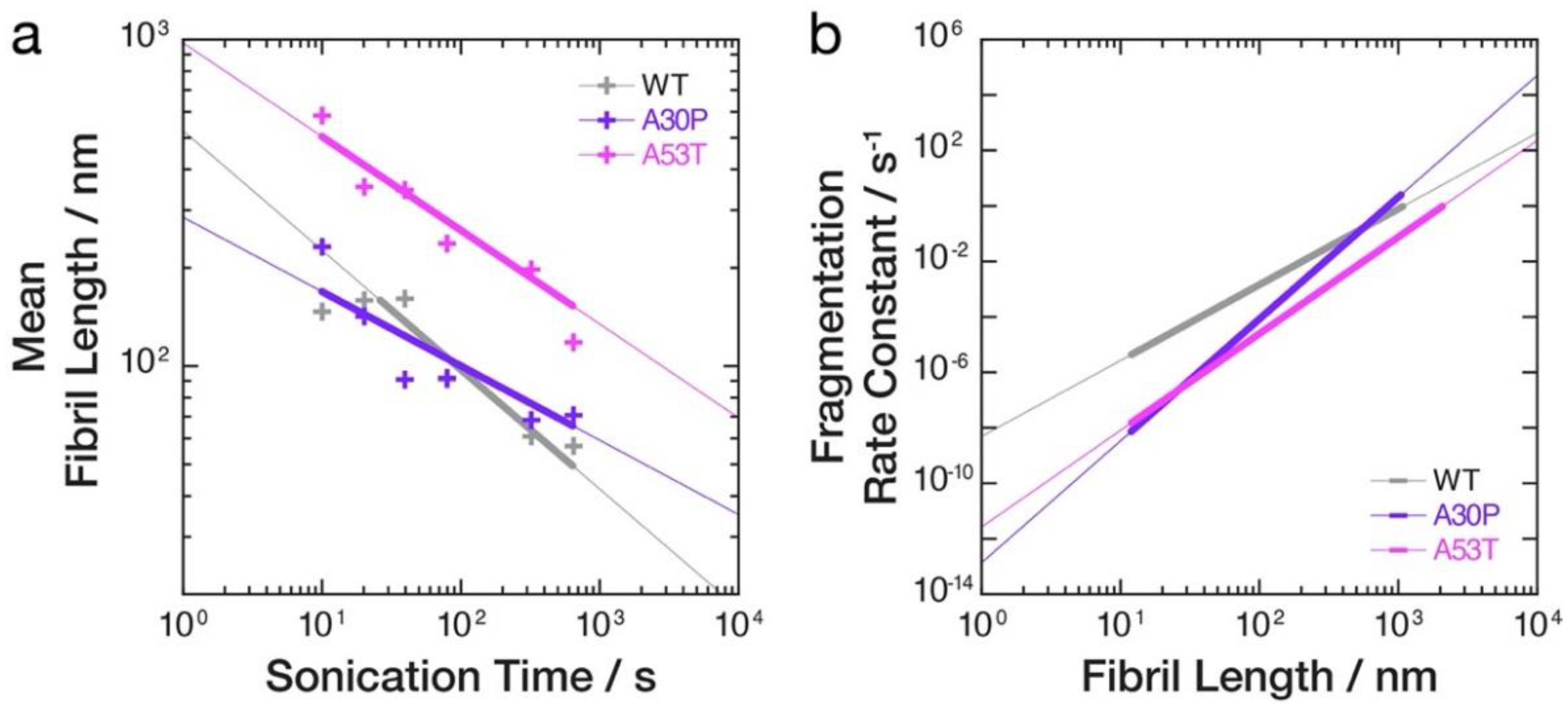
3.3. Quantitative Image Analysis of α-Synuclein Amyloid Fibrils Undergoing Fragmentation by Controlled Sonication Show Polymorph-Dependent Length Distribution Decays
3.4. The Rates of Fibril Fragmentation Promoted by Controlled Sonication Are Distinct for the Different Fibril Polymorphs Formed from WT, A30P and A53T α-Synuclein Variants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ke, P.C.; Zhou, R.; Serpell, L.C.; Riek, R.; Knowles, T.P.J.; Lashuel, H.A.; Gazit, E.; Hamley, I.W.; Davis, T.P.; Fandrich, M.; et al. Half a century of amyloids: Past, present and future. Chem. Soc. Rev. 2020, 49, 5473–5509. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Vendruscolo, M.; Dobson, C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Boyer, D.R.; Li, B.; Sun, C.; Fan, W.; Sawaya, M.R.; Jiang, L.; Eisenberg, D.S. Structures of fibrils formed by Alpha-Synuclein hereditary disease mutant H50q reveal new polymorphs. Nat. Struct. Mol. Biol. 2019, 26, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Lovestam, S.; Schweighauser, M.; Matsubara, T.; Murayama, S.; Tomita, T.; Ando, T.; Hasegawa, K.; Yoshida, M.; Tarutani, A.; Hasegawa, M.; et al. Seeded assembly in vitro does not replicate the structures of alpha-synuclein filaments from multiple system atrophy. FEBS Open Biol. 2021, 11, 999–1013. [Google Scholar] [CrossRef]
- Peelaerts, W.; Bousset, L.; van der Perren, A.; Moskalyuk, A.; Pulizzi, R.; Giugliano, M.; van den Haute, C.; Melki, R.; Baekelandt, V. Alpha-synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 2015, 522, 340–344. [Google Scholar] [CrossRef]
- Walker, L.C. Proteopathic strains and the heterogeneity of neurodegenerative diseases. Annu. Rev. Genet. 2016, 50, 329–346. [Google Scholar] [CrossRef] [PubMed]
- Singleton, A.B.; Farrer, M.; Johnson, J.; Singleton, A.; Hague, S.; Kachergus, J.; Hulihan, M.; Peuralinna, T.; Dutra, A.; Nussbaum, R.; et al. Alpha-synuclein locus triplication causes Parkinson’s disease. Science 2003, 302, 841. [Google Scholar] [CrossRef] [Green Version]
- Chartier-Harlin, M.C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M.; et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364, 1167–1169. [Google Scholar] [CrossRef]
- Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Graeber, M.; Kosel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. Ala30pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [Green Version]
- Flagmeier, P.; Meisl, G.; Vendruscolo, M.; Knowles, T.P.; Dobson, C.M.; Buell, A.K.; Galvagnion, C. Mutations associated with familial Parkinson’s disease alter the initiation and amplification steps of alpha-synuclein aggregation. Proc. Natl. Acad. Sci. USA 2016, 113, 10328–10333. [Google Scholar] [CrossRef] [Green Version]
- Ruggeri, F.S.; Flagmeier, P.; Kumita, J.R.; Meisl, G.; Chirgadze, D.Y.; Bongiovanni, M.N.; Knowles, T.P.J.; Dobson, C.M. The influence of pathogenic mutations in alpha-synuclein on biophysical and structural characteristics of amyloid fibrils. ACS Nano 2020, 14, 5213–5222. [Google Scholar] [CrossRef]
