High-Throughput Proteomic Analysis of Human Dermal Fibroblast Response to Different Blood Derivatives: Autologous Topical Serum Derived from Plasma Rich in Growth Factors (PRGF) versus Leukocyte- and Platelet-Rich Plasma (L-PRP)
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Characterization of Blood Derivatives
2.2. Cell Culture
2.3. Protein Extraction
2.4. Proteomic Analysis
2.5. Functional Analysis
3. Results
3.1. Characterization of Blood Derivatives
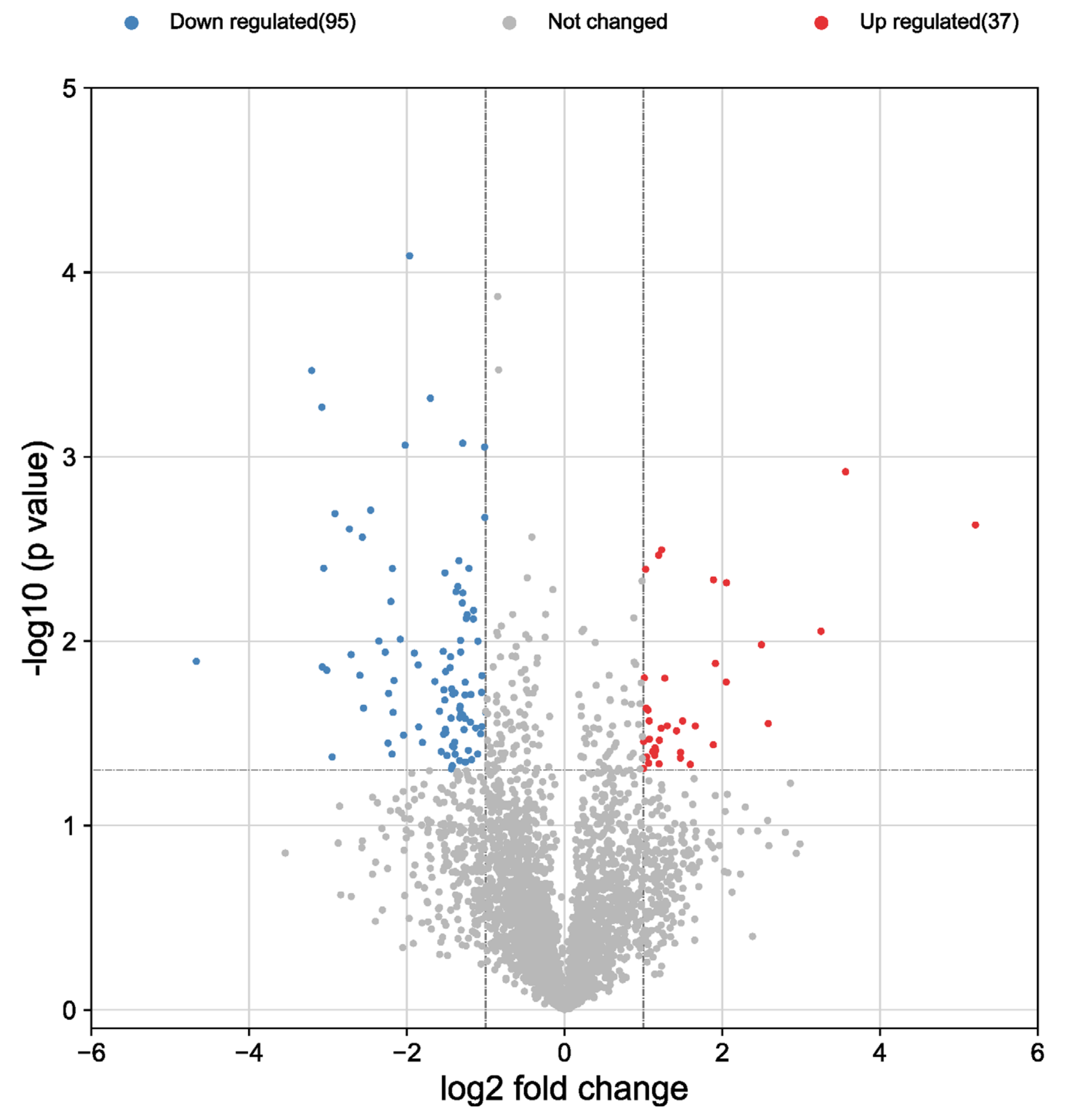
3.2. Proteomic Characterization
3.3. Functional Analysis: Gene Ontology
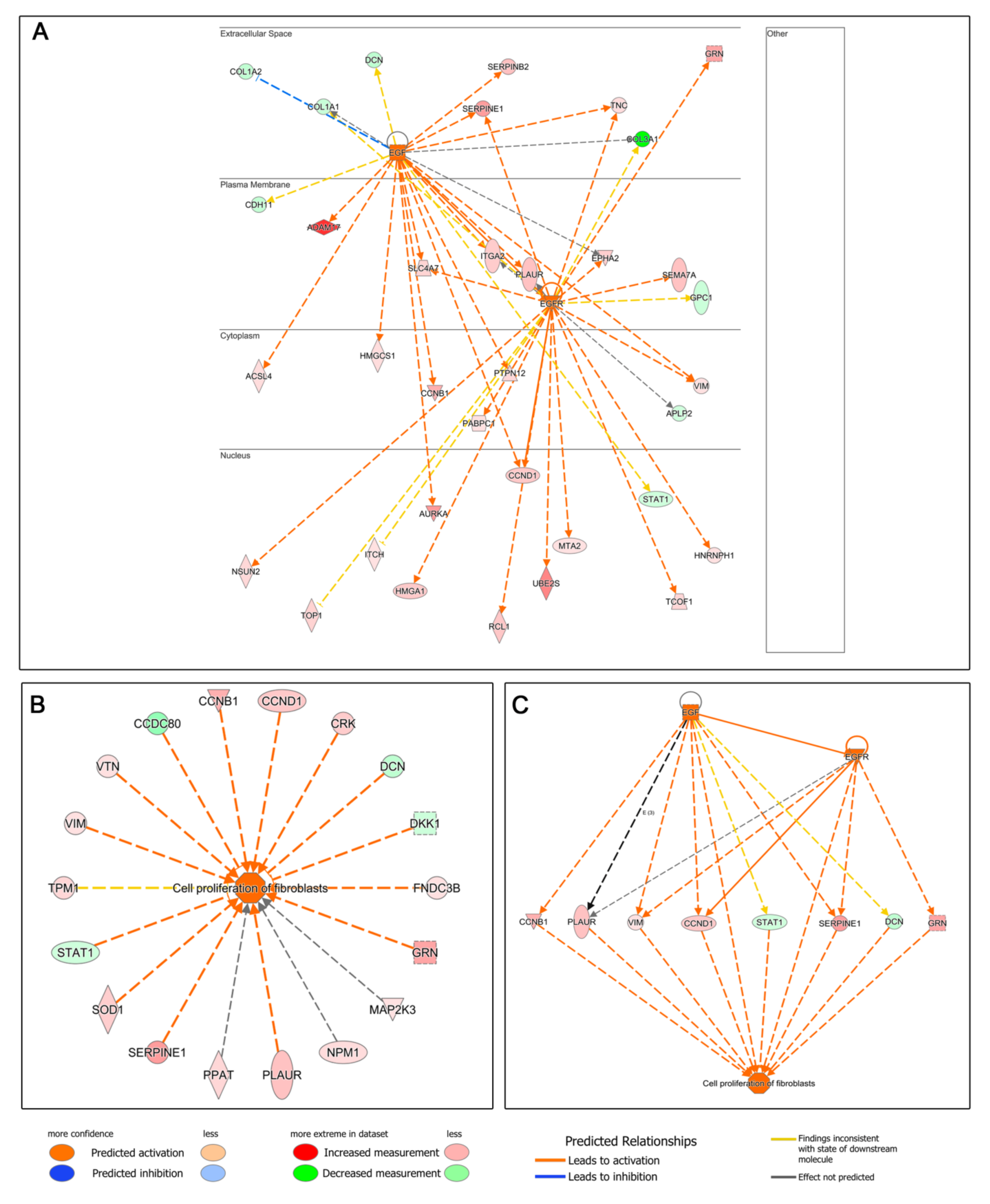
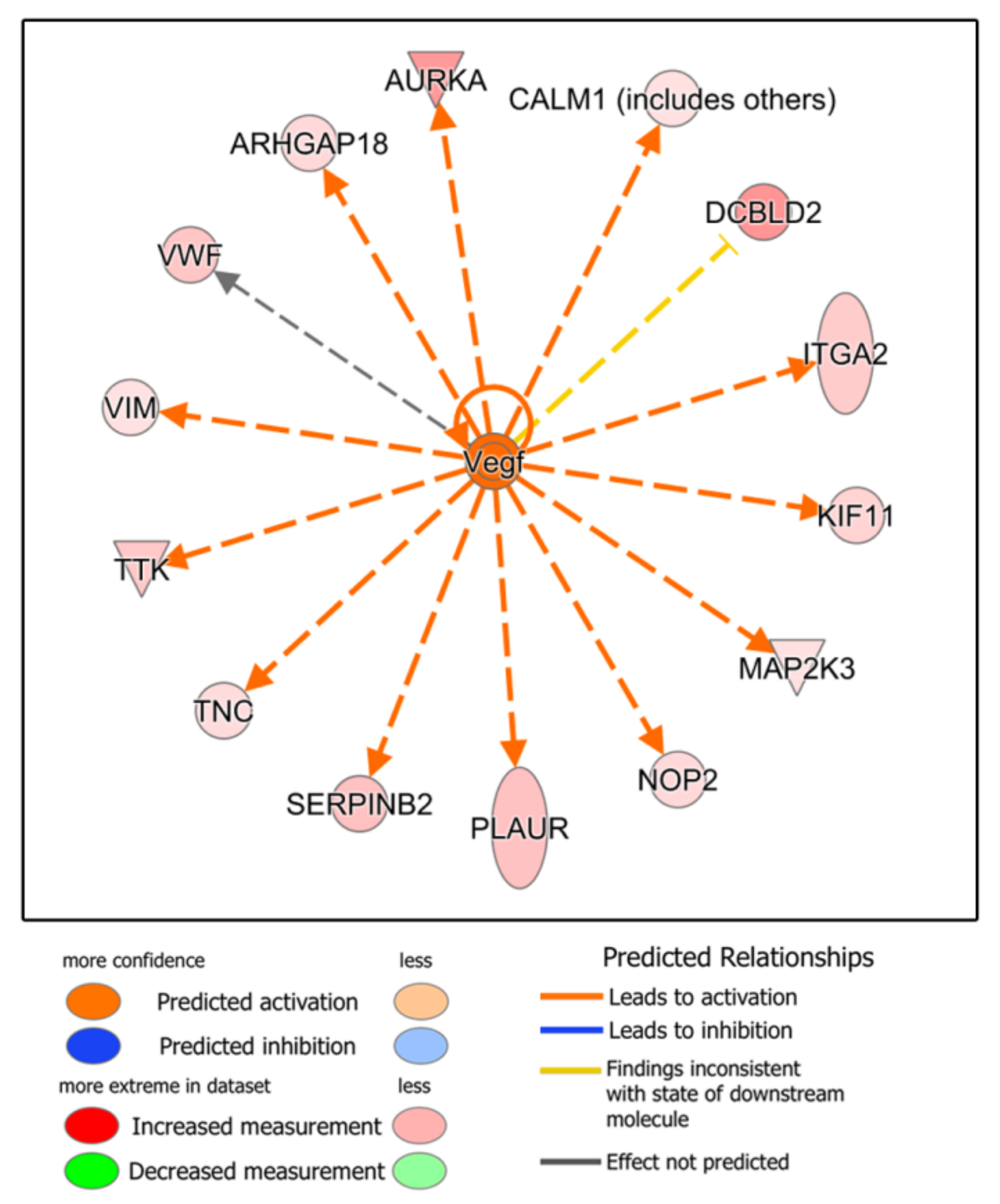
3.4. Functional Analysis: Ingenuity Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nilforoushzadeh, M.A.; Amirkhani, M.A.; Khodaverdi, E.; Razzaghi, Z.; Afzali, H.; Izadpanah, S.; Zare, S. Tissue engineering in dermatology from lab to market. Tissue Cell 2022, 74, 101717. [Google Scholar] [CrossRef] [PubMed]
- Sorg, H.; Tilkorn, D.J.; Hager, S.; Hauser, J.; Mirastschijski, U. Skin Wound Healing: An Update on the Current Knowledge and Concepts. Eur. Surg. Res. 2017, 58, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Nourian Dehkordi, A.; Mirahmadi Babaheydari, F.; Chehelgerdi, M.; Raeisi Dehkordi, S. Skin tissue engineering: Wound healing based on stem-cell-based therapeutic strategies. Stem Cell. Res. Ther. 2019, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- Takeo, M.; Lee, W.; Ito, M. Wound healing and skin regeneration. Cold Spring Harb. Perspect. Med. 2015, 5, a023267. [Google Scholar] [CrossRef]
- Than, P.A.; Davis, C.R.; Gurtner, G.C. Chapter 4—Clinical Management of Wound Healing and Hypertrophic Scarring. In Skin Tissue Engineering and Regenerative Medicine; Albanna, M.Z., Holmes Iv, J.H., Eds.; Academic Press: Boston, MA, USA, 2016; pp. 61–81. [Google Scholar]
- Chen, D.; Hou, Q.; Zhong, L.; Zhao, Y.; Li, M.; Fu, X. Bioactive Molecules for Skin Repair and Regeneration: Progress and Perspectives. Stem Cells Int. 2019, 2019, 6789823. [Google Scholar] [CrossRef]
