Host-Genome Similarity Characterizes the Adaption of SARS-CoV-2 to Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. SARS-CoV-2 Genomes
2.2. Host-Genome-Similarity (HGS) Definition
- Prepare the subgenomic CDS sequence (such as the sequence of ORF 6) of virus strain.
- Choose human genomic database (Homo sapiens GRCh38.p12 chromosomes) database.
- Set BLASTn program parameters. The expect threshold as 10, word size as 11, penalty for a nucleotide mismatch as −3, reward for a nucleotide match as 2, gap costs are chosen as existence = 5 and extension = 2, max matches in a query range as 1.
- Run the BLASTn matching analysis.
- The HGS value is obtained through the matching scores produced by BLASTn analysis and the length of target sequence.
3. Results and Discussions
3.1. SARS-CoV-2 Delta Variant Shares More Genomic Similarity to Human
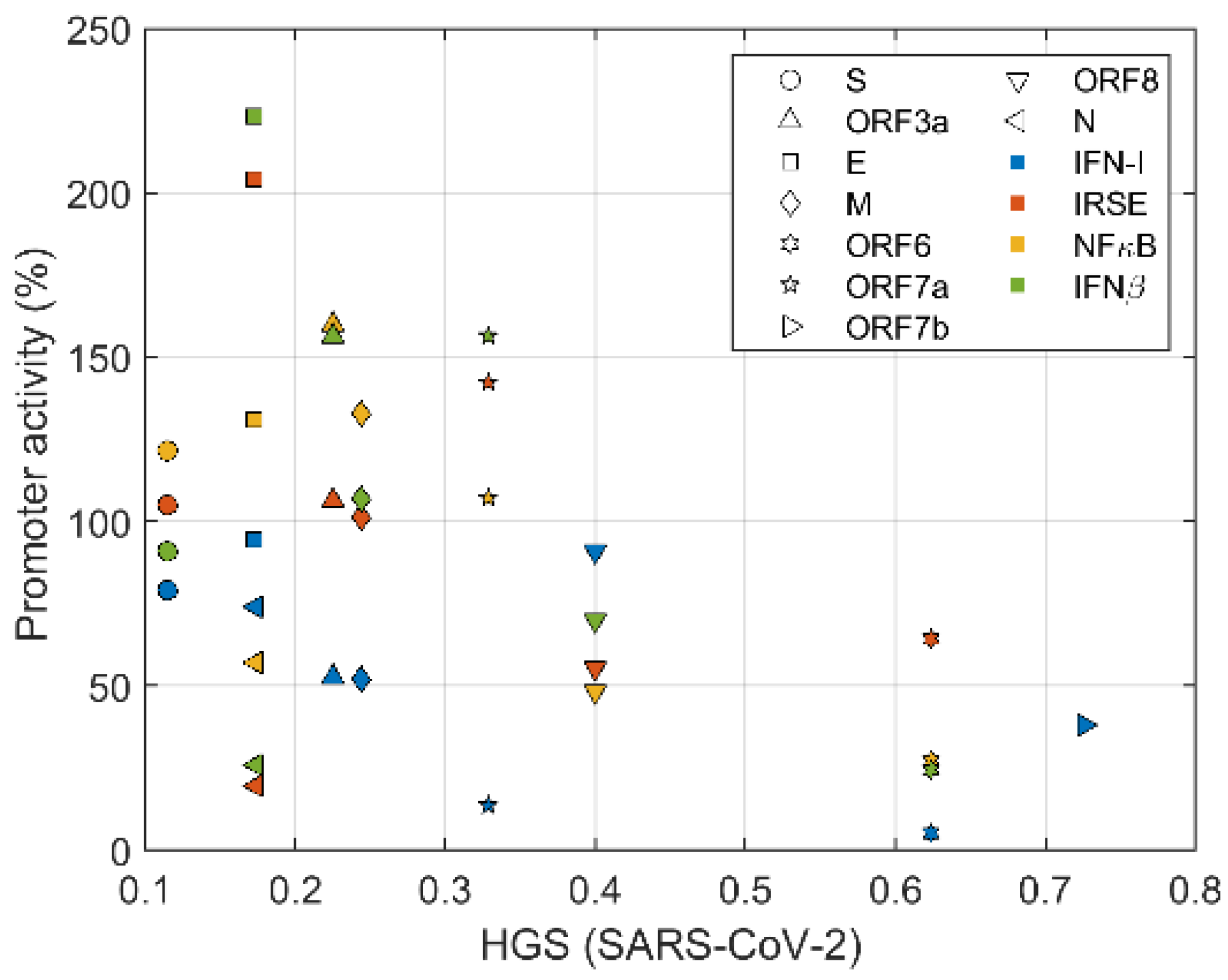
3.2. Virus Genes Related in Suppressing Innate Immune Tend to Have High HGS
3.3. Suppression of IFN-I Signaling by ORFs with Different HGS Values
3.4. The High HGS Mutation (ACT > ATT) Is Shared by Alpha, Delta and Omicron Variants
4. Conclusions
- There is correlation between the HGS and the inhibition of antivirus productions, which is a direct indicator of the virus to inhibit interferon synthesis.