- Jarrett, J.T.; Lansbury, P.T., Jr. Amyloid fibril formation requires a chemically discriminating nucleation event: Studies of an amyloidogenic sequence from the bacterial protein Osmb. Biochemistry 1992, 31, 12345–12352. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-F. Nucleation: The birth of a new protein phase. Biophys. J. 2015, 109, 1999–2000. [Google Scholar] [CrossRef] [Green Version]
- Collins, S.R.; Douglass, A.; Vale, R.D.; Weissman, J.S. Mechanism of Prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol 2004, 2, e321. [Google Scholar] [CrossRef] [Green Version]
- Tornquist, M.; Michaels, T.C.T.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.A.; Knowles, T.P.J.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.-F.; Hellewell, A.L.; Gosal, W.S.; Homans, S.W.; Hewitt, E.W.; Radford, S.E. Fibril fragmentation enhances amyloid cytotoxicity. J. Biol. Chem. 2009, 284, 34272–34282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.-F.; Homans, S.W.; Radford, S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 8926–8931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, Y.; Shen, H.C.; Komi, Y.; Sugiyama, S.; Kurinomaru, T.; Tomabechi, Y.; Krayukhina, E.; Okamoto, K.; Yokoyama, T.; Shirouzu, M.; et al. Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system. Nat. Chem. Biol. 2022, 18, 321–331. [Google Scholar] [CrossRef] [PubMed]
- De Giorgi, F.; Laferriere, F.; Zinghirino, F.; Faggiani, E.; Lends, A.; Bertoni, M.; Yu, X.; Grelard, A.; Morvan, E.; Habenstein, B.; et al. Novel self-replicating alpha-synuclein polymorphs that escape tht monitoring can spontaneously emerge and acutely spread in neurons. Sci. Adv. 2020, 6, eabc4364. [Google Scholar] [CrossRef] [PubMed]
- Buell, A.K.; Galvagnion, C.; Gaspar, R.; Sparr, E.; Vendruscolo, M.; Knowles, T.P.; Linse, S.; Dobson, C.M. Solution conditions determine the relative importance of nucleation and growth processes in alpha-synuclein aggregation. Proc. Natl. Acad. Sci. USA 2014, 111, 7671–7676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarutani, A.; Suzuki, G.; Shimozawa, A.; Nonaka, T.; Akiyama, H.; Hisanaga, S.; Hasegawa, M. The effect of fragmented pathogenic alpha-synuclein seeds on prion-like propagation. J. Biol. Chem. 2016, 291, 18675–18688. [Google Scholar] [CrossRef] [Green Version]
- Cappai, R.; Leck, S.L.; Tew, D.J.; Williamson, N.A.; Smith, D.P.; Galatis, D.; Sharples, R.A.; Curtain, C.C.; Ali, F.E.; Cherny, R.A.; et al. Dopamine promotes alpha-synuclein aggregation into sds-resistant soluble oligomers via a distinct folding pathway. FASEB J. 2005, 19, 1377–1379. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-F.; Homans, S.W.; Radford, S.E. Amyloid fibril length distribution quantified by atomic force microscopy single-particle image analysis. Protein Eng. Des. Select. 2009, 22, 489–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, W.-F. amyloid fibril length quantification by atomic force microscopy. In Bio-Nanoimaging Protein Misfolding & Aggregation; Uversky, V.N., Lyubchenko, Y.L., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 17–25. [Google Scholar]
