Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Chitosan Beads Preparation and Immobilization of Man/Cel5B
2.3. Activity of Immobilized Man/Cel5B Enzyme
2.4. Estimation of Protein Loading and Immobilization Yield
2.5. Biochemical Characterization of Immobilized Man/Cel5B
2.5.1. Effect of Temperature on Immobilized Man/Cel5B Activity
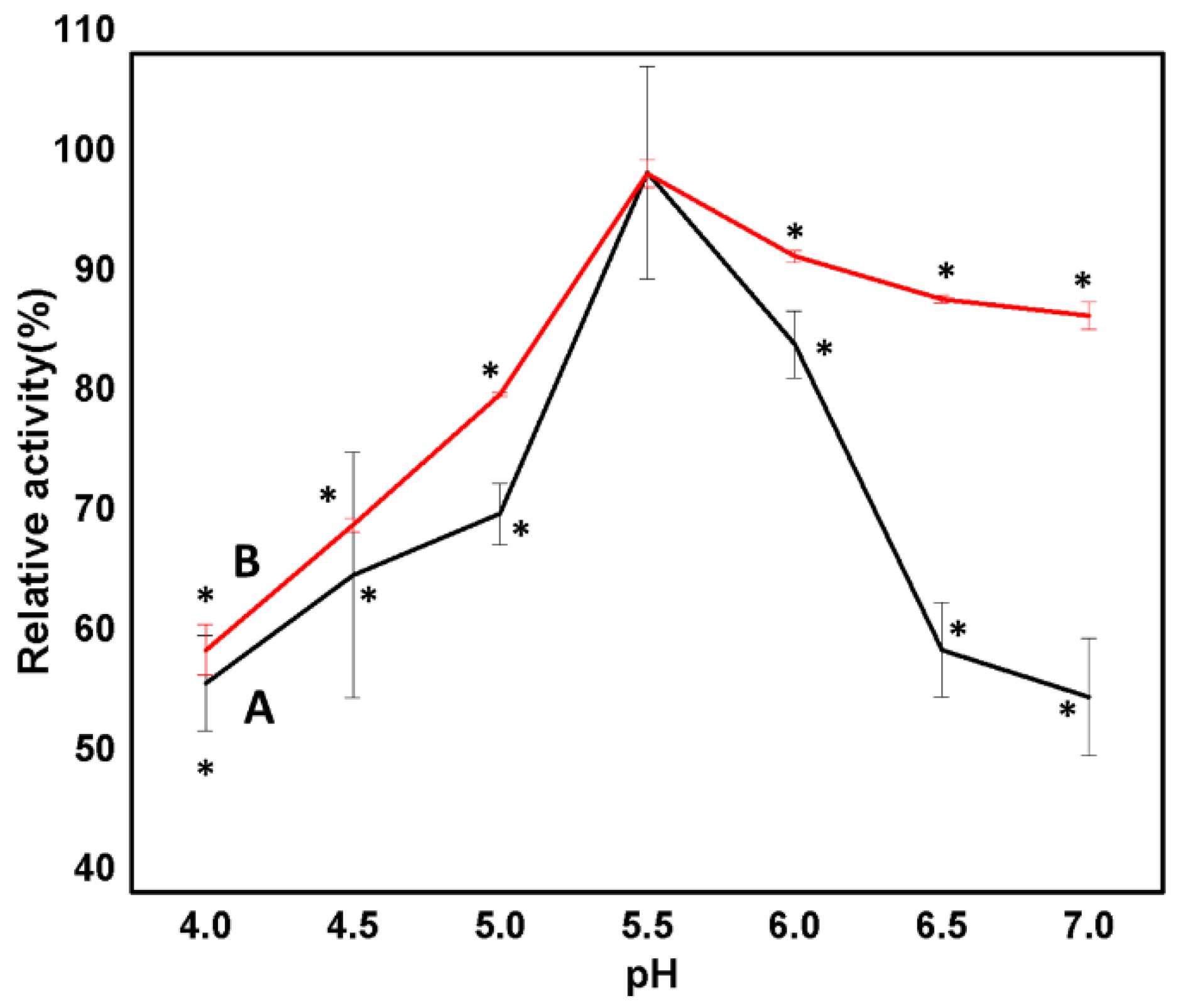
2.5.2. Effect of pH on Enzyme Activity
2.5.3. Kinetics and Thermodynamics of Substrate Hydrolysis
2.5.4. Thermostability of Immobilized Enzyme and Kinetics of Enzyme Denaturation
2.6. Reusability of Immobilized Beads
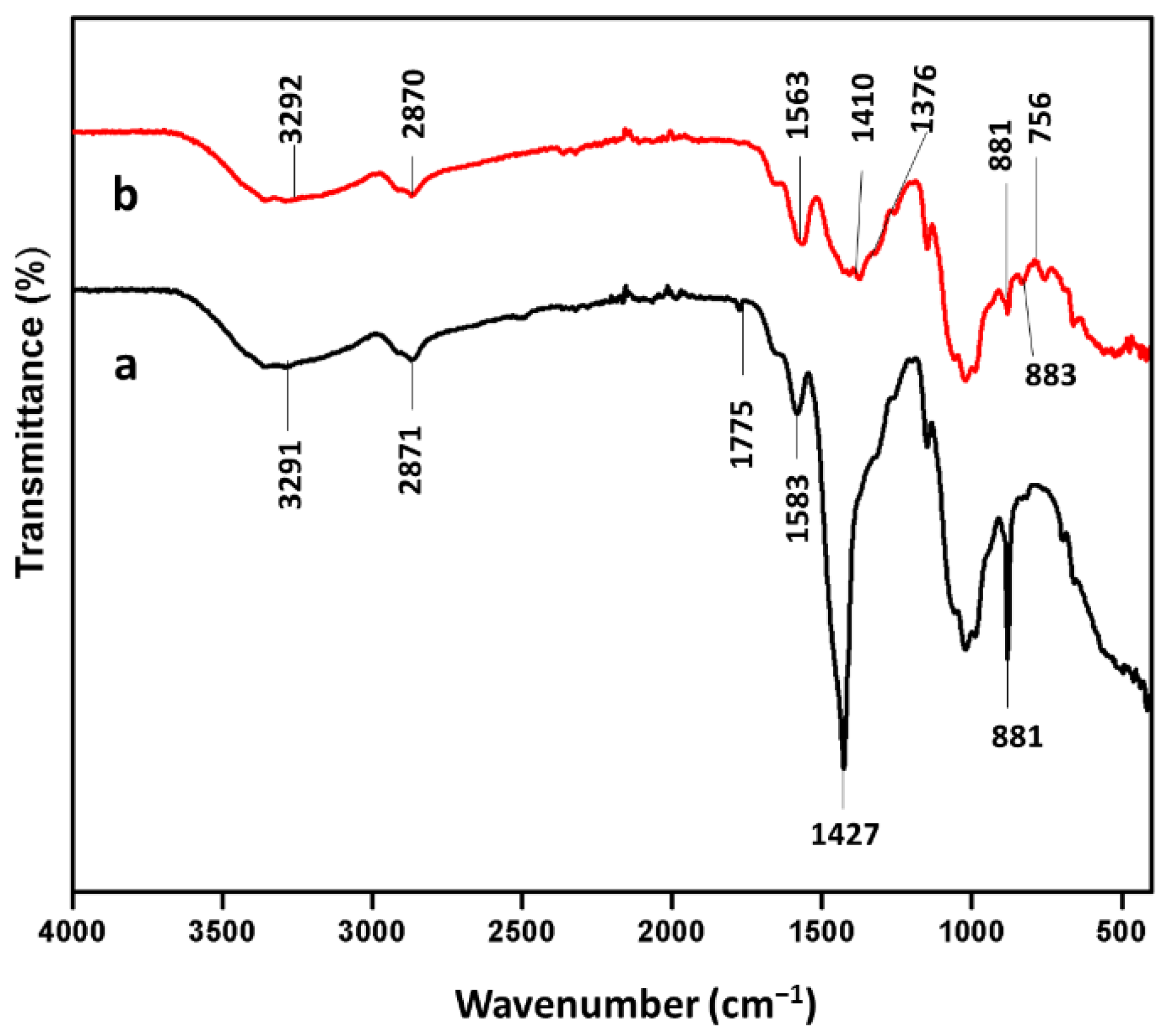
2.7. Analysis of Chitosan and Enzyme Immobilized Chitosan Beads
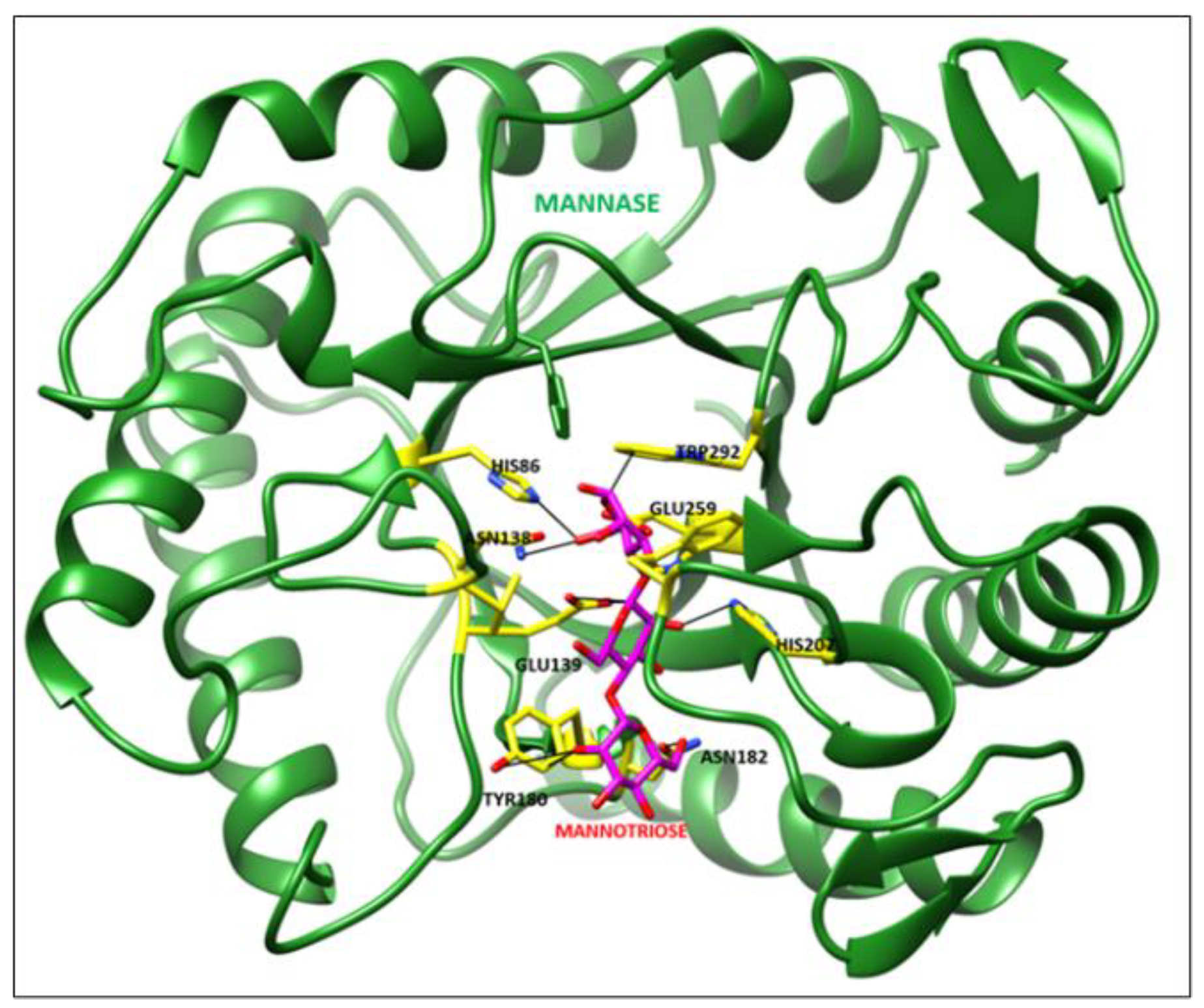
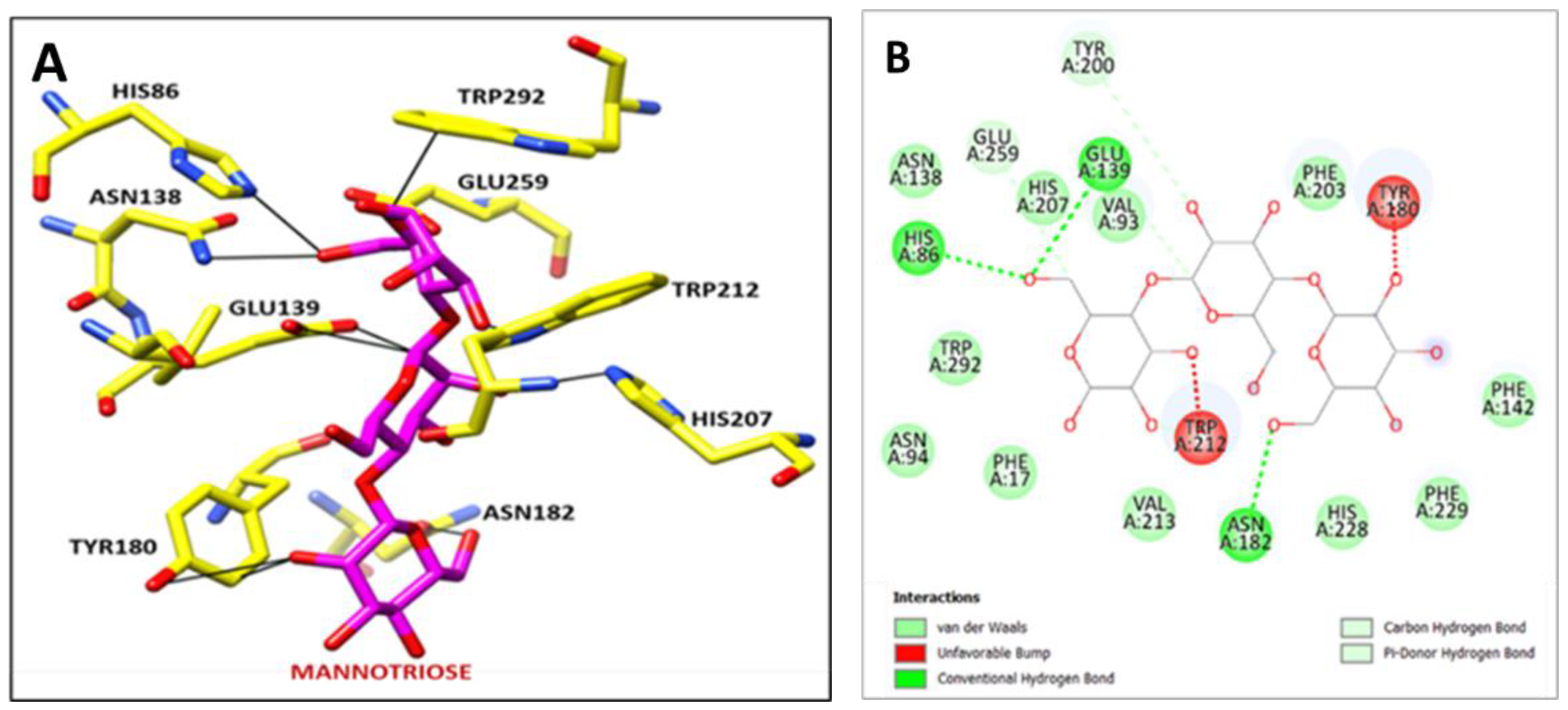
2.8. Molecular Docking Study
2.9. Statistical Analysis
3. Results and Discussion
3.1. Surface Morphology of Chitosan Beads
3.2. Biochemical Characterization of Free and Immobilized Enzyme
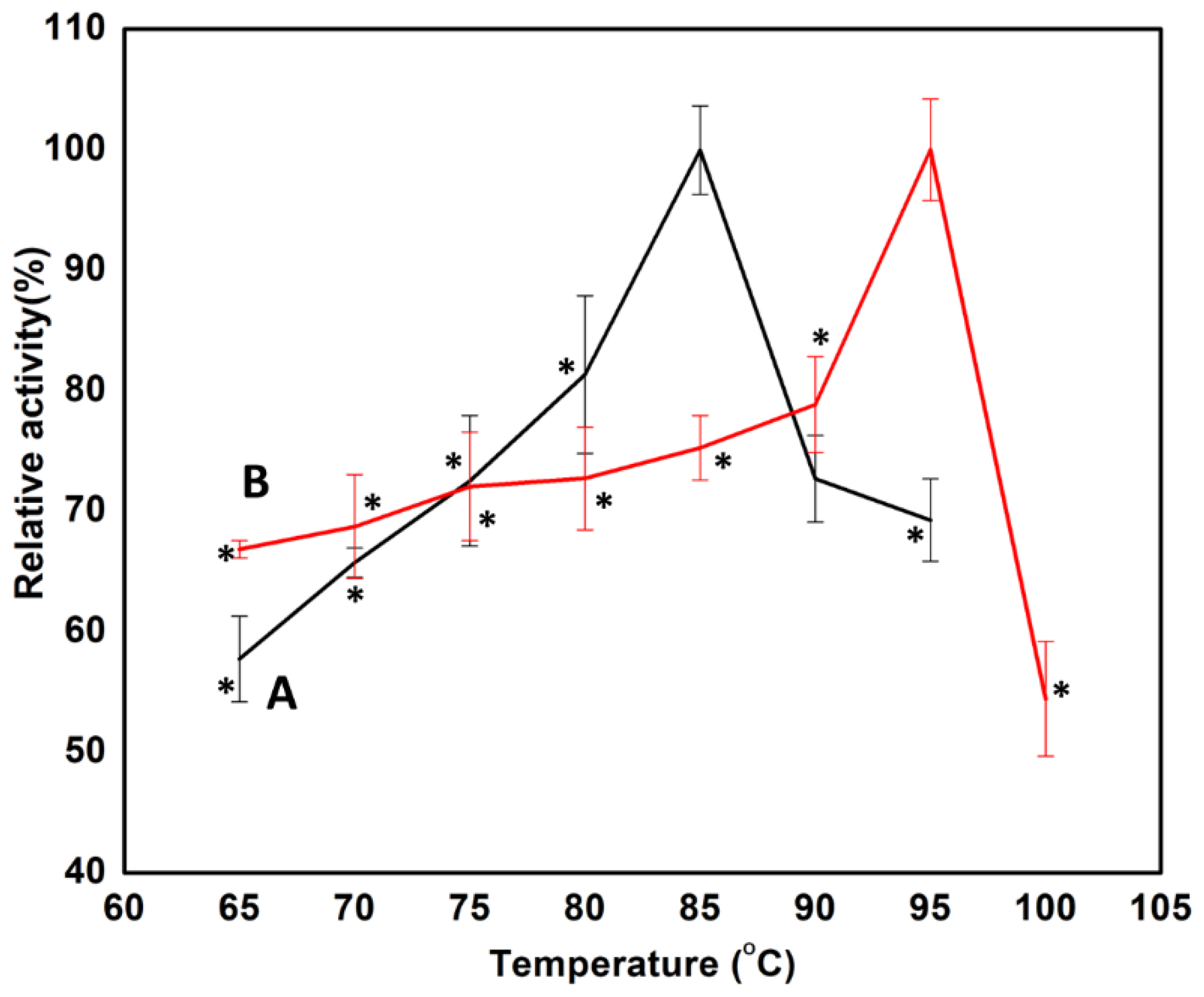
3.2.1. Effect of Temperature on Man/Cel5 B Immobilized Enzyme
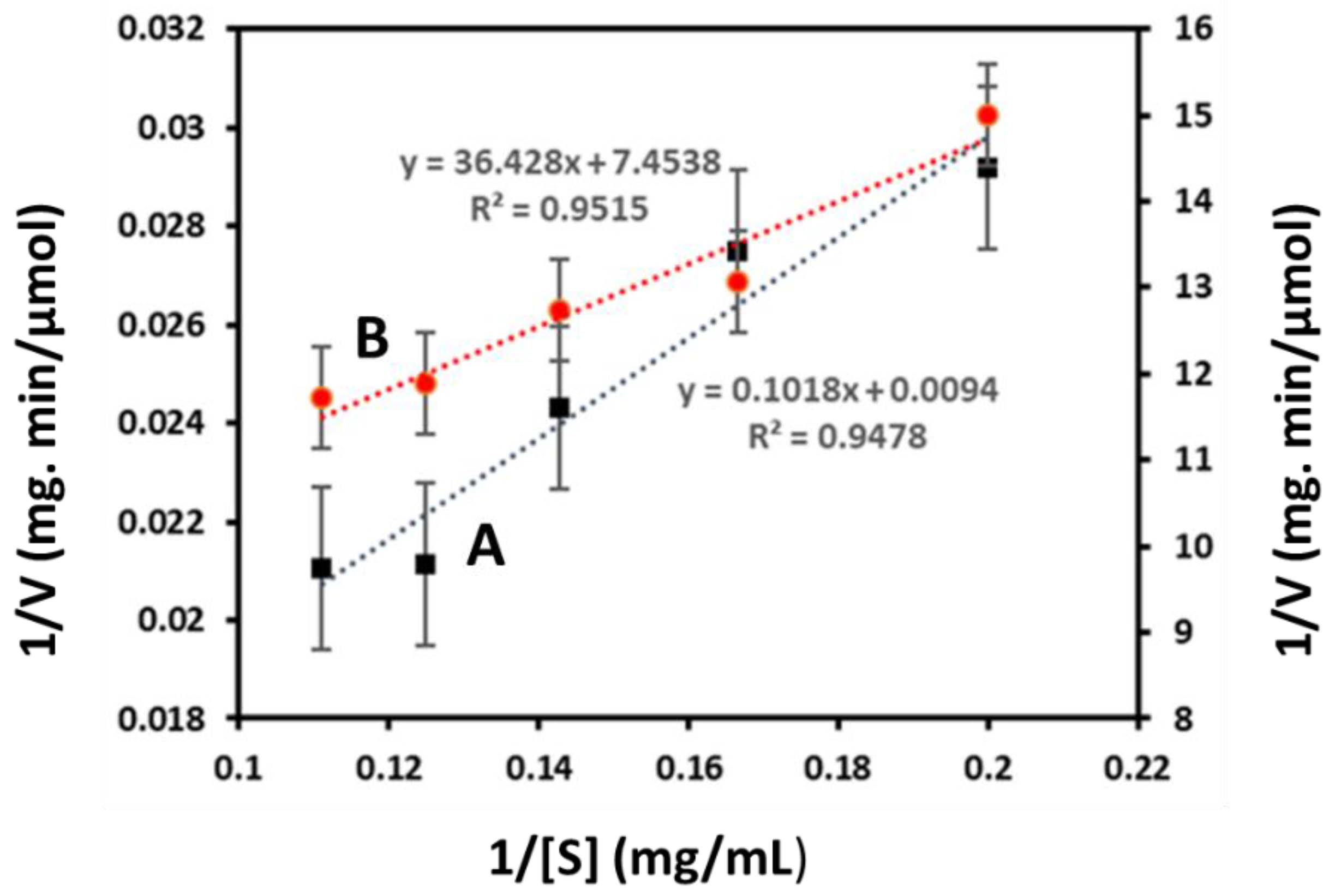
3.2.2. Kinetic Paraments of Free and Immobilized Enzyme
3.2.3. Thermodynamics of LBG Hydrolysis
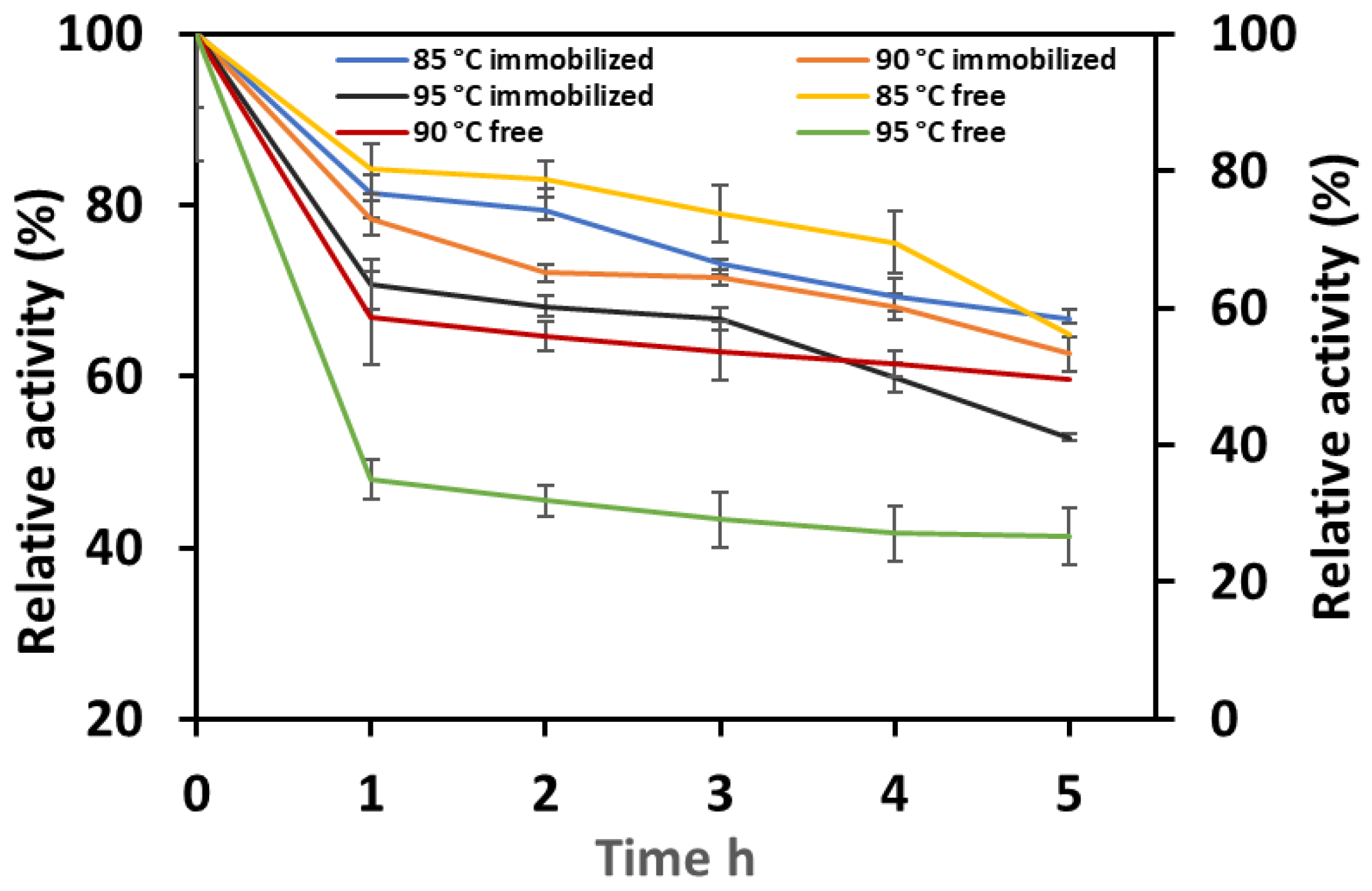
3.2.4. Thermostability of Free and Immobilized Man/Cel5B
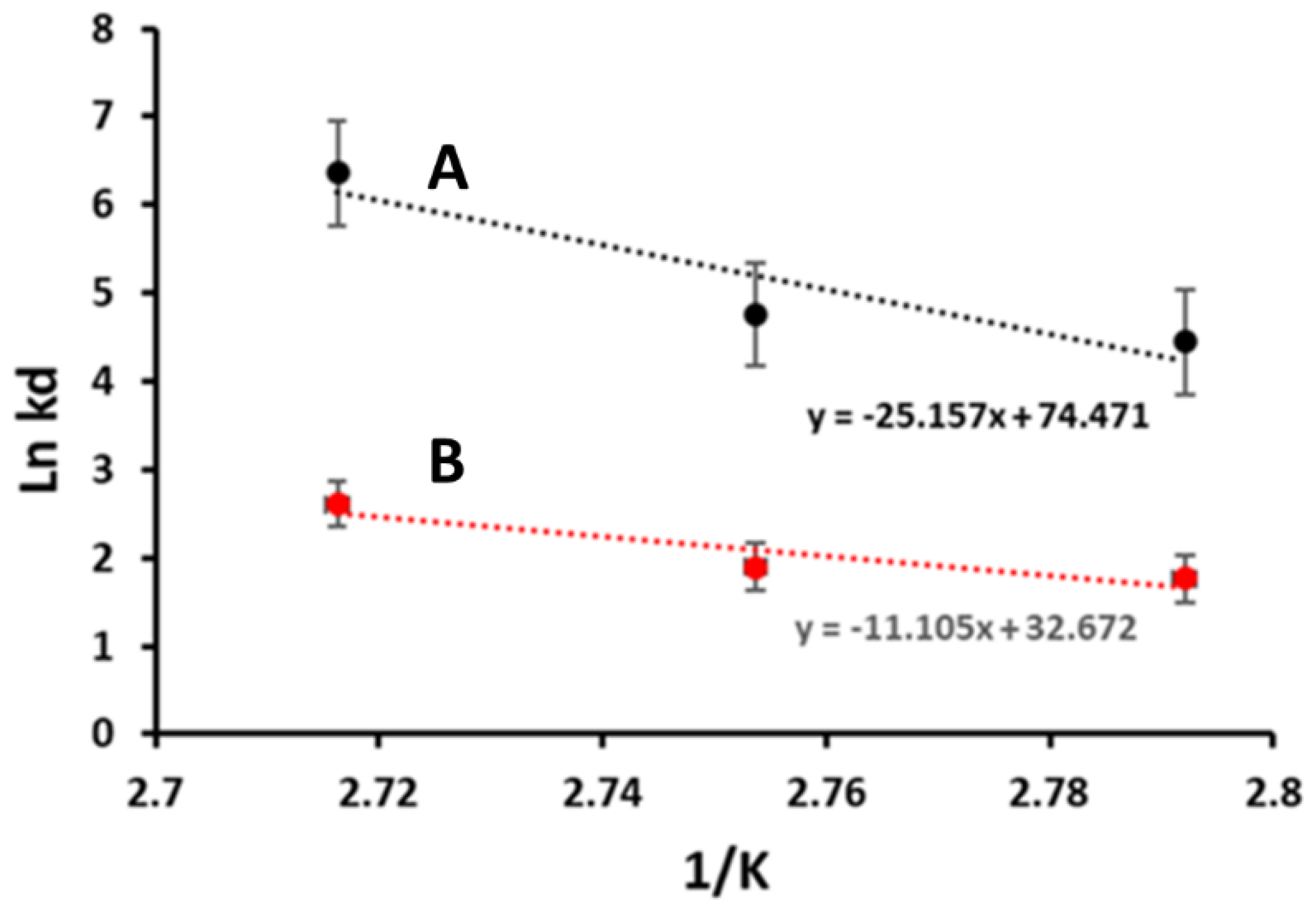
3.2.5. Thermodynamics of Enzyme Stability after Immobilization
3.2.6. Effect of pH on Free and Immobilized Man/Cel5B
3.3. Reusability of Immobilized Man/Cel5B
3.4. Fourier-Transform Infrared Spectroscopy (FTIR) Spectrum of Chitosan and Chitosan Immobilized Beads
3.5. X-ray Powder Diffraction (XRD) of Chitosan and Chitosan Immobilized Beads
3.6. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.S.; Puri, N.; Sharma, P.; Gupta, N. Mannanases: Microbial sources, production, properties and potential biotechnological applications. Appl. Microbiol. Biotechnol. 2012, 93, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, D.; Costa, L.; Ximenes, E.; Filho, F. Microbial β-mannosidases and their industrial applications. Appl. Microbiol. Biotechnol. 2018, 103, 535–547. [Google Scholar]
