Mammalian Melatonin Agonist Pharmaceuticals Stimulate Rhomboid Proteins in Plants
Abstract
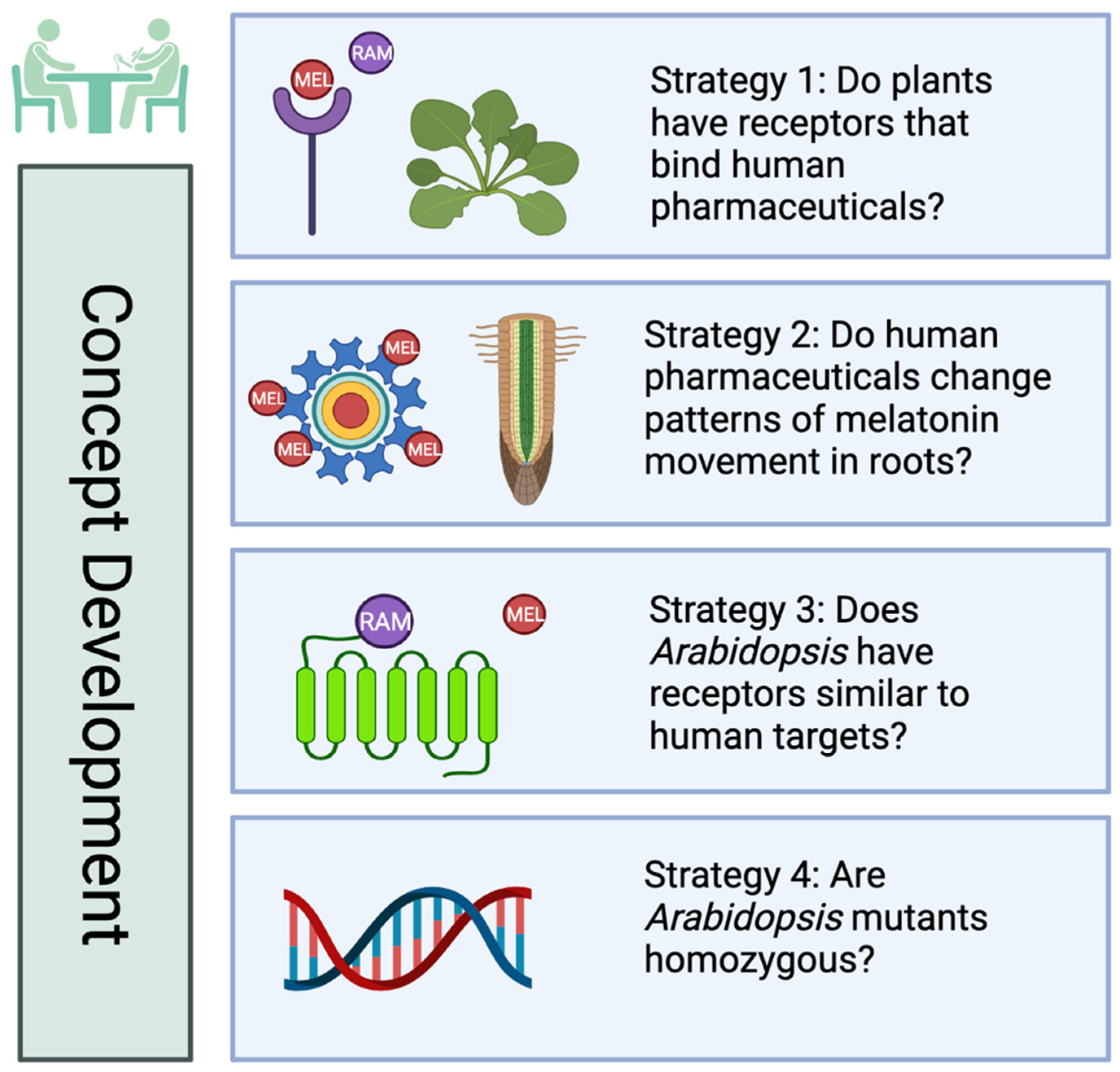
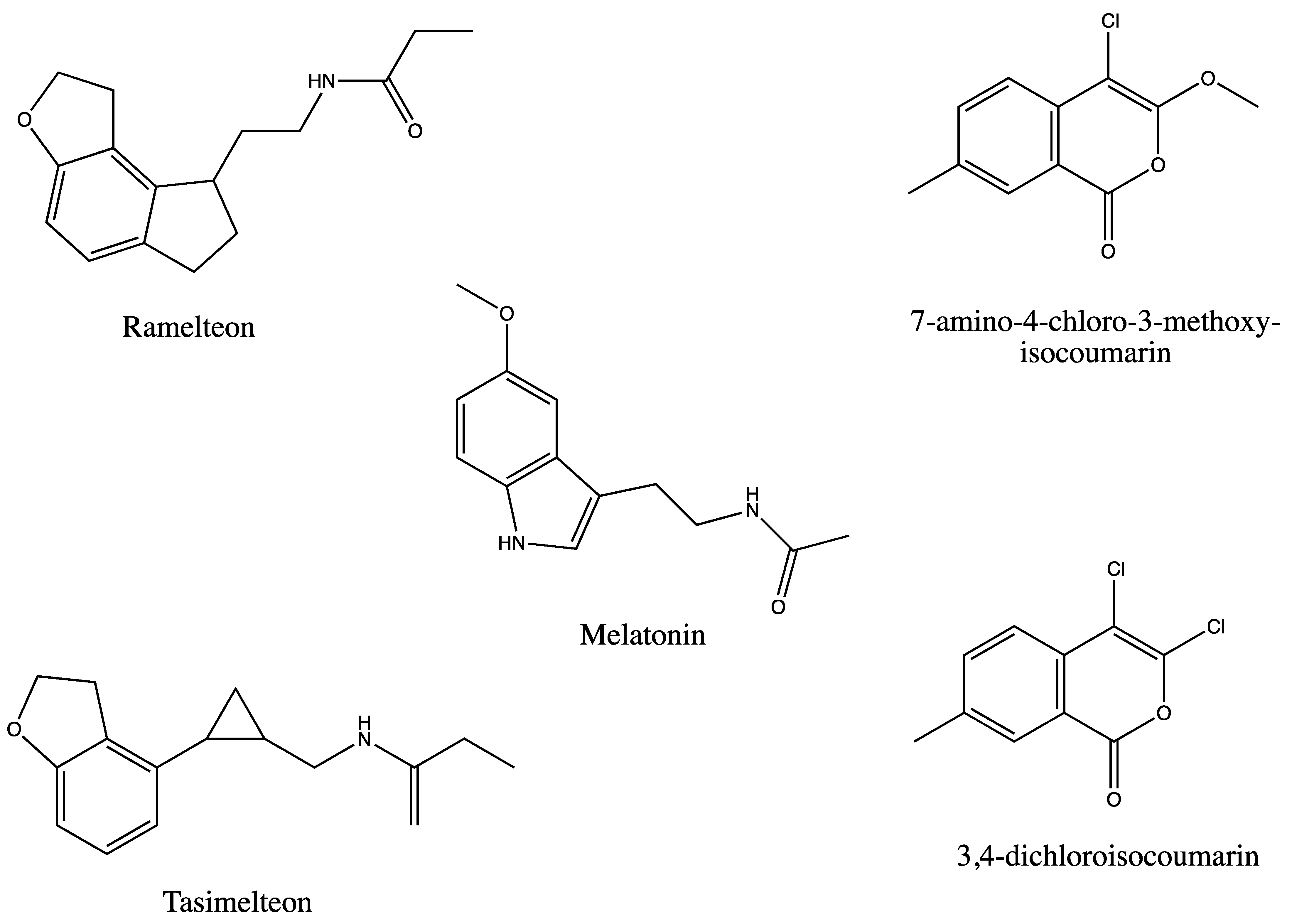
:1. Introduction
2. Materials and Methods
2.1. Growth Experiments
2.2. Growth Conditions
2.3. Gravitropism Assays
2.4. Quantum Dot (QD) Conjugation
2.5. QD Exposure
2.6. QD Imaging and Processing
2.7. Phytochemical Analysis
2.8. Sequence Alignment and Identification of RBL7 as a Candidate for a MEL Interacting Protein
2.9. Response of RBL7 Mutants to RAM or MEL Treatment
2.10. Confirmation of Insert in rbl7 Mutant Lines
2.11. Statistical Analysis
3. Results
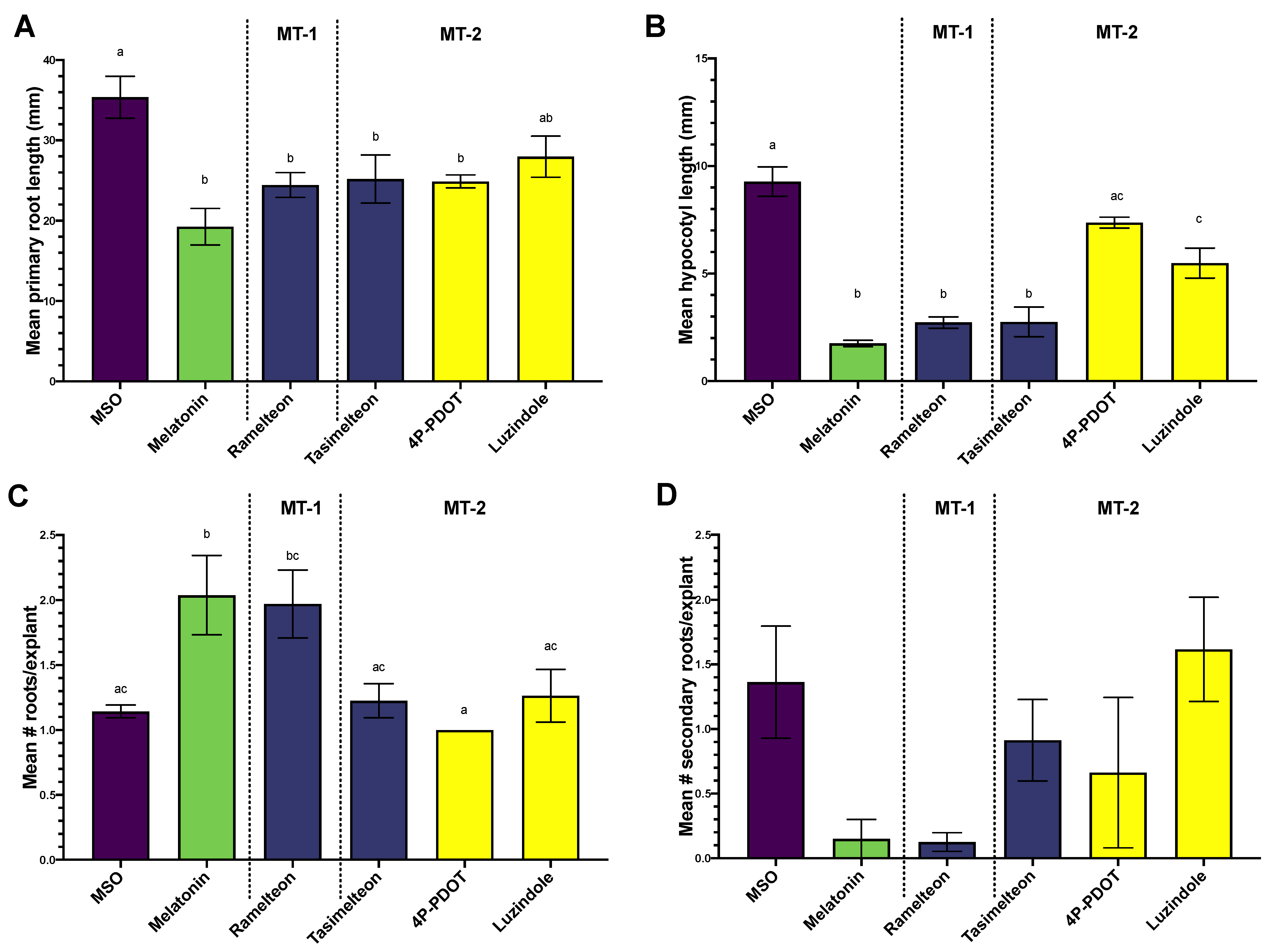
3.1. Effects of MEL, Agonist and Antagonist Treatment on Growth and Regeneration in Col-0
3.2. Phytochemical Quantification
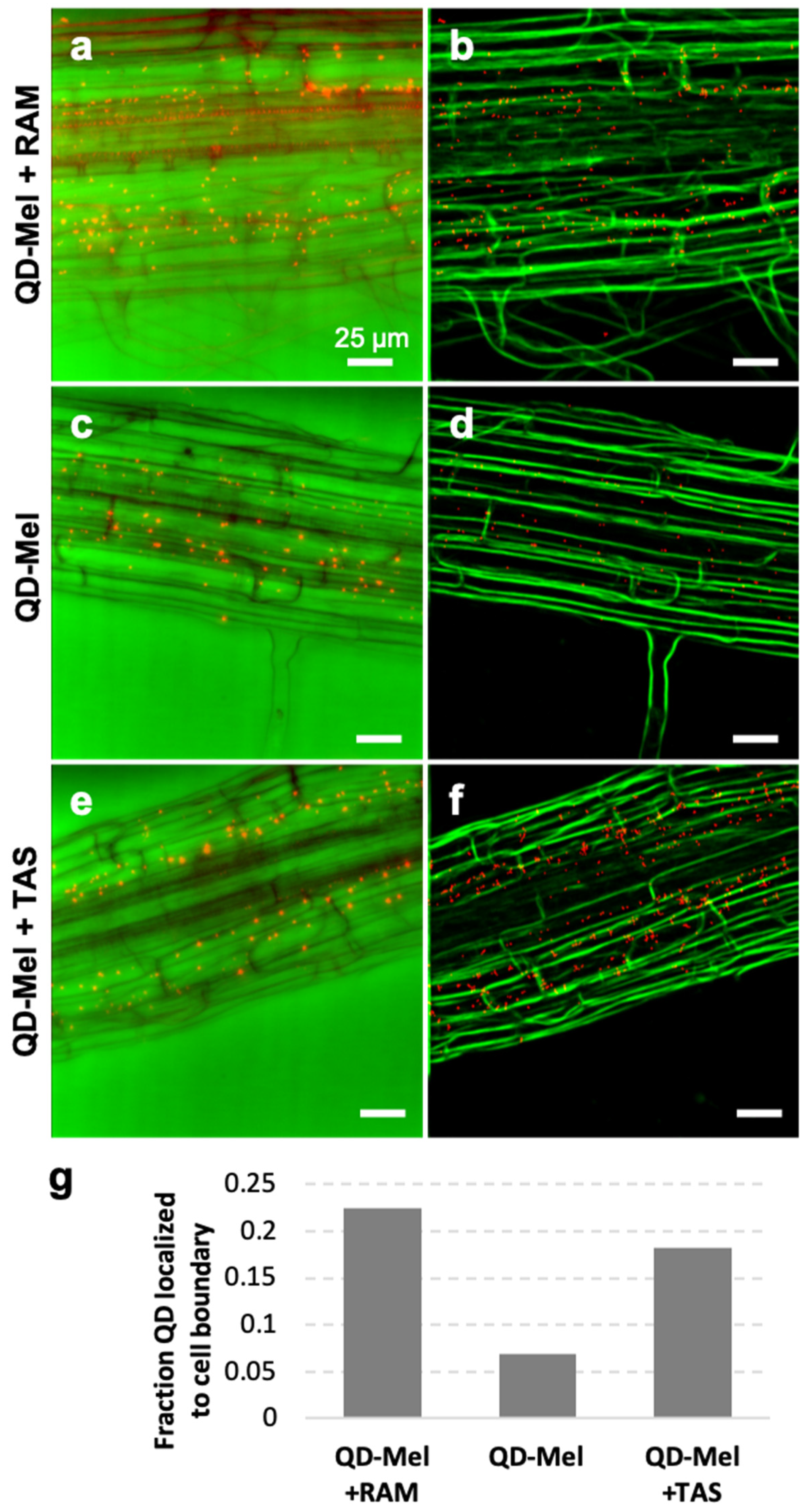
3.3. Localization
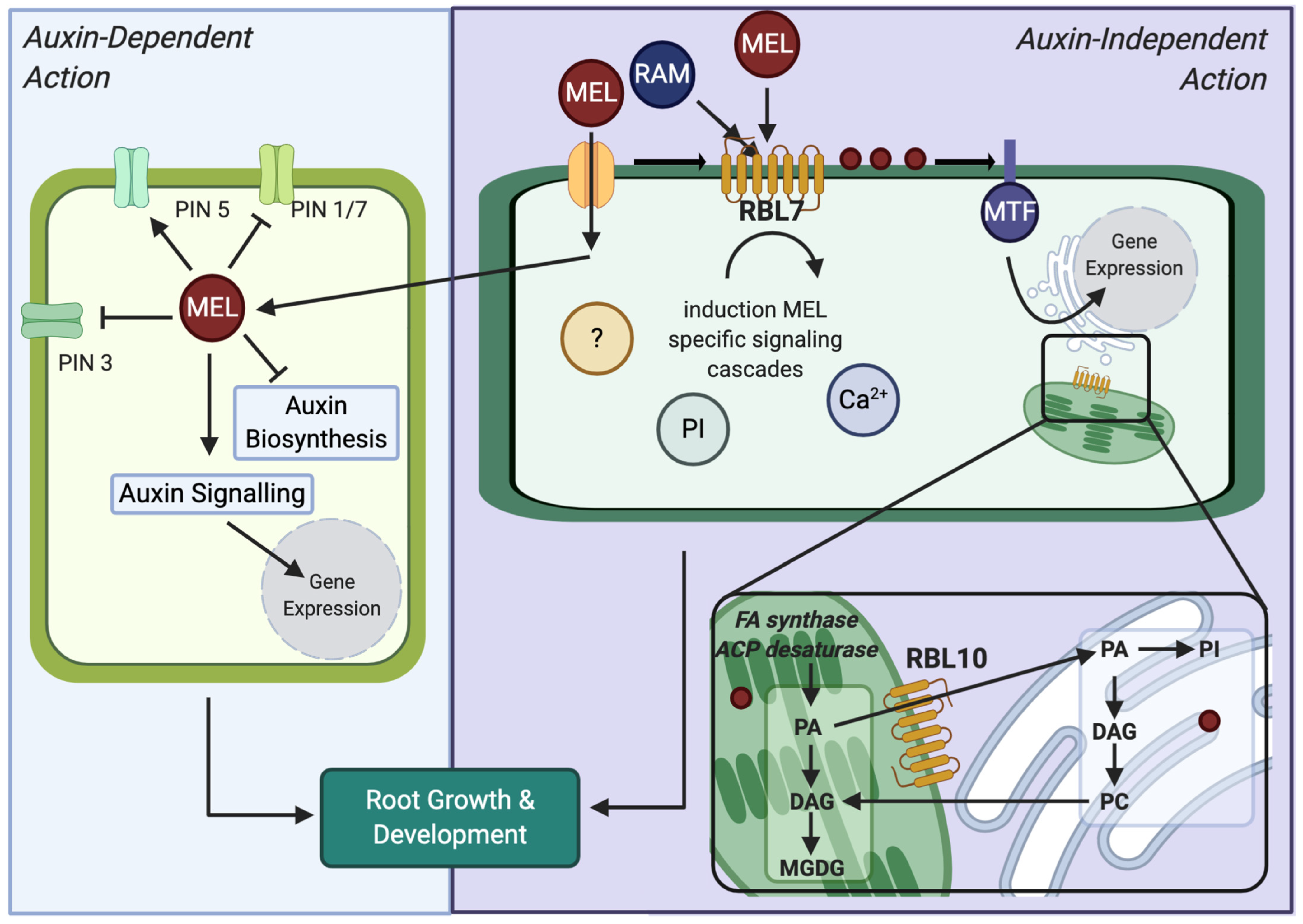
3.4. Identification of RBL7 as a Candidate for a MEL Interacting Protein
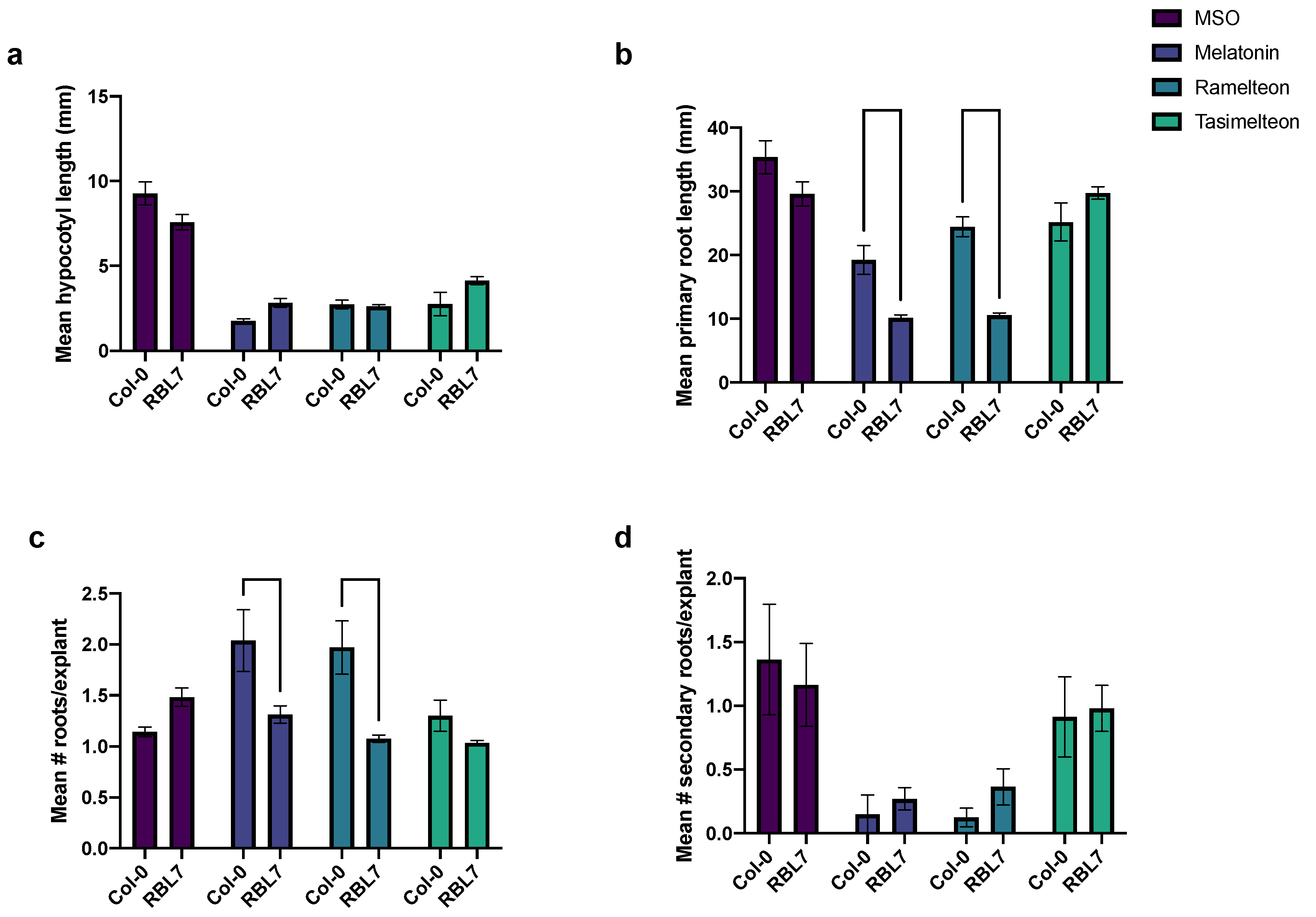
3.5. Effects of MEL and Agonist Treatment in rbl7 Mutants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murch, S.J.; Simmons, C.B.; Saxena, P.K. Melatonin in feverfew and other medicinal plants. Lancet 1997, 350, 1598–1599. [Google Scholar] [CrossRef]
- Murch, S.J.; Erland, L.A.E. A Systematic Review of Melatonin in Plants: An example of evolution of literature. Front. Plant Sci. 2021, 12, 683047. [Google Scholar] [CrossRef] [PubMed]
- Pelagio-Flores, R.; Munoz-Parra, E.; Ortiz-Castro, R.; Lopez-Bucio, J. Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling. J. Pineal Res. 2012, 53, 279–288. [Google Scholar] [CrossRef]