- Anitua, E.; Pino, A.; Orive, G. Opening new horizons in regenerative dermatology using platelet-based autologous therapies. Int. J. Dermatol. 2017, 56, 247–251. [Google Scholar] [CrossRef]
- Burnouf, T.; Goubran, H.A.; Chen, T.M.; Ou, K.L.; El-Ekiaby, M.; Radosevic, M. Blood-derived biomaterials and platelet growth factors in regenerative medicine. Blood Rev. 2013, 27, 77–89. [Google Scholar] [CrossRef]
- Cobos, R.; Aizpuru, F.; Parraza, N.; Anitua, E.; Orive, G. Effectiveness and efficiency of platelet rich plasma in the treatment of diabetic ulcers. Curr. Pharm. Biotechnol. 2015, 16, 630–634. [Google Scholar] [CrossRef]
- Linertova, R.; Del Pino-Sedeno, T.; Perez, L.G.; Aragon-Sanchez, J.; Andia-Ortiz, I.; Trujillo-Martin, M.; Iruzubieta-Barragan, F.J.; Serrano-Aguilar, P. Cost-effectiveness of Platelet-Rich Plasma for Diabetic Foot Ulcer in Spain. Int. J. Low Extrem. Wounds 2021, 20, 119–127. [Google Scholar] [CrossRef]
- Anitua, E.; Prado, R.; Nurden, A.T.; Nurden, P. Characterization of Plasma Rich in Growth Factors (PRGF): Components and formulations. In Platelet Rich Plasma in Orthopaedics and Sports Medicine; Anitua, E., Cugat, R., Sánchez, M., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 29–45. [Google Scholar]
- Anitua, E. Plasma rich in growth factors: Preliminary results of use in the preparation of future sites for implants. Int. J. Oral Maxillofac. Implants 1999, 14, 529–535. [Google Scholar]
- Anitua, E.; Nurden, P.; Prado, R.; Nurden, A.T.; Padilla, S. Autologous fibrin scaffolds: When platelet- and plasma-derived biomolecules meet fibrin. Biomaterials 2019, 192, 440–460. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Munoz, V.; Aspe, L.; Tierno, R.; Garcia-Salvador, A.; Goni-de-Cerio, F.; Pino, A. In Vitro and In Vivo Effect of Platelet-Rich Plasma-Based Autologous Topical Serum on Cutaneous Wound Healing. Skin Pharmacol. Physiol. 2022, 35, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Troya, M.; Pino, A. A novel protein-based autologous topical serum for skin regeneration. J. Cosmet. Dermatol. 2020, 19, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Prado, R.; Orive, G. Platelet-rich plasma and myofibroblasts: Is the composition the key to success? Adv. Skin Wound Care 2015, 28, 198–199. [Google Scholar] [PubMed]