- Lesage, G.; Bussey, H. Cell Wall Assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2006, 70, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Tu, T.; Gu, Y.; Wang, Y.; Zheng, F.; Zheng, J.; Wang, Y.; Su, X.; Yao, B.; Luo, H. Insight into the Thermophilic Mechanism of a Glycoside Hydrolase Family 5 β-Mannanase. J. Agric. Food Chem. 2019, 67, 473–483. [Google Scholar] [CrossRef]
- Sadaqat, B.; Sha, C.; Rupani, P.F.; Wang, V.; Zuo, W.; Shao, W. Man/Cel5B, a Bifunctional Enzyme Having the Highest Mannanase Activity in the Hyperthermic Environment. Front. Bioeng. Biotechnol. 2021, 9, 637649. [Google Scholar] [CrossRef]
- Dawood, A.; Ma, K. Applications of microbial β-mannanases. Front. Bioeng. Biotechnol. 2020, 8, 598630. [Google Scholar] [CrossRef]
- Mohapatra, B.R. Characterization of β-mannanase extracted from a novel Streptomyces species Alg-S25 immobilized on chitosan nanoparticles. Biotechnol. Biotechnol. Equip. 2021, 35, 150–161. [Google Scholar] [CrossRef]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters necessary to define an immobilized enzyme preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Lopes, L.A.; Dias, L.P.; da Costa, H.P.S.; da Silva Neto, J.X.; Morais, E.G.; de Oliveira, J.T.A.; Vasconcelos, I.M.; de Sousa, D.D.O.B. Immobilization of a peroxidase from Moringa oleifera Lam. roots (MoPOX) on chitosan beads enhanced the decolorization of textile dyes. Process Biochem. 2021, 110, 129–141. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, S.; Srivastava, B.; Singh, G.; Khatri, M.; Arya, S.K. Immobilization of mannanase on sodium alginate-grafted-β-cyclodextrin: An easy and cost-effective approach for the improvement of enzyme properties. Int. J. Biol. Macromol. 2020, 156, 1347–1358. [Google Scholar] [CrossRef] [PubMed]