- Ren, S.; Rutto, L.; Katuuramu, D. Melatonin acts synergistically with auxin to promote lateral root development through fine tuning auxin transport in Arabidopsis thaliana. PLoS ONE 2019, 14, e0221687. [Google Scholar]
- Bajwa, V.S.; Shukla, M.R.; Sherif, S.M.; Murch, S.J.; Saxena, P.K. Role of melatonin in alleviating cold stress in Arabidopsis thaliana. J. Pineal Res. 2014, 56, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Reiter, R.J.; Tan, D.-X.; Zhou, Z.; Cruz, M.H.C.; Fuentes-Broto, L.; Galano, A. Phytomelatonin: Assisting plants to survive and thrive. Molecules 2015, 20, 7396–7437. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.; Li, D.-X.; Zhang, J.-R.; Shan, C.; Rengel, Z.; Song, Z.-B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef]
- Wang, L.-F.; Lu, K.-K.; Li, T.-T.; Zhang, Y.; Guo, J.-X.; Song, R.-F.; Liu, W.-C. Maize Phytomelatonin Receptor1 functions in plant tolerance to osmotic and drought stress. J. Exp. Bot. 2021, erab553. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Murch, S.J.; Reiter, R.J.; Saxena, P.K. A new balancing act: The many roles of melatonin and serotonin in plant growth and development. Plant Signal. Behav. 2015, 10, e1096469-15. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Sun, Q.; Zhang, H.; Cao, Y.; Weeda, S.; Ren, S.; Guo, Y.-D. Roles of melatonin in abiotic stress resistance in plants. J. Exp. Bot. 2015, 66, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Erland, L.A.E.; Yasunaga, A.; Li, I.T.S.; Murch, S.J.; Saxena, P.K. Direct visualization of location and uptake of applied melatonin and serotonin in living tissues and their redistribution in plants in response to thermal stress. J. Pineal Res. 2019, 66, e12527. [Google Scholar] [CrossRef] [Green Version]