- ORF 7b and ORF 8 of SARS-CoV-2 have increased HGS in the last 2 years, reaching 114% and 110% of the original value, respectively.
- By HGS and mutation analysis of genomes of Alpha, Delta, and Omicron variants, it is found that the mutation ACT > ATT is commonly shared in the high HGS strain population.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020, bioRxiv:2020.01.22.914952. [Google Scholar]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.; et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv 2020, medRxiv:2020.02.06.20020974. [Google Scholar] [CrossRef]
- Chang, Y.; Ma, J.; Zhang, M.; Yu, Y. Preliminary study on genome homology of viruses and human. J. Norman Bethune Univ. Med. Sci. 1997, 23, 242–244. [Google Scholar]
- Senkevich, T.G.; Bugert, J.J.; Sisler, J.R.; Koonin, E.V.; Darai, G.; Moss, B. Genome sequence of a human tumorigenic poxvirus: Prediction of specific host response-evasion genes. Science 1996, 273, 813–816. [Google Scholar] [CrossRef]
- Vilchez, R.A.; Butel, J.S. Emergent human pathogen simian virus 40 and its role in cancer. Clin. Microbiol. Rev. 2004, 17, 495–508. [Google Scholar] [CrossRef]
- Martini, F.; Corallini, A.; Balatti, V.; Sabbioni, S.; Pancaldi, C.; Tognon, M. Simian virus 40 in humans. Infect. Agents Cancer 2007, 2, 13. [Google Scholar] [CrossRef]
- Fiers, W.; Contreras, R.; Haegeman, G.; Rogiers, R.; Van de Voorde, A.; Van Heuverswyn, H.; Van Herreweghe, J.; Volckaert, G.; Ysebaert, M. Complete nucleotide sequence of SV40 DNA. Nature 1978, 273, 113–120. [Google Scholar] [CrossRef]
- Rosenberg, M.; Segal, S.; Kuff, E.L.; Singer, M.F. The nucleotide sequence of repetitive monkey DNA found in defective simian virus 40. Cell 1977, 11, 845–857. [Google Scholar] [CrossRef]
- Shackelton, L.A.; Holmes, E.C. The evolution of large DNA viruses: Combining genomic information of viruses and their hosts. Trends Microbiol. 2004, 12, 458–465. [Google Scholar] [CrossRef]
- Enard, D.; Petrov, D.A. Evidence that RNA Viruses Drove Adaptive Introgression between Neanderthals and Modern Humans. Cell 2018, 175, 360–371.e313. [Google Scholar] [CrossRef] [PubMed]
- Enard, D.; Cai, L.; Gwennap, C.; Petrov, D.A. Viruses are a dominant driver of protein adaptation in mammals. eLife 2016, 5, e12469. [Google Scholar] [CrossRef] [PubMed]
- Raftery, M.; Müller, A.; Schönrich, G. Herpesvirus Homologues of Cellular Genes. In Molecular Evolution of Viruses—Past and Present: Evolution of Viruses by Acquisition of Cellular RNA and DNA; Becker, Y., Darai, G., Eds.; Springer: Boston, MA, USA, 2000; pp. 65–75. [Google Scholar]
- Fleming, S.B.; Haig, D.M.; Nettleton, P.; Reid, H.W.; McCaughan, C.A.; Wise, L.M.; Mercer, A.A. Sequence and Functional Analysis of a Homolog of Interleukin-10 Encoded by the Parapoxvirus Orf Virus. In Molecular Evolution of Viruses—Past and Present: Evolution of Viruses by Acquisition of Cellular RNA and DNA; Becker, Y., Darai, G., Eds.; Springer: Boston, MA, USA, 2000; pp. 85–95. [Google Scholar]
- Spencer, J.V.; Lockridge, K.M.; Barry, P.A.; Lin, G.; Tsang, M.; Penfold, M.E.T.; Schall, T.J. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 2002, 76, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Powell, P.P.; Dixon, L.K.; Parkhouse, R.M. An IkappaB homolog encoded by African swine fever virus provides a novel mechanism for downregulation of proinflammatory cytokine responses in host macrophages. J. Virol. 1996, 70, 8527–8533. [Google Scholar] [CrossRef] [PubMed]
- Revilla, Y.; Callejo, M.; Rodrıguez, J.M.; Culebras, E.; Nogal, M.L.; Salas, M.L.; Viñuela, E.; Fresno, M. Inhibition of Nuclear Factor κB Activation by a Virus-encoded IκB-like Protein. J. Biol. Chem. 1998, 273, 5405–5411. [Google Scholar] [CrossRef]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Peñaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-h.; et al. Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome. Science 2003, 300, 1394. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats Are Natural Reservoirs of SARS-Like Coronaviruses. Science 2005, 310, 676. [Google Scholar] [CrossRef]