- Panwar, D.; Kaira, G.S.; Kapoor, M. Cross-linked enzyme aggregates (CLEAs) and magnetic nanocomposite grafted CLEAs of GH26 endo-β-1,4-mannanase: Improved activity, stability and reusability. Int. J. Biol. Macromol. 2017, 105, 1289–1299. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.; Dev, M.J.; Singhal, R.S. Cross-linked beta-Mannanase Aggregates: Preparation, Characterization, and Application for Producing Partially Hydrolyzed Guar Gum. Appl. Biochem. Biotechnol. 2022, 194, 1981–2004. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, M.Y.; El-Aassar, S.A.; Youssef, A.S.; El-Sersy, N.A.; Beltagy, E.A. Extracellular β-mannanase production by the immobilization of the locally isolated Aspergillus niger. Int. J. Agric. Biol. 2006, 8, 57–62. [Google Scholar]
- Seenuvasan, M.; Kumar, K.S.; Kumar, A.; Parthiban, R. Review on surface modification of nanocarriers to overcome diffusion limitations: An enzyme immobilization aspect. Biochem. Eng. J. 2020, 158, 107574. [Google Scholar]
- Machado, N.B.; Miguez, J.P.; Bolina, I.C.; Salviano, A.B.; Gomes, R.A.; Tavano, O.L.; Luiz, J.H.H.; Tardioli, P.W.; Cren, E.C.; Mendes, A.A. Preparation, functionalization and characterization of rice husk silica for lipase immobilization via adsorption. Enzym. Microb. Technol. 2019, 128, 9–21. [Google Scholar] [CrossRef]
- Nunes, Y.L.; de Menezes, F.L.; de Sousa, I.G.; Cavalcante, A.L.G.; Cavalcante, F.T.T.; da Silva Moreira, K.; de Oliveira, A.L.B.; Mota, G.F.; Souza, J.E.D.S.; Falcão, I.R.D.A.; et al. Chemical and physical Chitosan modification for designing enzymatic industrial biocatalysts: How to choose the best strategy? Int. J. Biol. Macromol. 2021, 181, 1124–1170. [Google Scholar] [CrossRef]
- Mo, H.; Qiu, J.; Yang, C.; Zang, L.; Sakai, E.; Chen, J. Porous biochar/chitosan composites for high performance cellulase immobilization by glutaraldehyde. Enzym. Microb. Technol. 2020, 138, 109561. [Google Scholar] [CrossRef]
- Suryawanshi, R.K.; Jana, U.K.; Prajapati, B.P.; Kango, N. Immobilization of Aspergillus quadrilineatus RSNK-1 multi-enzymatic system for fruit juice treatment and mannooligosaccharide generation. Food Chem. 2019, 289, 95–102. [Google Scholar] [CrossRef]