- Erland, L.A.E.; Shukla, M.R.; Singh, A.S.; Murch, S.J.; Saxena, P.K. Melatonin and serotonin: Mediators in the symphony of plant morphogenesis. J. Pineal Res. 2018, 64, e12452. [Google Scholar] [CrossRef] [PubMed]
- Murch, S.J.; Saxena, P.K. Role of indoleamines in regulation of morphogenesis in in vitro cultures of St. John’s wort (Hypericum perforatum L.). Acta Hortic. 2004, 629, 425–432. [Google Scholar] [CrossRef]
- Li, H.; Chang, J.; Zheng, J.; Dong, Y.; Liu, Q.; Yang, X.; Wei, C.; Zhang, Y.; Ma, J.; Zhang, X. Local melatonin application induces cold tolerance in distant organs of Citrullus lanatus L. via long distance transport. Sci. Rep. 2017, 7, 40858. [Google Scholar] [CrossRef] [Green Version]
- Vafadar, F.; Amooaghaie, R.; Ehsanzadeh, P.; Ghanati, F.; Sajedi, R.H. Crosstalk between melatonin and Ca2+/CaM evokes systemic salt tolerance in Dracocephalum kotschyi. J. Plant Physiol. 2020, 252, 153237. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Labani, N.; Cecon, E.; Jockers, R. Melatonin Target Proteins: Too many or not enough? Front. Endocrinol. 2019, 10, 791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jockers, R.; Delagrange, P.; Dubocovich, M.L.; Markus, R.P.; Renault, N.; Tosini, G.; Cecon, E.; Zlotos, D.P. Update on melatonin receptors: IUPHAR Review 20. Br. J. Pharm. 2016, 173, 2702–2725. [Google Scholar] [CrossRef]
- Emet, M.; Ozcan, H.; Ozel, L.; Yayla, M.; Halici, Z.; Hacimuftuoglu, A. A review of melatonin, its receptors and drugs. Eurasian J. Med. 2016, 48, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Pala, D.; Lodola, A.; Bedini, A.; Spadoni, G.; Rivara, S. Homology Models of Melatonin Receptors: Challenges and recent advances. Int. J. Mol. Sci. 2013, 14, 8093–8121. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, M. Pharmacology of ramelteon, a selective MT1/MT2 receptor agonist: A novel therapeutic drug for sleep disorders. CNS Neurosci. 2009, 15, 32–51. [Google Scholar] [CrossRef] [Green Version]