- Giusti, I.; Di Francesco, M.; D’Ascenzo, S.; Palumbo, P.; Rughetti, A.; Dell’Orso, L.; Varasano, P.A.; Pressanti, G.L.; Dolo, V. Leukocyte depletion does not affect the in vitro healing ability of platelet rich plasma. Exp. Ther. Med. 2018, 15, 4029–4038. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Saita, Y.; Nishio, H.; Ikeda, H.; Takazawa, Y.; Nagao, M.; Takaku, T.; Komatsu, N.; Kaneko, K. Leukocyte concentration and composition in platelet-rich plasma (PRP) influences the growth factor and protease concentrations. J. Orthop. Sci. 2016, 21, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R. Filter Aided Sample Preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucl. Acids Res. 2022, 10, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Prado, R.; Troya, M.; Zalduendo, M.; de la Fuente, M.; Pino, A.; Muruzabal, F.; Orive, G. Implementation of a more physiological plasma rich in growth factor (PRGF) protocol: Anticoagulant removal and reduction in activator concentration. Platelets 2016, 27, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Fuente, M.; Muruzabal, F.; Merayo-Lloves, J. Development and optimization of a personalized fibrin membrane derived from the plasma rich in growth factors technology. Exp. Eye Res. 2021, 203, 108402. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Zalduendo, M.M.; Prado, R.; Alkhraisat, M.H.; Orive, G. Morphogen and proinflammatory cytokine release kinetics from PRGF-Endoret fibrin scaffolds: Evaluation of the effect of leukocyte inclusion. J. Biomed. Mater. Res. A 2015, 103, 1011–1020. [Google Scholar] [CrossRef]
- Rico-Leo, E.M.; Lorenzo-Martín, L.F.; Román, Á.C.; Bustelo, X.R.; Merino, J.M.; Fernández-Salguero, P.M. Aryl hydrocarbon receptor controls skin homeostasis, regeneration, and hair follicle cycling by adjusting epidermal stem cell function. Stem Cells 2021, 39, 1733–1750. [Google Scholar] [CrossRef]