- Raharjo, T.J.; Febrina, L.; Wardoyo, F.A.; Swasono, R.T. Research Article Effect of Deacetylation Degree of Chitosan as Solid Support in Lipase Immobilization by Glutaraldehyde Crosslink. J. Biochem. 2016, 11, 127–134. [Google Scholar]
- Işık, M. High stability of immobilized acetylcholinesterase on chitosan beads. ChemistrySelect 2020, 5, 4623–4627. [Google Scholar] [CrossRef]
- Kaushal, J.; Singh, G.; Arya, S.K. Immobilization of catalase onto chitosan and chitosan–bentonite complex: A comparative study. Biotechnol. Rep. 2018, 18, e00258. [Google Scholar] [CrossRef] [PubMed]
- Hermanto, D.; Mudasir, M.; Siswanta, D.; Kuswandi, B.; Ismillayli, N. Polyelectrolyte Complex (PEC) of the Alginate-Chitosan Membrane for Immobilizing Urease. J. Math. Fundam. Sci. 2019, 51, 309–319. [Google Scholar] [CrossRef]
- 27Lever, M. A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 1972, 47, 273–279. [Google Scholar]
- Simonian, M.H. Spectrophotometric determination of protein concentration. Curr. Protoc. Toxicol. 2004, 3, 1–7. [Google Scholar] [CrossRef]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar]
- Laskowaski, R.A.; McArther, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK a program to check sterio-chemical quality of a protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- D. Systèmes. Discovery Studio Visualizer; BIOVIA, Dassault Systèmes: San Diego, CA, USA, 2021. [Google Scholar]
- Bao, Y.; Shao, L.; Xing, G.; Qi, C. Cobalt, nickel and iron embedded chitosan microparticles as efficient and reusable catalysts for Heck cross-coupling reactions. Int. J. Biol. Macromol. 2019, 130, 203–212. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, A.R.; Jeon, S.J. Immobilization on chitosan of a thermophilic trehalose synthase from Thermus thermophilus HJ6. J. Microbiol. Biotechnol. 2010, 20, 513–517. [Google Scholar] [PubMed]