- Lavedan, C.; Forsberg, M.; Gentile, A.J. Tasimelteon: A selective and unique receptor binding profile. Neuropharmacol 2015, 91, 142–147. [Google Scholar] [CrossRef] [Green Version]
- Legros, C.; Devavry, S.; Caignard, S.; Tessier, C.; Delagrange, P.; Ouvry, C.; Boutin, J.A.; Nosjean, O. Melatonin MT1 and MT2 receptors display different molecular pharmacologies only in the G-protein coupled state. Br. J. Pharm. 2013, 171, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Dubocovich, M.L.; Masana, M.I.; Iacob, S.; Sauri, D.M. Melatonin receptor antagonists that differentiate between the human Mel1a and Mel1b recombinant subtypes are used to assess the pharmacological profile of the rabbit retina ML1 presynaptic heteroreceptor. Naunyn-Schmiedeberg’s Arch. Pharm. 1997, 355, 365–375. [Google Scholar] [CrossRef]
- Constantinescu, C.S.; Hilliard, B.; Ventura, E.; Rostami, A. Luzindole, a melatonin receptor antagonist, suppresses experimental autoimmune encephalomyelitis. Pathobiol 1997, 65, 190–194. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Murch, S.J.; Campbell, S.S.B.; Saxena, P.K. The role of serotonin and melatonin in plant morphogenesis: Regulation of auxin-induced root organogenesis in in vitro-cultured explants of St. John’s Wort (Hypericum perforatum L.). Vitr. Cell. Dev. Biol.—Plant 2001, 37, 786–793. [Google Scholar] [CrossRef]
- Erland, L.A.E.; Chattopadhyay, A.; Jones, A.M.P.; Saxena, P.K. Melatonin in Plants and Plant Culture Systems: Variability, stability and efficient quantification. Front. Plant Sci. 2016, 7, 1721. [Google Scholar] [CrossRef]
