Abstract
1,3-diaryl-2-propanone derivatives are synthetic compounds used as building blocks for the realization not only of antimicrobial drugs but also of new nanomaterials thanks to their ability to self-assemble in solution and interact with nucleopeptides. However, their ability to interact with proteins is a scarcely investigated theme considering the therapeutic importance that 1,3-diaryl-2-propanones could have in the modulation of protein-driven processes. Within this scope, we investigated the protein binding ability of 1,3-bis(1′-uracilyl)-2-propanone, which was previously synthesized in our laboratory utilizing a Dakin–West reaction and herein indicated as U2O, using bovine serum albumin (BSA) as the model protein. Through circular dichroism (CD) and UV spectroscopy, we demonstrated that the compound, but not the similar thymine derivative T2O, was able to alter the secondary structure of the serum albumin leading to significant consequences in terms of BSA structure with respect to the unbound protein (Δβ-turn + Δβ-sheet = +23.6%, Δα = −16.7%) as revealed in our CD binding studies. Moreover, molecular docking studies suggested that U2O is preferentially housed in the domain IIIB of the protein, and its affinity for the albumin is higher than that of the reference ligand HA 14−1 (HDOCK score (top 1–3 poses): −157.11 ± 1.38 (U2O); −129.80 ± 6.92 (HA 14−1); binding energy: −7.6 kcal/mol (U2O); −5.9 kcal/mol (HA 14−1)) and T2O (HDOCK score (top 1–3 poses): −149.93 ± 2.35; binding energy: −7.0 kcal/mol). Overall, the above findings suggest the ability of 1,3-bis(1′-uracilyl)-2-propanone to bind serum albumins and the observed reduction of the α-helix structure with the concomitant increase in the β-structure are consistent with a partial protein destabilization due to the interaction with U2O.
1. Introduction
Among the several families of proteins, albumins [1,2,3] are characterized by high peptide sequence homology, with human and bovine albumins sharing more than 75% identity [4]. Abundantly present in the circulatory system, albumins help maintain the osmotic blood pressure between the tissues and blood vessels [5]. Albumins, particularly bovine serum albumin (BSA), are often employed as protein models [6,7,8], and their interaction with most diverse ligands has been investigated for various applications in biomedicine and industrial areas [5,9,10,11,12,13,14,15]. The crystal structures of human serum albumin (HSA) and BSA, which share ~80% identity [16] and ~90% homology similarity [17] in the amino acid sequence, are shown superimposed in Figure 1a together with the sequence alignments. The albumin structure shows three domains with two main high-affinity binding sites alongside several others to which ligands bind with lower affinity [18].

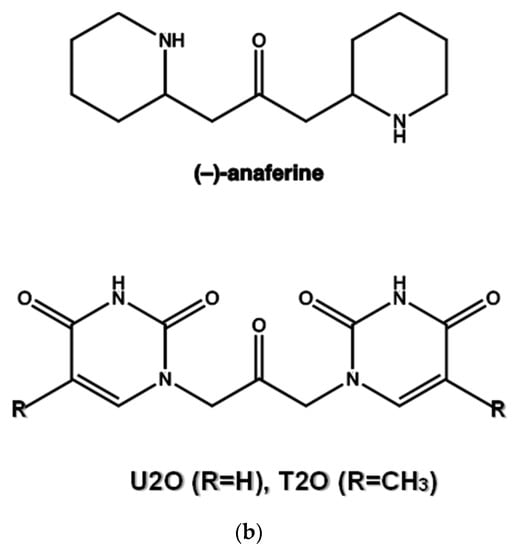
Figure 1.