- Dinçer, A.; Becerik, S.; Aydemir, T. Immobilization of tyrosinase on chitosan–clay composite beads. Int. J. Biol. Macromol. 2012, 50, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Khanum, F. Covalent immobilization of xylanase on glutaraldehyde activated alginate beads using response surface methodology: Characterization of immobilized enzyme. Process Biochem. 2011, 46, 1315–1322. [Google Scholar] [CrossRef]
- Tripathi, P.; Kumari, A.; Rath, P.; Kayastha, A.M. Immobilization of α-amylase from mung beans (Vigna radiata) on Amberlite MB 150 and chitosan beads: A comparative study. J. Mol. Catal. B Enzym. 2007, 49, 69–74. [Google Scholar] [CrossRef]
- Akkuş Çetinus, Ş.; Öztop, H.N. Immobilization of catalase into chemically crosslinked chitosan beads. Enzym. Microb. Technol. 2003, 32, 889–894. [Google Scholar] [CrossRef]
- Lin, K.P.; Feng, G.J.; Pu, F.L.; Hou, X.D.; Cao, S.L. Enhancing the Thermostability of Papain by Immobilizing on Deep Eutectic Solvents-Treated Chitosan With Optimal Microporous Structure and Catalytic Microenvironment. Front. Bioeng. Biotechnol. 2020, 8, 576266. [Google Scholar] [CrossRef]
- Sohrabi, N.; Rasouli, N.; Torkzadeh, M. Enhanced stability and catalytic activity of immobilized α-amylase on modified Fe3O4 nanoparticles. Chem. Eng. J. 2014, 240, 426–433. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Mostafa, F.A.; Ouis, M.A. Enhancement stability and catalytic activity of immobilized α-amylase using bioactive phospho-silicate glass as a novel inorganic support. Int. J. Biol. Macromol. 2018, 112, 371–382. [Google Scholar] [CrossRef]
- Akkuş Çetinus, Ş.; Öztop, H.N. Immobilization of catalase on chitosan film. Enzyme Microb. Technol. 2000, 26, 497–501. [Google Scholar] [CrossRef]
- Shafei, M.S.; Allam, R.F. Production and immobilization of partially purified lipase from Penicillium chrysogenum. Malays. J. Microbiol. 2010, 6, 196–202. [Google Scholar]
- Altun, G.D.; Cetinus, S.A. Immobilization of pepsin on chitosan beads. Food Chem. 2007, 100, 964–971. [Google Scholar] [CrossRef]