- Gao, Y.H.; Yu, Y.; Hu, X.G.; Cao, Y.J.; Wu, J.Z. Imaging of jasmonic acid binding sites in tissue. Anal. Biochem. 2013, 440, 205–211. [Google Scholar] [CrossRef]
- Whiteside, M.D.; Treseder, K.K.; Atsatt, P.R. The brighter side of soils: Quantum dots track organic nitrogen through fungi and plants. Ecology 2009, 90, 100–108. [Google Scholar] [CrossRef] [Green Version]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; et al. ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef]
- Saremba, B.M.; Tymm, F.J.M.; Baethke, K.; Rheault, M.; Sherif, S.; Saxena, P.K.; Murch, S.J. Plant signals during beetle (Scolytus multistriatus) feeding in American elm (Ulmus americana Planch). Plant Signal. Behav. 2017, 12, e1296997. [Google Scholar] [CrossRef] [Green Version]
- The Uniprot Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Back, K. Mitogen-activated protein kinase pathways are required for melatonin-mediated defense responses in plants. J. Pineal Res. 2016, 60, 327–335. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin is required for H2O2- and NO-mediated defense signaling through MAPKKK3 and OXI1 in Arabidopsis thaliana. J. Pineal Res. 2017, 62, e12379. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. The phytomelatonin receptor (PMRT1) Arabidopsis Cand2 is not a bona fide G protein–coupled melatonin receptor. Melatonin Res. 2020, 3, 177–186. [Google Scholar] [CrossRef]
- Sliwiak, J.; Dauter, Z.; Jaskolski, M. Crystal structure of Hyp-1, a Hypericum perforatum PR-10 protein, in complex with melatonin. Front. Plant Sci. 2016, 7, 668. [Google Scholar] [CrossRef] [Green Version]
