Peptidomimetics Based on C-Terminus of Blm10 Stimulate Human 20S Proteasome Activity and Promote Degradation of Proteins
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
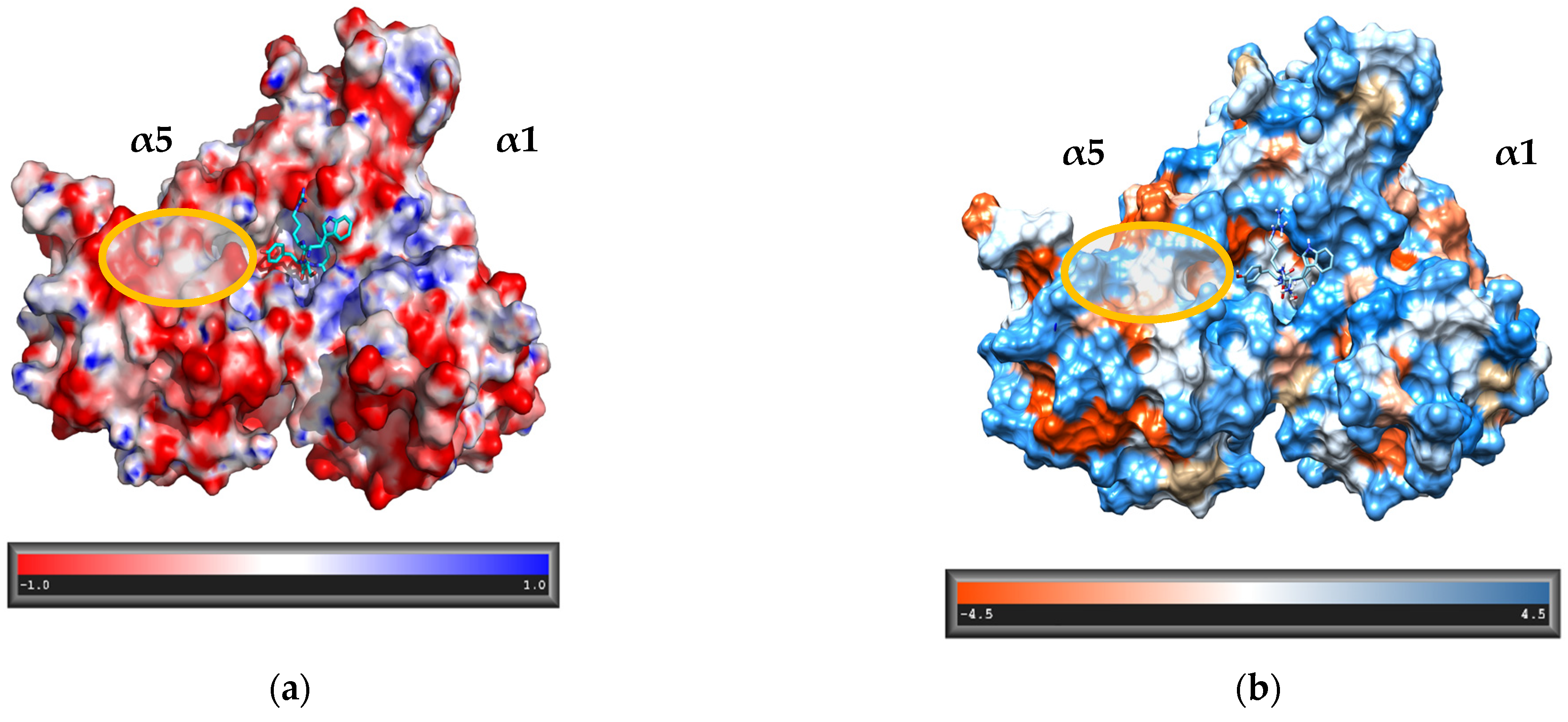
3.1. Design of the Peptidomimetics
3.2. Stimulating Capacity of M-1–M-12 Analogs
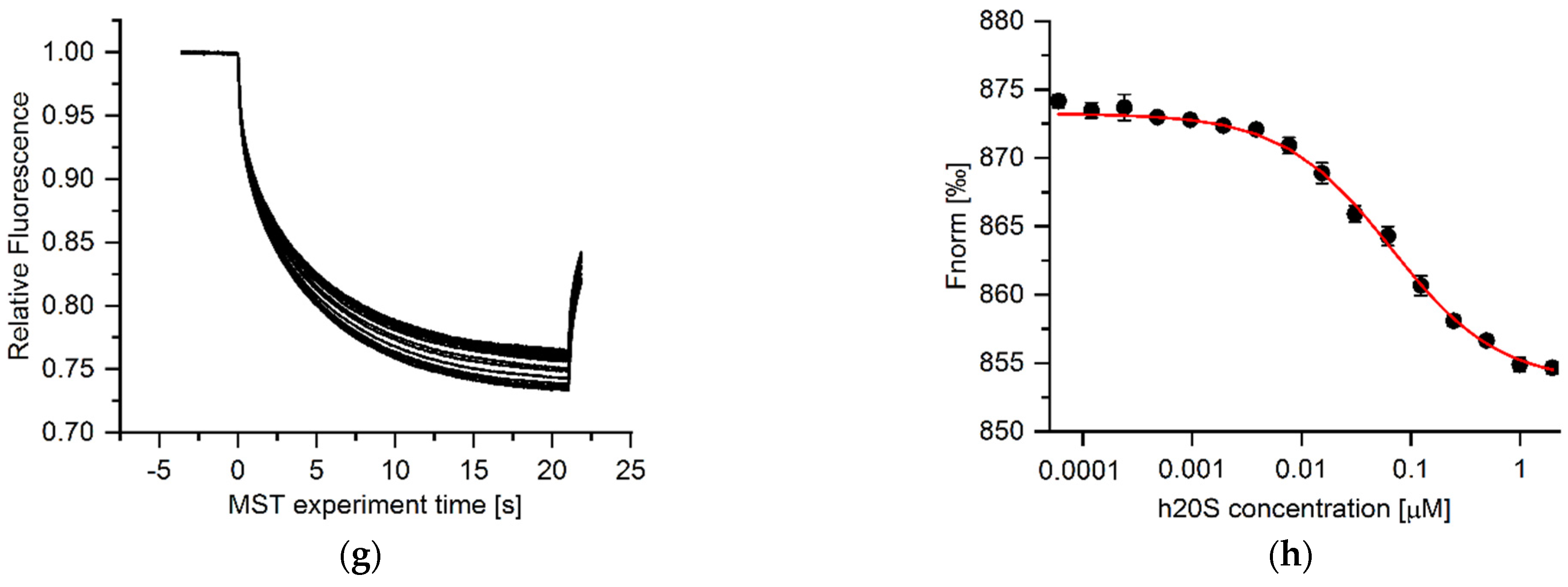
3.3. Microscale Thermophoresis (MST) Results
3.4. Degradation of Model Proteins
3.5. Stimulation of Proteasome Activity in Cell Culture
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, T.; Hohn, A.; Grune, T. The proteasome and the degradation of oxidized proteins: Part II—Protein oxidation and proteasomal degradation. Redox Biol. 2014, 2, 99–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadtmueller, B.M.; Hill, C.P. Proteasome activators. Mol. Cell 2011, 41, 8–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T.; Hill, C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 2010, 37, 728–735. [Google Scholar] [CrossRef] [Green Version]
- Whitby, F.G.; Masters, E.I.; Kramer, L.; Knowlton, J.R.; Yao, Y.; Wang, C.C.; Hill, C.P. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 2000, 408, 115–120. [Google Scholar] [CrossRef]
- Ruschak, A.M.; Kay, L.E. Proteasome allostery as a population shift between interchanging conformers. Proc. Natl. Acad. Sci. USA 2012, 109, E3454–E3462. [Google Scholar] [CrossRef] [Green Version]
- Saez, I.; Vilchez, D. The mechanistic links between proteasome activity, aging and age-related diseases. Curr. Genom. 2014, 15, 38–51. [Google Scholar] [CrossRef] [Green Version]
- Ciechanover, A.; Kwon, Y.T. Degradation of misfolded proteins in neurodegenerative diseases: Therapeutic targets and strategies. Exp. Mol. Med. 2015, 47, e147. [Google Scholar] [CrossRef] [Green Version]
- Chondrogianni, N.; Sakellari, M.; Lefaki, M.; Papaevgeniou, N.; Gonos, E.S. Proteasome activation delays aging in vitro and in vivo. Free Radic. Biol. Med. 2014, 71, 303–320. [Google Scholar] [CrossRef]