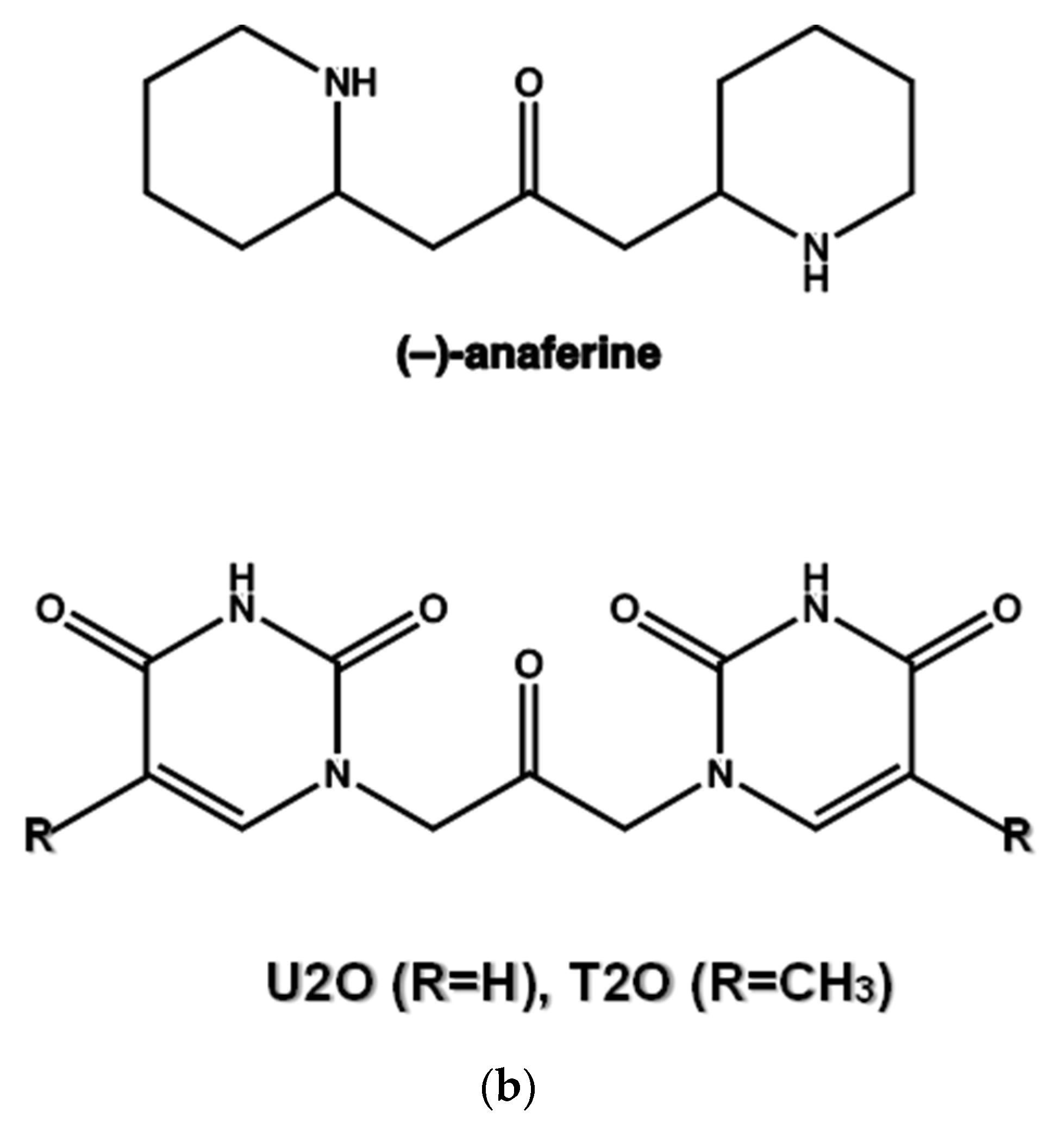
(a) Superimposed structures of HSA (PDB ID: 1n5u, light brown) and BSA (PDB ID: 4f5s, cyan) as visualized by UCSF Chimera software (Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, CA, USA, v.1.14) [49] and protein sequence alignment for HSA (P02768) and BSA (P02769) as obtained by Expasy software (Swiss Institute of Bioinformatics, Geneva, Switzerland, SIM-Alignment Tool for protein sequences) (https://web.expasy.org/sim/, accessed on 14 July 2022) with conserved binding site residues evidenced in red (https://www.uniprot.org/uniprotkb/P02768/entry and https://www.uniprot.org/uniprotkb/P02769/entry, accessed on 14 July 2022). (b) Chemical structures of the compounds studied as the protein ligands in the current work compared to (–)-anaferine.
In their primary physiological role, albumins transport many classes of molecules, such as metal ions, steroids, fatty acids, and amino acids, from the bloodstream to their specific target organs [4,19]. Remarkably, this function and the albumin’s ability to bind molecules with high affinity led to the pharmaceutical application of albumins as drug-carrying systems [20,21,22,23].