- Yount, B.; Roberts, R.S.; Sims, A.C.; Deming, D.; Frieman, M.B.; Sparks, J.; Denison, M.R.; Davis, N.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus Group-Specific Open Reading Frames Encode Nonessential Functions for Replication in Cell Cultures and Mice. J. Virol. 2005, 79, 14909. [Google Scholar] [CrossRef]
- Haan, C.A.M.d.; Masters, P.S.; Shen, X.; Weiss, S.; Rottier, P.J.M. The Group-Specific Murine Coronavirus Genes Are Not Essential, but Their Deletion, by Reverse Genetics, Is Attenuating in the Natural Host. Virology 2002, 296, 177–189. [Google Scholar] [CrossRef]
- Haijema, B.J.; Volders, H.; Rottier, P.J.M. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004, 78, 3863–3871. [Google Scholar] [CrossRef]
- Hodgson, T.; Britton, P.; Cavanagh, D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006, 80, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Casais, R.; Davies, M.; Cavanagh, D.; Britton, P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005, 79, 8065–8078. [Google Scholar] [CrossRef] [PubMed]
- Frieman, M.; Yount, B.; Heise, M.; Kopecky-Bromberg, S.A.; Palese, P.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J. Virol. 2007, 81, 9812. [Google Scholar] [CrossRef] [PubMed]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.-L.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Obitsu, S.; Ahmed, N.; Nishitsuji, H.; Hasegawa, A.; Nakahama, K.-i.; Morita, I.; Nishigaki, K.; Hayashi, T.; Masuda, T.; Kannagi, M. Potential enhancement of osteoclastogenesis by severe acute respiratory syndrome coronavirus 3a/X1 protein. Arch. Virol. 2009, 154, 1457–1464. [Google Scholar] [CrossRef]
- Kanzawa, N.; Nishigaki, K.; Hayashi, T.; Ishii, Y.; Furukawa, S.; Niiro, A.; Yasui, F.; Kohara, M.; Morita, K.; Matsushima, K.; et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-κB activation. FEBS Lett. 2006, 580, 6807–6812. [Google Scholar] [CrossRef]
- McBride, R.; Fielding, B.C. The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis. Viruses 2012, 4, 2902–2923. [Google Scholar] [CrossRef]
- Nelemans, T.; Kikkert, M. Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections. Viruses 2019, 11, 961. [Google Scholar] [CrossRef]
- Totura, A.L.; Baric, R.S. SARS coronavirus pathogenesis: Host innate immune responses and viral antagonism of interferon. Curr. Opin. Virol. 2012, 2, 264–275. [Google Scholar] [CrossRef]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.-Y.R.; Lui, P.-Y.; Jin, D.-Y. A molecular arms race between host innate antiviral response and emerging human coronaviruses. Virol. Sin. 2016, 31, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557. [Google Scholar] [CrossRef]
- Ferran, M.C.; Skuse, G.R. Evasion of Host Innate Immunity by Emerging Viruses: Antagonizing Host RIG-I Pathways. J. Emerg. Dis. Virol. 2017, 3, 1–8. [Google Scholar]
- Zhang, Q.; Yoo, D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016, 226, 128–141. [Google Scholar] [CrossRef]
- Netland, J.; Ferraro, D.; Pewe, L.; Olivares, H.; Gallagher, T.; Perlman, S. Enhancement of Murine Coronavirus Replication by Severe Acute Respiratory Syndrome Coronavirus Protein 6 Requires the N-Terminal Hydrophobic Region but Not C-Terminal Sorting Motifs. J. Virol. 2007, 81, 11520. [Google Scholar] [CrossRef] [PubMed]
- Pewe, L.; Zhou, H.; Netland, J.; Tangadu, C.; Olivares, H.; Shi, L.; Look, D.; Gallagher, T.; Perlman, S. A SARS-CoV-specific protein enhances virulence of an attenuated strain of mouse hepatitis virus. Adv. Exp. Med. Biol. 2006, 581, 493–498. [Google Scholar]