- Njomen, E.; Tepe, J.J. Proteasome activation as a new therapeutic approach to target proteotoxic disorders. J. Med. Chem. 2019, 62, 6469–6481. [Google Scholar] [CrossRef]
- Huang, L.; Ho, P.; Chen, C.H. Activation and inhibition of proteasomes by betulinic acid and its derivatives. FEBS Lett. 2007, 581, 4955–4959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, C.L.; Njomen, E.; Sjögren, B.; Dexheimer, T.S.; Tepe, J.J. Small molecule enhancement of 20S proteasome activity targets intrinsically disordered proteins. ACS Chem. Biol. 2017, 12, 2240–2247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiolek, T.J.; Magyar, C.L.; Wall, T.J.; Davies, S.B.; Campbell, M.V.; Savich, C.J.; Tepe, J.J.; Mosey, R.A. Dihydroquinazolines enhance 20S proteasome activity and induce degradation of α-synuclein, an intrinsically disordered protein associated with neurodegeneration. Bioorg. Med. Chem. Lett. 2021, 36, 127821. [Google Scholar] [CrossRef]
- Fiolek, T.J.; Keel, K.L.; Tepe, J.J. Fluspirilene analogs activate the 20S proteasome and overcome proteasome impairment by intrinsically disordered protein oligomers. ACS Chem. Neurosci. 2021, 12, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Dal Vechio, F.H.; Cerqueira, F.; Augusto, O.; Lopes, R.; Demasi, M. Peptides that activate the 20S proteasome by gate opening increased oxidized protein removal and reduced protein aggregation. Free Radic. Biol. Med. 2014, 67, 304–313. [Google Scholar] [CrossRef]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–741. [Google Scholar] [CrossRef] [Green Version]
- Gillette, T.G.; Kumar, B.; Thompson, D.; Slaughter, C.A.; DeMartino, G.N. Differential roles of the COOH termini of AAA subunits of PA700 (19S regulator) in asymmetric assembly and activation of the 26S proteasome. J. Biol. Chem. 2008, 283, 31813–31822. [Google Scholar] [CrossRef] [Green Version]
- Thibaudeau, T.A.; Anderson, R.T.; Smith, D.M. A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 2018, 9, 1097. [Google Scholar] [CrossRef]