- Riaz, M.; Perveen, R.; Javed, M.R.; Nadeem, H.; Rashid, M.H. Kinetic and thermodynamic properties of novel glucoamylase from Humicola sp. Enzym. Microb. Technol. 2007, 41, 558–564. [Google Scholar] [CrossRef]
- Hameed, U.; Ul-Haq, I. Kinetics and thermodynamics of catalysis and thermal inactivation of a novel α-amylase (Tp-AmyS) from Thermotoga petrophila. Biocatal. Biotransform. 2020, 38, 227–233. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszyńska, D. Immobilization as a strategy for improving enzyme properties-application to oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, E.A.L.; Lima, Á.S.; Soares, C.M.F.; de Aquino Santana, L.C.L. Lipase from Aspergillus niger obtained from mangaba residue fermentation: Biochemical characterization of free and immobilized enzymes on a sol-gel matrix. Acta Scientiarum. Technol. 2017, 39, 1–8. [Google Scholar] [CrossRef][Green Version]
- Sel, E.; Ulu, A.; Ateş, B.; Köytepe, S. Comparative study of catalase immobilization via adsorption on P (MMA-co-PEG500MA) structures as an effective polymer support. Polym. Bull. 2021, 78, 2663–2684. [Google Scholar] [CrossRef]
- Zheng, F.; Cui, B.-K.; Wu, X.-J.; Meng, G.; Liu, H.-X.; Jing, S. Immobilization of laccase onto chitosan beads to enhance its capability to degrade synthetic dyes. Int. Biodeterior. 2016, 110, 69–78. [Google Scholar] [CrossRef]
- Wehaidy, H.R.; Abdel-Naby, M.A.; El-Hennawi, H.M.; Youssef, H.F. Nanoporous Zeolite-X as a new carrier for laccase immobilization and its application in dyes decolorization. Biocatal. Agric. Biotechnol. 2019, 19, 101135. [Google Scholar] [CrossRef]
- Worthen, A.J.; Irving, K.S.; Lapitsky, Y. Supramolecular strategy effects on chitosan bead stability in acidic media: A comparative study. Gels 2019, 5, 11. [Google Scholar] [CrossRef]
- Wu, S.C.; Lia, Y.K. Application of bacterial cellulose pellets in enzyme immobilization. J. Mol. Catal. B Enzym. 2008, 54, 103–108. [Google Scholar] [CrossRef]