- Konstantinova, N.; Korbei, B.; Luschnig, C. Auxin and Root Gravitropism: Addressing basic cellular processes by exploiting a defined growth response. Int. J. Mol. Sci. 2021, 22, 2749. [Google Scholar] [CrossRef]
- Stauch, B.; Johansson, L.C.; McCorvy, J.D.; Patel, N.; Han, G.W.; Huang, X.-P.; Gati, C.; Batyuk, A.; Slocum, S.T.; Ishchenko, A.; et al. Structural basis of ligand recognition at the human MT1 melatonin receptor. Nature 2019, 569, 284–288. [Google Scholar] [CrossRef]
- Urban, S. Rhomboid proteins: Conserved membrane proteases with divergent biological functions. Gene Dev. 2006, 20, 3054–3068. [Google Scholar] [CrossRef] [Green Version]
- Knopf, R.R.; Adam, Z. Rhomboid proteases in plants—Still in square one? Physiol. Plant. 2012, 145, 41–51. [Google Scholar] [CrossRef]
- Adamiec, M.; Ciesielska, M.; Zalaś, P.; Luciński, R. Arabidopsis thaliana intramembrane proteases. Acta Physiol. Plant 2017, 39, 146. [Google Scholar] [CrossRef] [Green Version]
- Lemberg, M.K.; Freeman, M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007, 17, 1634–1646. [Google Scholar] [CrossRef] [Green Version]
- Seo, P.J.; Kim, S.-G.; Park, C.-M. Membrane-bound transcription factors in plants. Trend Plant Sci. 2008, 13, 550–556. [Google Scholar] [CrossRef]
- Weeda, S.; Na Zhang, N.; Zhao, X.; Ndip, G.; Guo, Y.; Buck, G.A.; Fu, C.; Ren, S. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE 2014, 9, e93462. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; You, J.; Li, J.; Wang, Y.; Chan, Z. Melatonin promotes Arabidopsis primary root growth in an IAA-dependent manner. J. Exp. Bot. 2021, 72, 5599–5611. [Google Scholar] [CrossRef]
- Kreutzberger, A.J.B.; Ji, M.; Aaron, J.; Mihaljević, L.; Urban, S. Rhomboid distorts lipids to break the viscosity-imposed speed limit of membrane diffusion. Science 2019, 363, eaao0076. [Google Scholar] [CrossRef]