- Giżyńska, M.; Witkowska, J.; Karpowicz, P.; Rostankowski, R.; Chocron, E.S.; Pickering, A.M.; Osmulski, P.; Gaczynska, M.; Jankowska, E. Proline- and arginine-rich peptides as flexible allosteric modulators of human proteasome activity. J. Med. Chem. 2019, 62, 359–370. [Google Scholar] [CrossRef]
- Witkowska, J.; Giżyńska, M.; Grudnik, P.; Golik, P.; Karpowicz, P.; Giełdoń, A.; Dubin, G.; Jankowska, E. Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome—Insights into the enzyme activation mechanism. Sci. Rep. 2017, 7, 6177. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pikora, M.; Gieldon, A. RASMOL AB—New functionalities in the program for structure analysis. Acta Biochim. Pol. 2015, 62, 629–631. [Google Scholar] [CrossRef] [Green Version]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydrophatic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Deshmukh, F.K.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The contribution of the 20S proteasome to proteostasis. Biomolecules 2019, 16, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nussbaum, A.K.; Dick, T.P.; Keilholz, W.; Schirle, M.; Stevanovic, S.; Dietz, K.; Heinemever, W.; Groll, M.; Wolf, D.H.; Huber, R.; et al. Cleavage motifs of the yeast 20S proteasome β subunits deduced from digests of enolase 1. Proc. Natl. Acad. Sci. USA 1998, 95, 12504–12509. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Castelao, B.; Goethals, M.; Vandekerckhove, J.; Castaño, J.G. Mechanism of cleavage of alpha-synuclein by the 20S proteasome and modulation of its degradation by the RedOx state of the N-terminal methionines. Biochim. Biophys. Acta 2014, 1843, 352–365. [Google Scholar] [CrossRef]




| Compound | Sequence | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| M-1 | Y | S | Q | E | Peg2 | L | W | R | S | Y | Y | S |
| M-2 | E | N | S | K | Peg2 | L | W | R | S | Y | Y | S |