- Elentner, A.; Schmuth, M.; Yannoutsos, N.; Eichmann, T.O.; Gruber, R.; Radner, F.P.W.; Hermann, M.; Del Frari, B.; Dubrac, S. Epidermal Overexpression of Xenobiotic Receptor PXR Impairs the Epidermal Barrier and Triggers Th2 Immune Response. J. Investig. Dermatol. 2018, 138, 109–120. [Google Scholar] [CrossRef]
- Oetjen, L.K.; Trier, A.M.; Kim, B.S. PXR: A New Player in Atopic Dermatitis. J. Investig. Dermatol. 2018, 138, 8–10. [Google Scholar] [CrossRef]
- Anitua, E.; Zalduendo, M.; Troya, M.; Padilla, S.; Orive, G. Leukocyte inclusion within a platelet rich plasma-derived fibrin scaffold stimulates a more pro-inflammatory environment and alters fibrin properties. PLoS ONE 2015, 10, e0121713. [Google Scholar] [CrossRef]
- Anitua, E.; Zalduendo, M.; Troya, M.; Tierno, R.; Alkhraisat, M.H. The inclusion of leukocytes into platelet rich plasma reduces scaffold stability and hinders extracellular matrix remodelling. Ann. Anat. Anat. Anz. 2022, 240, 151853. [Google Scholar] [CrossRef]
- Anitua, E.; Alonso, R.; Girbau, C.; Aguirre, J.J.; Muruzabal, F.; Orive, G. Antibacterial effect of plasma rich in growth factors (PRGF(R)-Endoret(R)) against Staphylococcus aureus and Staphylococcus epidermidis strains. Clin. Exp. Dermatol. 2012, 37, 652–657. [Google Scholar] [CrossRef]
- Mariani, E.; Canella, V.; Berlingeri, A.; Bielli, A.; Cattini, L.; Landini, M.P.; Kon, E.; Marcacci, M.; Di Matteo, B.; Filardo, G. Leukocyte presence does not increase microbicidal activity of Platelet-rich Plasma in vitro. BMC Microbiol. 2015, 15, 149. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Drago, L.; Bortolin, M.; Vassena, C.; Romano, C.L.; Taschieri, S.; Fabbro, M.D. Plasma Components and Platelet Activation Are Essential for the Antimicrobial Properties of Autologous Platelet-Rich Plasma: An In Vitro Study. PLoS ONE 2014, 9, e107813. [Google Scholar]