- Beal, D.M.; Tournus, M.; Marchante, R.; Purton, T.J.; Smith, D.P.; Tuite, M.F.; Doumic, M.; Xue, W.-F. The division of amyloid fibrils: Systematic comparison of fibril fragmentation stability by linking theory with experiments. iScience 2020, 23, 101512. [Google Scholar] [CrossRef] [PubMed]
- Tournus, M.; Escobedo, M.; Xue, W.-F.; Doumic, M. Insights into the dynamic trajectories of protein filament division revealed by numerical investigation into the mathematical model of pure fragmentation. PLoS Comput. Biol. 2021, 17, e1008964. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Williams, J.K.; Yan, R.; Mouradian, M.M.; Baum, J. Increased dynamics of alpha-synuclein fibrils by beta-synuclein leads to reduced seeding and cytotoxicity. Sci. Rep. 2019, 9, 17579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutter, L.; Aubrey, L.D.; Xue, W.-F. On the structural diversity and individuality of polymorphic amyloid protein assemblies. J. Mol. Biol. 2021, 433, 167124. [Google Scholar] [CrossRef] [PubMed]
- Lutter, L.; Serpell, C.J.; Tuite, M.F.; Serpell, L.C.; Xue, W.-F. Three-dimensional reconstruction of individual helical nano-filament structures from atomic force microscopy topographs. Biomol. Concepts 2020, 11, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, L.D.; Blakeman, B.J.F.; Lutter, L.; Serpell, C.J.; Tuite, M.F.; Serpell, L.C.; Xue, W.-F. Quantification of amyloid fibril polymorphism by nano-morphometry reveals the individuality of filament assembly. Commun. Chem. 2020, 3, 125. [Google Scholar] [CrossRef]
- Marchante, R.; Beal, D.M.; Koloteva-Levine, N.; Purton, T.J.; Tuite, M.F.; Xue, W.-F. The physical dimensions of amyloid aggregates control their infective potential as prion particles. eLife 2017, 6, e27109. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Knowles, T.P.J.; Terentjev, E.M. Strength of nanotubes, filaments, and nanowires from sonication-induced scission. Adv. Mater. 2009, 21, 3945–3948. [Google Scholar] [CrossRef] [Green Version]
- Ohhashi, Y.; Kihara, M.; Naiki, H.; Goto, Y. Ultrasonication-Induced Amyloid Fibril Formation of Beta2-Microglobulin. J. Biol. Chem. 2005, 280, 32843–32848. [Google Scholar] [CrossRef] [Green Version]
- Xue, W.-F.; Radford, S.E. An imaging and systems modeling approach to fibril breakage enables prediction of amyloid behavior. Biophys. J. 2013, 105, 2811–2819. [Google Scholar] [CrossRef] [Green Version]
- Kakuda, K.; Ikenaka, K.; Araki, K.; So, M.; Aguirre, C.; Kajiyama, Y.; Konaka, K.; Noi, K.; Baba, K.; Tsuda, H.; et al. Ultrasonication-based rapid amplification of alpha-synuclein aggregates in cerebrospinal fluid. Sci. Rep. 2019, 9, 6001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakajima, K.; Noi, K.; Yamaguchi, K.; So, M.; Ikenaka, K.; Mochizuki, H.; Ogi, H.; Goto, Y. Optimized Sonoreactor for accelerative amyloid-fibril assays through enhancement of primary nucleation and fragmentation. Ultrason. Sonochem. 2021, 73, 105508. [Google Scholar] [CrossRef] [PubMed]