| M-3 | K | N | S | N | Peg2 | L | W | R | S | Y | Y | S |
| M-4 | N | N | S | E | Peg2 | L | W | R | S | Y | Y | S |
| M-5 | Har * | N | S | N | Peg2 | D | W | R | S | Y | Y | S |
| M-6 | K | N | E | N | Peg2 | L | W | R | S | Y | Y | S |
| M-7 | K | N | S | N | Peg2 | D | W | R | S | Y | Y | S |
| M-8 | L | N | S | N | Peg2 | L | W | R | S | Y | Y | S |
| M-9 | E | N | S | N | Peg2 | L | W | R | S | Y | Y | S |
| M-10 | K | N | S | N | Peg2 | D | W | Cit * | S | Y | Y | S |
| M-11 | K | N | S | K | Peg2 | L | W | R | S | Y | Y | S |
| M-12 | K | N | S | N | Peg2 | D | W | R | S | dY * | Y | S |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cekała, K.; Trepczyk, K.; Sowik, D.; Karpowicz, P.; Giełdoń, A.; Witkowska, J.; Giżyńska, M.; Jankowska, E.; Wieczerzak, E. Peptidomimetics Based on C-Terminus of Blm10 Stimulate Human 20S Proteasome Activity and Promote Degradation of Proteins. Biomolecules 2022, 12, 777. https://doi.org/10.3390/biom12060777
Cekała K, Trepczyk K, Sowik D, Karpowicz P, Giełdoń A, Witkowska J, Giżyńska M, Jankowska E, Wieczerzak E. Peptidomimetics Based on C-Terminus of Blm10 Stimulate Human 20S Proteasome Activity and Promote Degradation of Proteins. Biomolecules. 2022; 12(6):777. https://doi.org/10.3390/biom12060777
Chicago/Turabian StyleCekała, Katarzyna, Karolina Trepczyk, Daria Sowik, Przemysław Karpowicz, Artur Giełdoń, Julia Witkowska, Małgorzata Giżyńska, Elżbieta Jankowska, and Ewa Wieczerzak. 2022. "Peptidomimetics Based on C-Terminus of Blm10 Stimulate Human 20S Proteasome Activity and Promote Degradation of Proteins" Biomolecules 12, no. 6: 777. https://doi.org/10.3390/biom12060777
APA StyleCekała, K., Trepczyk, K., Sowik, D., Karpowicz, P., Giełdoń, A., Witkowska, J., Giżyńska, M., Jankowska, E., & Wieczerzak, E. (2022). Peptidomimetics Based on C-Terminus of Blm10 Stimulate Human 20S Proteasome Activity and Promote Degradation of Proteins. Biomolecules, 12(6), 777. https://doi.org/10.3390/biom12060777