Heterocyclic compounds are an important class of molecules endowed with interesting biological functions and therapeutical potential, which are structurally related to different natural compounds [24,25,26]. Among others, benzofurans are potential anti-inflammatory [27], antibiotic [28,29,30], anticancer [31,32,33], neuroprotective and analgesic [34], and antiparasitic [35] compounds. The ability to bind nucleic acids [36] and inhibit specific serine/threonine kinases implied in cancer development (thus affecting the cancer cell cycle) were proposed as some of the most probable anticancer mechanisms for this class of molecules [32,37].
1,3-diaryl-2-propanone derivatives [38,39] are synthetic compounds used as building blocks for the realization not only of antimicrobial drugs [40] but also of new nanomaterials thanks to their ability to self-assemble in solution and interact with nucleopeptides [39,41]. Structurally, they are heteroaromatic analogs of (−)-anaferine, an alkaloid with sedative-hypnotic and anticancer properties extracted from Asiatic solanaceous plants (Figure 1b) [42]. Structural analogies can also be seen with certain 2-propanone derivatives 1,3-disubstituted with six-term rings (phenyl and piperazine rings) endowed with antimuscarinic properties [43]. In addition to the vast number of biomedical applications of 1,3-diaryl-2-propanones, our compound can be seen as an analog of diaryl urea derivatives that constitute an important family of anticancer drugs thanks to their ability to interact with specific proteins involved in the disease [44]. However, differently from diaryl ureas, the ability of 1,3-diaryl-2-propanone derivatives to interact with proteins remains a theme still scarcely explored and worthy of further research for the therapeutic importance that 1,3-diaryl-2-propanones could have in the modulation of protein-driven processes. With this in mind, we decided to investigate the protein binding ability of 1,3-bis(1′-uracilyl)-2-propanone, which was previously synthesized in our laboratory [41] utilizing the Dakin–West reaction [45,46,47,48] and herein indicated as U2O [41] (Figure 1b), using BSA as the model protein. Moreover, a comparison with T2O [39], a closely related derivative carrying thymine moieties in place of uracil (Figure 1b), was performed by spectroscopy, as described in the sections below.
2. Materials and Methods
2.1. Synthesis of 1,3-Bis(1′-uracilyl)-2-propanone (U2O) and 1,3-Bis(1′-thyminyl)-2-propanone (T2O)
U2O and T2O were obtained with high purity (≥95%) as ascertained by HPLC analysis following a synthetic procedure described in the literature based on the Dakin–West reaction [39,41]. All the intermediates used in the synthesis were from Acros Organics (Thermo Scientific Chemicals, Waltham, MA, USA) and Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and used without further purification steps.
2.2. CD and UV Binding Studies
Circular dichroism (CD) spectra were recorded on a Jasco J-715 spectropolarimeter equipped with a Jasco Peltier PTC-423S/15 temperature controller (Jasco Europe S.r.l, Cremella, Italy) using a Hellma-238-QS tandem quartz cell (2 × 0.4375 cm from Hellma Italia S.r.l., Milano, Italy) according to the previous literature experiments [50,51,52,53,54,55,56,57,58,59] with a response time of 1 s, a scanning speed of 100 nm min−1, and a bandwidth of 2.0 nm in the 205–260 nm wavelength range. All the spectra were averaged over three scans. UV absorption spectra were collected simultaneously to CD on the same instrument [60] to minimize possible errors induced by separate measurements on different instruments [61]. CD (Δε) values (M−1 cm−1) were calculated according to the equation: Δε = θ/(32980 × C × l), with θ being the measured ellipticity (mdeg), C the concentration (M), and l the optical path length (cm). All experiments were performed at 10 °C in 10 mM of sodium phosphate buffer at pH 7.4. The concentration of BSA (Sigma-Aldrich) was 1.5 μM.
2.3. CD Spectra Deconvolution
For the deconvolution of the circular dichroism spectra, CD (mdeg) and wavelength (nm) data were given as input to the program CD3 (http://lucianoabriata.altervista.org/jsinscience/cd/cd3.html, accessed on 30 May 2022) [62,63]. Only data corresponding to positive coefficient values were selected for the protein structure analysis, choosing the “Fit 4 components” option.