- Blibech, M.; Chaari, F.; Bhiri, F.; Dammak, I.; Ghorbel, R.E.; Chaabouni, S.E. Production of manno-oligosaccharides from locust bean gum using immobilized Penicillium occitanis mannanase. J. Mol. Catal. B Enzym. 2011, 73, 111–115. [Google Scholar] [CrossRef]
- Patel, S.K.; Kalia, V.C.; Choi, J.H.; Haw, J.R.; Kim, I.W.; Lee, J.K. Immobilization of laccase on SiO2 nanocarriers improves its stability and reusability. J. Microbiol. Biotechnol. 2014, 24, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Shan, C.L.; Zhou, Q.; Fang, Y.; Wang, Y.L.; Xu, F.; Han, L.; Ibrahim, M.; Guo, L.; Xie, G.; et al. Synthesis, characterization, and antibacterial activity of cross-linked chitosan-glutaraldehyde. Mar. Drugs 2013, 11, 1534–1552. [Google Scholar] [CrossRef]
- Klein, M.P.; Nunes, M.R.; Rodrigues, R.C.; Benvenutti, E.V.; Costa, T.M.; Hertz, P.F.; Ninow, J.L. Effect of the support size on the properties of β-galactosidase immobilized on chitosan: Advantages and disadvantages of macro and nanoparticles. Biomacromolecules 2012, 13, 2456–2464. [Google Scholar] [CrossRef]
- Chagas, P.M.B.; Caetano, A.A.; Tireli, A.A.; Cesar, P.H.S.; Corrêa, A.D.; do Rosário Guimarães, I. Use of an Environmental Pollutant from Hexavalent Chromium Removal as a Green Catalyst in The Fenton Process. Sci. Rep. 2019, 9, 1–15. [Google Scholar]




| Thermodynamic Parameter | Immobilized Enzyme | Free Enzyme |
|---|---|---|
| Eact (KJmol−1) | 6.78 ± 0.88 | 26.35 ± 3.33 |
| ΔH (KJmol−1) | 3.72 ± 0.26 | 23.37 ± 2.68 |
| ΔG (KJmol−1) | 85.17 ± 3.26 | 72.68 ± 4.21 |
| ΔS (J·mol−1·K−1) | −221.33 ± 5.13 | −137.74 ± 4.49 |
| Parameter | Immobilized Enzyme | Free Enzyme | ||||
|---|---|---|---|---|---|---|
| Temperature (°C) | 85 | 90 | 95 | 85 | 90 | 95 |
| Kd (min−1) | 4.45 ± 0.03 | 4.75 ± 0.99 | 6.63 ± 1.23 | 5.82 ± 0.42 | 6.66 ± 0.24 | 13.5 ± 0.44 |
| t1/2 (min−1) | 561 ± 2.12 | 524 ± 3.30 | 392 ± 3.63 | 428 ± 1.35 | 374 ± 2.63 | 184 ± 3.33 |
| D-value | 1864 ± 8.53 | 1742 ± 4.12 | 1303 ± 1.67 | 1424 ± 3.33 | 1244 ± 6.55 | 611 ± 5.45 |