- Kumar, P.; Gunalan, V.; Liu, B.; Chow, V.T.K.; Druce, J.; Birch, C.; Catton, M.; Fielding, B.C.; Tan, Y.-J.; Lal, S.K. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology 2007, 366, 293–303. [Google Scholar] [CrossRef]
- Gunalan, V.; Mirazimi, A.; Tan, Y.J. A putative diacidic motif in the SARS-CoV ORF6 protein influences its subcellular localization and suppression of expression of co-transfected expression constructs. BMC Res. Notes 2011, 4, 446. [Google Scholar] [CrossRef]
- Zhao, J.; Falcón, A.; Zhou, H.; Netland, J.; Enjuanes, L.; Breña, P.P.; Perlman, S. Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication. J. Virol. 2009, 83, 2368–2373. [Google Scholar] [CrossRef]
- Geng, H.; Liu, Y.-M.; Chan, W.-S.; Lo, A.W.-I.; Au, D.M.-Y.; Waye, M.M.-Y.; Ho, Y.-Y. The putative protein 6 of the severe acute respiratory syndrome-associated coronavirus: Expression and functional characterization. FEBS Lett. 2005, 579, 6763–6768. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Feng, Y.; Chen, H.; Luk, H.K.H.; Yang, W.-H.; Li, K.S.M.; Zhang, Y.-Z.; Huang, Y.; Song, Z.-Z.; Chow, W.-N.; et al. Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination. J. Virol. 2015, 89, 10532–10547. [Google Scholar] [CrossRef] [PubMed]
- Keng, C.-T.; Choi, Y.-W.; Welkers, M.R.A.; Chan, D.Z.L.; Shen, S.; Gee Lim, S.; Hong, W.; Tan, Y.-J. The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells. Virology 2006, 354, 132–142. [Google Scholar] [CrossRef]
- Wong, H.H.; Fung, T.S.; Fang, S.; Huang, M.; Le, M.T.; Liu, D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 2018, 515, 165–175. [Google Scholar] [CrossRef]
- Shi, C.-S.; Nabar, N.R.; Huang, N.-N.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019, 5, 101. [Google Scholar] [CrossRef]
- Rothenburg, S.; Brennan, G. Species-Specific Host–Virus Interactions: Implications for Viral Host Range and Virulence. Trends Microbiol. 2020, 28, 46–56. [Google Scholar] [CrossRef]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef]
- Marra, M.A.; Jones, S.J.M.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.N.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y.; et al. The Genome Sequence of the SARS-Associated Coronavirus. Science 2003, 300, 1399. [Google Scholar] [CrossRef]
- Narayanan, K.; Maeda, A.; Maeda, J.; Makino, S. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 2000, 74, 8127–8134. [Google Scholar] [CrossRef]
- Karlin, S.; Altschul, S.F. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc. Natl. Acad. Sci. USA 1990, 87, 2264–2268. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sun, W. Discovering SARS-CoV-2 genes and mutations adapted for humans in 2594 genomes. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Virtually, 9–12 December 2021; pp. 2566–2573. [Google Scholar]
- Sun, W. The discovery of gene mutations making SARS-CoV-2 well adapted for humans: Host-genome similarity analysis of 2594 genomes from China, the USA and Europe. bioRxiv 2020, bioRxiv:2020.09.03.280727. [Google Scholar]
- Hansen, C.H.; Schelde, A.B.; Moustsen-Helm, I.R.; Emborg, H.-D.; Krause, T.G.; Mølbak, K.; Valentiner-Branth, P.; on behalf of the Infectious Disease Preparedness Group at Statens Serum. Vaccine effectiveness against SARS-CoV-2 infection with the Omicron or Delta variants following a two-dose or booster BNT162b2 or mRNA-1273 vaccination series: A Danish cohort study. medRxiv 2021, medRxiv:2021.12.20.21267966. [Google Scholar]
- Miller, N.L.; Clark, T.; Raman, R.; Sasisekharan, R. Insights on the mutational landscape of the SARS-CoV-2 Omicron variant receptor-binding domain. Cell Rep. Med. 2022, 3, 100527. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Liu, H.; Wu, N.C.; Lee, C.-C.D.; Zhu, X.; Zhao, F.; Huang, D.; Yu, W.; Hua, Y.; Tien, H.; et al. Structural basis of a shared antibody response to SARS-CoV-2. Science 2020, 369, 1119–1123. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv 2021, bioRxiv:2021.01.18.427166. [Google Scholar] [CrossRef]