- Lavell, A.; Froehlich, J.E.; Baylis, O.; Rotondo, A.D.; Benning, C. A predicted plastid rhomboid protease affects phosphatidic acid metabolism in Arabidopsis thaliana. Plant J. 2019, 99, 978–987. [Google Scholar]
- Yu, Y.; Wang, A.; Li, X.; Kou, M.; Wang, W.; Chen, X.; Xu, T.; Zhu, M.; Ma, D.; Li, Z.; et al. Melatonin-stimulated triacylglycerol breakdown and energy turnover under salinity stress contributes to the maintenance of plasma membrane H+-ATPase activity and K+/Na+ homeostasis in sweet potato. Front. Plant Sci. 2018, 9, 256. [Google Scholar] [CrossRef] [Green Version]
- Chandok, M.R.; Sopory, S.K. 5-Hydroxytryptamine affects turnover of polyphosphoinositides in maize and stimulates nitrate reductase in the absence of light. FEBS Lett. 1994, 356, 39–42. [Google Scholar] [CrossRef] [Green Version]
- Raghuram, N.; Sopory, S.K. Evidence for some common signal-transduction events for opposite regulation of nitrate reductase and phytochrome-I gene-expression by light. Plant Mol. Biol. 1995, 29, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, J.A.; Erland, L.A.; Shipley, P.R.; Murch, S.J. Plant Perception of Light: The role of indoleamines in Scutellaria species. Melatonin Res. 2020, 3, 161–176. [Google Scholar] [CrossRef]
- Sanchez-Barcelo, E.J.; Mediavilla, M.D.; Vriend, J.; Reiter, R.J. COP1 and COP9 signalosome, evolutionarily conserved photomorphogenic proteins as possible targets of melatonin. J. Pineal Res. 2016, 61, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Li, A.; Yu, H.; Li, W.; Liang, C.; Guo, S.; Zhang, R.; Chu, C. Melatonin regulates root architecture by modulating auxin response in rice. Front. Plant Sci. 2017, 8, 134. [Google Scholar] [CrossRef] [Green Version]