2.4. Molecular Docking and In Silico Protein–Ligand Interaction Analysis
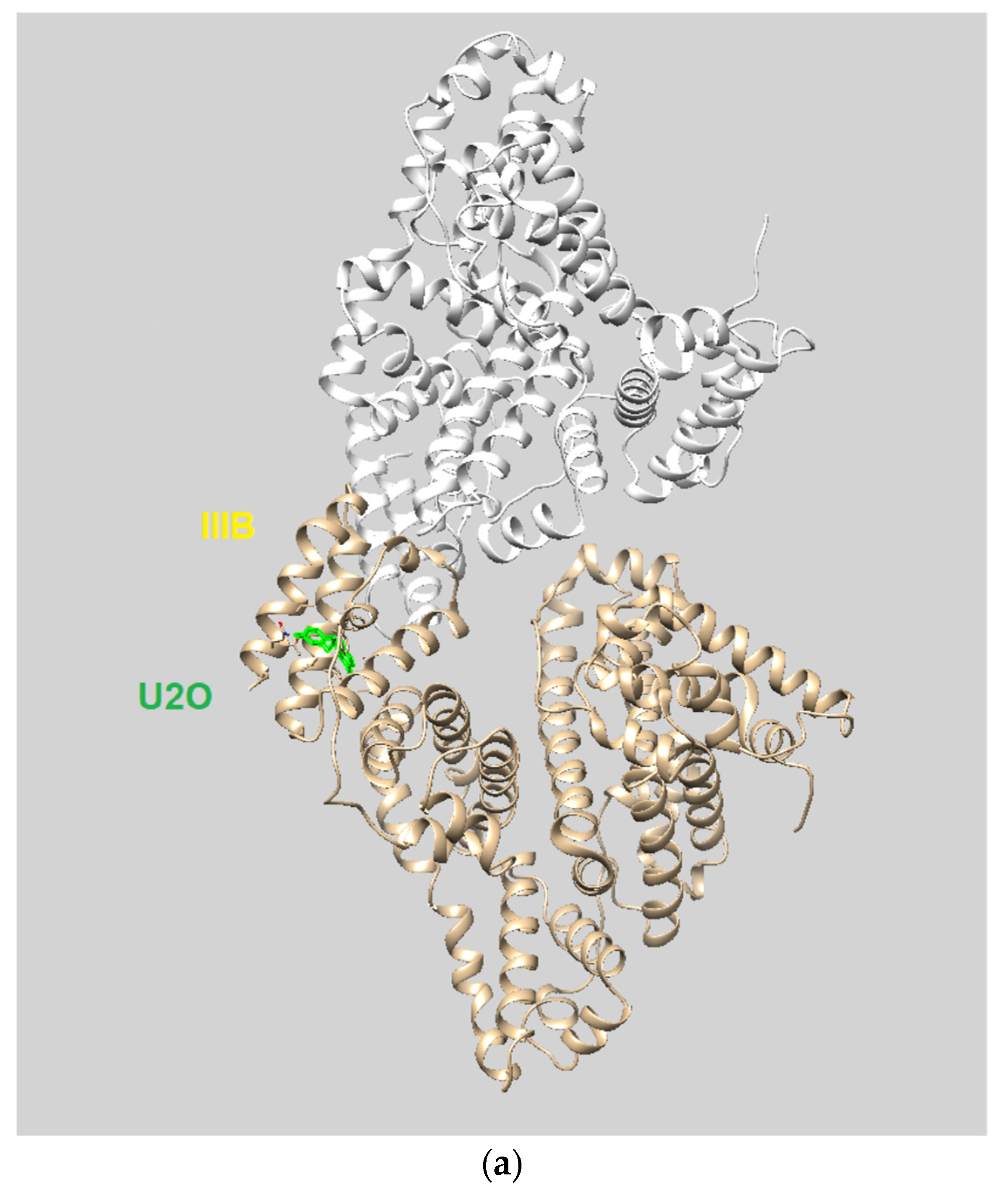
Molecular docking (MD) simulations [7,64,65,66,67,68] were performed running the HDOCK server (http://hdock.phys.hust.edu.cn, accessed on 30 May 2022) [69,70], suitable for both macromolecule-to-macromolecule [69] and macromolecules-to-small molecules [71] rigid dockings, using default parameters. The PDB entry 4f5s [72] and the energy-minimized 3D structure models of U2O and T2O (obtained by MOLVIEW (http://molview.org, accessed on 30 May 2022), and saved as .pdb files) were uploaded into the HDOCK server as the target and ligands, respectively. Interestingly, the HDOCK server predicts the protein/ligand interaction through a hybrid algorithm of template-free docking (when PDB IDs/structures are furnished such as in our case) and template-based (when only the target sequence is furnished) and specifically uses the PDB ID: 1n5u present in its database as a template for the 3D structure of the albumin, which is endowed with the highest homological identity with respect to 4f5s [73]. Based on the structural similarity of HSA to BSA, docking of ligands into possible binding sites of BSA was previously performed using the crystal structure of HSA [73]. However, caution should be paid when considering binding sites such as Site 1 that are less conserved among the different albumins than Site 2, because this can lead to different positions of the ligands within the site and different binding efficiencies for the two albumins [74,75,76]. Since we furnished the PDB ID: 4f5s as an input target, our dockings were based on BSA and not HSA structure. Thanks to the iterative knowledge-based scoring function ITScore-PP, the HDOCK server ranked the top ten poses obtained after each docking run. The program’s energy score (HDOCK score) values predicted by ITScore-PP are dimensionless, with larger negative numbers indicating higher affinity interactions between the interacting ligand and the target macromolecule, which was previously reported to correlate well to experimental binding affinities with a correlation coefficient of R = 0.71 [77]. More details on the HDOCK docking server, including the procedures for the docking, can be found at http://hdock.phys.hust.edu.cn (accessed on 30 May 2022). We analyzed the top-ranked pose (Top-1) and the top three and ten ranked poses for the complexes predicted by HDOCK according to the energy scores provided by the program, as explained in the Results section. The protein–ligand interaction diagrams reported in this work were obtained by ProteinsPlus (https://proteins.plus/, accessed