- Pulliam, J.R.C.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv 2021, medRxiv:2021.11.11.21266068. [Google Scholar]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Dosch, S.F.; Mahajan, S.D.; Collins, A.R. SARS coronavirus spike protein-induced innate immune response occurs via activation of the NF-κB pathway in human monocyte macrophages in vitro. Virus Res. 2009, 142, 19–27. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Usera, F.; Enjuanes, L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014, 194, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Akira, S. Regulation of innate immune responses by autophagy-related proteins. J. Cell Biol. 2010, 189, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus–host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liao, C.H.; Wang, Q.; Tan, Y.J.; Luo, R.; Qiu, Y.; Ge, X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated Type I Interferon and Inflammatory Monocyte-Macrophage Responses Cause Lethal Pneumonia in SARS-CoV-Infected Mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef]
- Thiel, V.; Weber, F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 2008, 19, 121–132. [Google Scholar] [CrossRef]
- Pichlmair, A.; Reis e Sousa, C. Innate Recognition of Viruses. Immunity 2007, 27, 370–383. [Google Scholar] [CrossRef]
- Payne, S. Chapter 6—Immunity and Resistance to Viruses. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 61–71. [Google Scholar]
- Randall, R.E.; Goodbourn, S. Interferons and viruses: An interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 2008, 89, 1–47. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immun. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I Inteferon Gene Induction by the Interferon Regulatory Factor Family of Transcription Factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Kindler, E.; Thiel, V.; Weber, F. Interaction of SARS and MERS Coronaviruses with the Antiviral Interferon Response. Adv. Virus Res. 2016, 96, 219–243. [Google Scholar] [CrossRef]
- Häcker, H.; Tseng, P.-H.; Karin, M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immun. 2011, 11, 457–468. [Google Scholar] [CrossRef]
- Cao, X. New DNA-sensing pathway feeds RIG-I with RNA. Nat. Immun. 2009, 10, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Takeda, K. Current views of toll-like receptor signaling pathways. Gastroenterol. Res. Pract. 2010, 2010, 240365. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Schwartz, O. (Eds.) HIV Interactions with Dendritic Cells Infection and Immunity; Springer: New York, NY, USA, 2013. [Google Scholar]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5′-Triphosphate RNA Is the Ligand for RIG-I. Science 2006, 314, 994. [Google Scholar] [CrossRef]
- Pichlmair, A.; Schulz, O.; Tan, C.P.; Näslund, T.I.; Liljeström, P.; Weber, F.; Reis e Sousa, C. RIG-I-Mediated Antiviral Responses to Single-Stranded RNA Bearing 5′-Phosphates. Science 2006, 314, 997. [Google Scholar] [CrossRef]
- Abbas, Y.M.; Pichlmair, A.; Górna, M.W.; Superti-Furga, G.; Nagar, B. Structural basis for viral 5’-PPP-RNA recognition by human IFIT proteins. Nature 2013, 494, 60–64. [Google Scholar] [CrossRef]
- Wu, B.; Peisley, A.; Richards, C.; Yao, H.; Zeng, X.; Lin, C.; Chu, F.; Walz, T.; Hur, S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 2013, 152, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Takeuchi, O.; Mikamo-Satoh, E.; Hirai, R.; Kawai, T.; Matsushita, K.; Hiiragi, A.; Dermody, T.S.; Fujita, T.; Akira, S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid–inducible gene-I and melanoma differentiation–associated gene 5. J. Exp. Med. 2008, 205, 1601–1610. [Google Scholar] [CrossRef] [PubMed]