- Koloteva-Levine, N.; Aubrey, L.D.; Marchante, R.; Purton, T.J.; Hiscock, J.R.; Tuite, M.F.; Xue, W.-F. Amyloid particles facilitate surface-catalyzed cross-seeding by acting as promiscuous nanoparticles. Proc. Natl. Acad. Sci. USA 2021, 118, e2104148118. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Ter-Avanesyan, M.D. Structure and replication of yeast prions. Cell 1998, 94, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Vaquer-Alicea, J.; Diamond, M.I. Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 2019, 88, 785–810. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Carroni, M.; Nussbaum-Krammer, C.; Mogk, A.; Nillegoda, N.B.; Szlachcic, A.; Guilbride, D.L.; Saibil, H.R.; Mayer, M.P.; Bukau, B. Human Hsp70 disaggregase reverses Parkinson’s-linked alpha-synuclein amyloid fibrils. Mol. Cell 2015, 59, 781–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tittelmeier, J.; Sandhof, C.A.; Ries, H.M.; Druffel-Augustin, S.; Mogk, A.; Bukau, B.; Nussbaum-Krammer, C. The Hsp110/Hsp70 disaggregation system generates spreading-competent toxic alpha-synuclein species. EMBO J. 2020, 39, e103954. [Google Scholar] [CrossRef]
- Tariq, A.; Lin, J.; Jackrel, M.E.; Hesketh, C.D.; Carman, P.J.; Mack, K.L.; Weitzman, R.; Gambogi, C.; Murillo, O.A.H.; Sweeny, E.A.; et al. Mining disaggregase sequence space to safely counter Tdp-43, fus, and alpha-synuclein proteotoxicity. Cell Rep. 2019, 28, 2080–2095.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| γ (±SE) | α/nm−1 (log α ± SE) | B100/s−1 (log B100 ± SE) | Mean Fibril Height (±SEM)/nm | |
|---|---|---|---|---|
| WT | 2.7 ± 0.4 | 9.1·10−4 (−3.0 ± 0.1) | 140·10−5 (−2.8 ± 0.2) | 7.8 ± 0.1 |
| A30P | 4.4 ± 0.9 | 12·10−4 (−2.9 ± 0.1) | 8.2·10−5 (−4.1 ± 0.6) | 6.4 ± 0.1 |
| A53T | 3.4 ± 0.5 | 4.8·10−4 (−3.3 ± 0.1) | 2.5·10−5 (−4.6 ± 0.5) | 8.2 ± 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanami, S.; Purton, T.J.; Smith, D.P.; Tuite, M.F.; Xue, W.-F. Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils. Biomolecules 2022, 12, 630. https://doi.org/10.3390/biom12050630
Sanami S, Purton TJ, Smith DP, Tuite MF, Xue W-F. Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils. Biomolecules. 2022; 12(5):630. https://doi.org/10.3390/biom12050630
Chicago/Turabian StyleSanami, Sarina, Tracey J. Purton, David P. Smith, Mick F. Tuite, and Wei-Feng Xue. 2022. "Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils" Biomolecules 12, no. 5: 630. https://doi.org/10.3390/biom12050630
APA StyleSanami, S., Purton, T. J., Smith, D. P., Tuite, M. F., & Xue, W.-F. (2022). Comparative Analysis of the Relative Fragmentation Stabilities of Polymorphic Alpha-Synuclein Amyloid Fibrils. Biomolecules, 12(5), 630. https://doi.org/10.3390/biom12050630