- Manchester, L.C.; Coto-Montes, A.; Boga, J.A.; Andersen, L.P.H.; Zhou, Z.; Galano, A.; Vriend, J.; Tan, D.-X.; Reiter, R.J. Melatonin: An ancient molecule that makes oxygen metabolically tolerable. J. Pineal Res. 2015, 59, 403–419. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Description | Sequence |
|---|---|---|
| HN48F | Gene-Specific Primer | 5′-ACAGTCCTAAAATCTCAAACCCAG-3′ |
| 8904R | TDNA Primer | 5′-ATATTGACCATCATACTCATTGC-3′ |
| 479R | WT Primer | 5′-GCACAATTCAACATGTTTCCA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erland, L.A.E.; Dumigan, C.R.; Forsyth, J.A.; Frolova, L.; Yasunaga, A.B.; Pun, W.; Li, I.T.S.; Deyholos, M.K.; Murch, S.J. Mammalian Melatonin Agonist Pharmaceuticals Stimulate Rhomboid Proteins in Plants. Biomolecules 2022, 12, 882. https://doi.org/10.3390/biom12070882
Erland LAE, Dumigan CR, Forsyth JA, Frolova L, Yasunaga AB, Pun W, Li ITS, Deyholos MK, Murch SJ. Mammalian Melatonin Agonist Pharmaceuticals Stimulate Rhomboid Proteins in Plants. Biomolecules. 2022; 12(7):882. https://doi.org/10.3390/biom12070882
Chicago/Turabian StyleErland, Lauren A. E., Christopher R. Dumigan, Jillian A. Forsyth, Liubov Frolova, Adam B. Yasunaga, Winnie Pun, Isaac T. S. Li, Michael K. Deyholos, and Susan J. Murch. 2022. "Mammalian Melatonin Agonist Pharmaceuticals Stimulate Rhomboid Proteins in Plants" Biomolecules 12, no. 7: 882. https://doi.org/10.3390/biom12070882
APA StyleErland, L. A. E., Dumigan, C. R., Forsyth, J. A., Frolova, L., Yasunaga, A. B., Pun, W., Li, I. T. S., Deyholos, M. K., & Murch, S. J. (2022). Mammalian Melatonin Agonist Pharmaceuticals Stimulate Rhomboid Proteins in Plants. Biomolecules, 12(7), 882. https://doi.org/10.3390/biom12070882