- Pichlmair, A.; Schulz, O.; Tan, C.-P.; Rehwinkel, J.; Kato, H.; Takeuchi, O.; Akira, S.; Way, M.; Schiavo, G.; Reis e Sousa, C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009, 83, 10761–10769. [Google Scholar] [CrossRef] [PubMed]
- Zielecki, F.; Weber, M.; Eickmann, M.; Spiegelberg, L.; Zaki, A.M.; Matrosovich, M.; Becker, S.; Weber, F. Human cell tropism and innate immune system interactions of human respiratory coronavirus EMC compared to those of severe acute respiratory syndrome coronavirus. J. Virol. 2013, 87, 5300–5304. [Google Scholar] [CrossRef]
- Spiegel, M.; Pichlmair, A.; Martínez-Sobrido, L.; Cros, J.; García-Sastre, A.; Haller, O.; Weber, F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 2005, 79, 2079–2086. [Google Scholar] [CrossRef]
- Pewe, L.; Zhou, H.; Netland, J.; Tangudu, C.; Olivares, H.; Shi, L.; Look, D.; Gallagher, T.; Perlman, S. A severe acute respiratory syndrome-associated coronavirus-specific protein enhances virulence of an attenuated murine coronavirus. J. Virol. 2005, 79, 11335–11342. [Google Scholar] [CrossRef][Green Version]
- Chau, T.L.; Gioia, R.; Gatot, J.S.; Patrascu, F.; Carpentier, I.; Chapelle, J.P.; O’Neill, L.; Beyaert, R.; Piette, J.; Chariot, A. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem. Sci. 2008, 33, 171–180. [Google Scholar] [CrossRef]
- Hacker, H.; Karin, M. Regulation and function of IKK and IKK-related kinases. Sci. STKE Signal Transduct. Knowl. Environ. 2006, 357, re13. [Google Scholar] [CrossRef]
- Le, T.M.; Wong, H.H.; Tay, F.P.L.; Fang, S.; Keng, C.-T.; Tan, Y.J.; Liu, D.X. Expression, post-translational modification and biochemical characterization of proteins encoded by subgenomic mRNA8 of the severe acute respiratory syndrome coronavirus. FEBS J. 2007, 274, 4211–4222. [Google Scholar] [CrossRef]










| Narayanan | Marra | Rota | Start SARS | End SARS | Length SARS | Start SARS2 | End SARS2 | Length SARS2 |
|---|---|---|---|---|---|---|---|---|
| ORF1a | ORF1a | 1a | 3361 | 13413 | 10053 | 266 | 13483 | 13218 |
| N/R | N/R | N/R | 13685 | 13759 | 75 | 13685 | 13759 | 75 |
| ORF1b | ORF1b | 1b | 13398 | 21485 | 8088 | 13768 | 21555 | 7788 |
| S | S-protein | S | 21492 | 25259 | 3768 | 21536 | 25384 | 3849 |
| N/R | N/R | N/R | 25207 | 25329 | 123 | 25332 | 25448 | 117 |
| ORF3a | ORF3 | X1 | 25268 | 26092 | 825 | 25393 | 26220 | 828 |
| E | E-protein | E | 26117 | 26347 | 231 | 26245 | 26472 | 228 |
| M | M-protein | M | 26398 | 27063 | 666 | 26523 | 27191 | 669 |
| ORF6 | ORF7 | X3 | 27074 | 27265 | 192 | 27202 | 27387 | 186 |
| ORF7a | ORF8 | X4 | 27273 | 27641 | 369 | 27394 | 27759 | 366 |
| ORF7b | ORF9 | N/R | 27638 | 27772 | 135 | 27756 | 27887 | 132 |
| ORF8a | ORF10 | N/R | 27779 | 27853 | 75 | 27894 | 28259 | 366 |
| ORF8b | ORF11 | X5 | 27862 | 28116 | 255 | N/R | N/R | N/R |
| N | N-protein | N | 28118 | 29386 | 1269 | 28274 | 29533 | 1260 |
| N/R | N/R | N/R | 29413 | 29490 | 78 | 29558 | 29674 | 117 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W. Host-Genome Similarity Characterizes the Adaption of SARS-CoV-2 to Humans. Biomolecules 2022, 12, 972. https://doi.org/10.3390/biom12070972
Sun W. Host-Genome Similarity Characterizes the Adaption of SARS-CoV-2 to Humans. Biomolecules. 2022; 12(7):972. https://doi.org/10.3390/biom12070972
Chicago/Turabian StyleSun, Weitao. 2022. "Host-Genome Similarity Characterizes the Adaption of SARS-CoV-2 to Humans" Biomolecules 12, no. 7: 972. https://doi.org/10.3390/biom12070972
APA StyleSun, W. (2022). Host-Genome Similarity Characterizes the Adaption of SARS-CoV-2 to Humans. Biomolecules, 12(7), 972. https://doi.org/10.3390/biom12070972