- Castillo, T.N.; Pouliot, M.A.; Kim, H.J.; Dragoo, J.L. Comparison of growth factor and platelet concentration from commercial platelet-rich plasma separation systems. Am. J. Sports Med. 2011, 39, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Oudelaar, B.W.; Peerbooms, J.C.; Huis In ’t Veld, R.; Vochteloo, A.J.H. Concentrations of Blood Components in Commercial Platelet-Rich Plasma Separation Systems: A Review of the Literature. Am. J. Sports Med. 2019, 47, 479–487. [Google Scholar] [CrossRef]
- Anitua, E.; Prado, R.; Azkargorta, M.; Rodriguez-Suarez, E.; Iloro, I.; Casado-Vela, J.; Elortza, F.; Orive, G. High-throughput proteomic characterization of plasma rich in growth factors (PRGF-Endoret)-derived fibrin clot interactome. J. Tissue Eng. Regen. Med. 2015, 9, E1–E12. [Google Scholar] [CrossRef]
- Anitua, E.; Muruzabal, F.; Pino, A.; Prado, R.; Azkargorta, M.; Elortza, F.; Merayo-Lloves, J. Proteomic Characterization of Plasma Rich in Growth Factors and Undiluted Autologous Serum. Int. J. Mol. Sci. 2021, 22, 12176. [Google Scholar] [CrossRef]
- Anitua, E.; de la Fuente, M.; Muruzabal, F.; Sanchez-Avila, R.M.; Merayo-Lloves, J.; Azkargorta, M.; Elortza, F.; Orive, G. Differential profile of protein expression on human keratocytes treated with autologous serum and plasma rich in growth factors (PRGF). PLoS ONE 2018, 13, e0205073. [Google Scholar] [CrossRef]
- Lee, H.W.; Choi, K.H.; Kim, J.Y.; Kim, K.O.; Haotian, B.; Yuxuan, L.; Noh, K.C. Proteomic Classification and Identification of Proteins Related to Tissue Healing of Platelet-Rich Plasma. Clin. Orthop. Surg. 2020, 12, 120–129. [Google Scholar] [CrossRef]
- Miroshnychenko, O.; Chalkley, R.J.; Leib, R.D.; Everts, P.A.; Dragoo, J.L. Proteomic analysis of platelet-rich and platelet-poor plasma. Regen. Ther. 2020, 15, 226–235. [Google Scholar] [CrossRef]



| Whole Blood | PRGF | L-PRP | |
|---|---|---|---|
| Leukocytes (×103/μL) | 5.65 ± 0.18 | 0.15 ± 0.08 | 8.59 ± 0.65 |
| Lymphocytes (%) | 38.1 ± 4.1 | n.d. | 50.3 ± 12.5 |
| Monocytes (%) | 3.6 ± 1.5 | n.d. | 2.1 ± 1.3 |