| ΔHd (KJmol−1) | 206.24 ± 2.04 | 206.20 ± 2.85 | 206.16 ± 1.35 | 89.41 ± 2.45 | 89.37 ± 1.84 | 89.33 ± 2.63 |
| ΔGd (KJmol−1) | 82.80 ± 1.48 | 84.00 ± 2.43 | 85.20 ± 1.67 | 72.71 ± 1.64 | 73.77 ± 2.50 | 74.82 ± 8.46 |
| ΔSd (J·mol−1·K−1) | −230.63 ± 1.90 | −230.75 ± 4.44 | −230.87 ± 3.24 | −202.76 ± 3.44 | −202.89 ± 1.53 | −203.01 ± 2.43 |
| Sr. No. | Interaction between Amino Acid Residues of Mannanase with Mannotriose | Distance in Å |
|---|---|---|
| 1 | BMA 1.C O5 ------ TRP 292.A CZ2: | 3.302 |
| 2 | HIS 86.A NE2 ------ BMA 1.C O6: | 2.645 |
| 3 | ASN 138.A ND2 ------ BMA 1.C O6: | 3.473 |
| 4 | BMA 1.C C6 ------ GLU 259.A OE1: | 3.319 |
| 5 | TRP 212.A NE1 ------ BMA 1.C O3: | 1.243 |
| 6 | BMA 2.C O2 ------ HIS 207.A NE2: | 3.039 |
| 7 | GLU 139.A OE1 ------ BMA 2.C C1: | 2.769 |
| 8 | TYR 180.A CE2 ------ BMA 3.C O2: | 1.721 |
| 9 | ASN 182.A OD1 ------ BMA 3.C O6: | 2.344 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadaqat, B.; Sha, C.; Dar, M.A.; Dhanavade, M.J.; Sonawane, K.D.; Mohamed, H.; Shao, W.; Song, Y. Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads. Biomolecules 2022, 12, 999. https://doi.org/10.3390/biom12070999
Sadaqat B, Sha C, Dar MA, Dhanavade MJ, Sonawane KD, Mohamed H, Shao W, Song Y. Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads. Biomolecules. 2022; 12(7):999. https://doi.org/10.3390/biom12070999
Chicago/Turabian StyleSadaqat, Beenish, Chong Sha, Mudasir Ahmad Dar, Maruti J. Dhanavade, Kailas D. Sonawane, Hassan Mohamed, Weilan Shao, and Yuanda Song. 2022. "Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads" Biomolecules 12, no. 7: 999. https://doi.org/10.3390/biom12070999
APA StyleSadaqat, B., Sha, C., Dar, M. A., Dhanavade, M. J., Sonawane, K. D., Mohamed, H., Shao, W., & Song, Y. (2022). Modifying Thermostability and Reusability of Hyperthermophilic Mannanase by Immobilization on Glutaraldehyde Cross-Linked Chitosan Beads. Biomolecules, 12(7), 999. https://doi.org/10.3390/biom12070999