| Neutrophils (%) | 53.8 ± 2.6 | n.d. | 44.3 ± 10.0 |
| Eosinophils (%) | 3.9 ± 1.6 | n.d. | 2.9 ± 2.2 |
| Basophils (%) | 0.7 ± 0.1 | n.d. | 0.5 ± 0.1 |
| Erythrocytes (×106/μL) | 5.03 ± 0.08 | 0.02 ± 0.02 | 1.69 ± 1.09 |
| Platelets (×103/μL) | 215 ± 39 | 431 ± 133 | 434 ± 164 |
| Leukocyte concentration factor | - | 0.03 ± 0.01 | 1.52 ± 0.15 |
| Erythrocyte concentration factor | - | 0 | 0.34 ± 0.22 |
| Platelet concentration factor | - | 2.0 ± 0.4 | 2.0 ± 0.4 |
| Platelet yield (%) | - | 68.1 ± 5.9 | 67.5 ± 2.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anitua, E.; Pino, A.; Azkargorta, M.; Elortza, F.; Prado, R. High-Throughput Proteomic Analysis of Human Dermal Fibroblast Response to Different Blood Derivatives: Autologous Topical Serum Derived from Plasma Rich in Growth Factors (PRGF) versus Leukocyte- and Platelet-Rich Plasma (L-PRP). Biomolecules 2022, 12, 1002. https://doi.org/10.3390/biom12071002
Anitua E, Pino A, Azkargorta M, Elortza F, Prado R. High-Throughput Proteomic Analysis of Human Dermal Fibroblast Response to Different Blood Derivatives: Autologous Topical Serum Derived from Plasma Rich in Growth Factors (PRGF) versus Leukocyte- and Platelet-Rich Plasma (L-PRP). Biomolecules. 2022; 12(7):1002. https://doi.org/10.3390/biom12071002
Chicago/Turabian StyleAnitua, Eduardo, Ander Pino, Mikel Azkargorta, Felix Elortza, and Roberto Prado. 2022. "High-Throughput Proteomic Analysis of Human Dermal Fibroblast Response to Different Blood Derivatives: Autologous Topical Serum Derived from Plasma Rich in Growth Factors (PRGF) versus Leukocyte- and Platelet-Rich Plasma (L-PRP)" Biomolecules 12, no. 7: 1002. https://doi.org/10.3390/biom12071002
APA StyleAnitua, E., Pino, A., Azkargorta, M., Elortza, F., & Prado, R. (2022). High-Throughput Proteomic Analysis of Human Dermal Fibroblast Response to Different Blood Derivatives: Autologous Topical Serum Derived from Plasma Rich in Growth Factors (PRGF) versus Leukocyte- and Platelet-Rich Plasma (L-PRP). Biomolecules, 12(7), 1002. https://doi.org/10.3390/biom12071002