on 30 May 2022) [78]. The FireDock software (Tel Aviv University, Tel Aviv, Israel) [79] was then used for rescoring and refinement because of its ability to improve the flexibility and correct scoring errors typically experienced during the molecular docking calculations by fast rigid-body docking tools [79]. The top 100 results for the rigid BSA/U2O docking previously obtained by PatchDock, a docking program based on ligand–receptor geometric shape complementarity [80], were transferred to FireDock for refinement. The top-ranked FireDock solutions, according to the contribution of the atomic contact energy (ACE), were chosen for the study of the complexes. Binding energies (kcal/mol) were computed by AutoDock Vina using the 1-Click Mcule online platform (https://mcule.com (accessed on 14 July 2022, Mcule Inc., Palo Alto, CA, USA)) [81,82]. Details on the procedure with 1-Click Mcule including a tutorial are available at https://mcule.com/apps/1-click-docking/ (accessed on 14 July 2022). The x, y, z coordinates for the binding centers were those corresponding to Phe 550 (34.799, 13.656, 124.426) for the subdomain IIIB (U2O), Val 432 (11.840, 24.326, 120.374) for the subdomain IIIA (T2O), and Ser 191 (59.980, 20.440, 89.569) for the subdomain IIA (HA 14−1).
2.5. Pharmacokinetic Properties
The SMILES (Simplified Molecular Input Line Entry System) codes of U2O (O=C(CN1C=CC(=O)NC1=O)CN2C=CC(=O)NC2=O) and T2O (Cc2cn(CC(=O)Cn1cc(C)c(=O)[nH]c1=O)c(=O)[nH]c2=O) were obtained with the MOLVIEW software (TU Delft, Deft, Netherlands, v2.4) and applied to calculate the logarithms of the partition coefficients (cLogP), blood-brain barrier (BBB) permeability, pan-assay interference compounds (PAINS) score, and druggability properties (Lipinski model) presented in this work by using the SwissADME web service (http://www.swissadme.ch/index.php, accessed on 30 May 2022).
3. Results and Discussion
Experimental and computational (docking, ligand–protein analysis) studies were conducted to achieve insights into the molecular recognition of the model protein BSA by the U2O and T2O ligands. CD and UV experiments were performed in a two-chamber cell (Figure 2a) [83,84,85] in which we placed the protein and an excess of ligand separately. After mixing the solutions, the CD spectra of the two components (BSA and U[T]2O) were measured. The concentrations of both protein and ligand were halved after mixing, but the path length (2 × 0.4375 cm) increased by a factor of two. Any spectral difference observed after mixing the two component solutions was indicative of interaction [83,84,85].
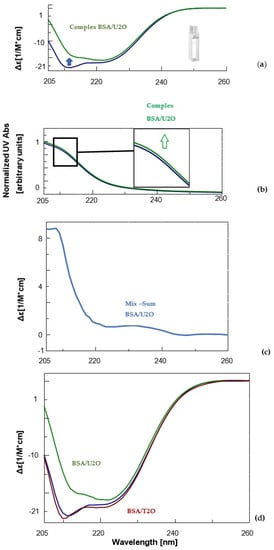
Figure 2.
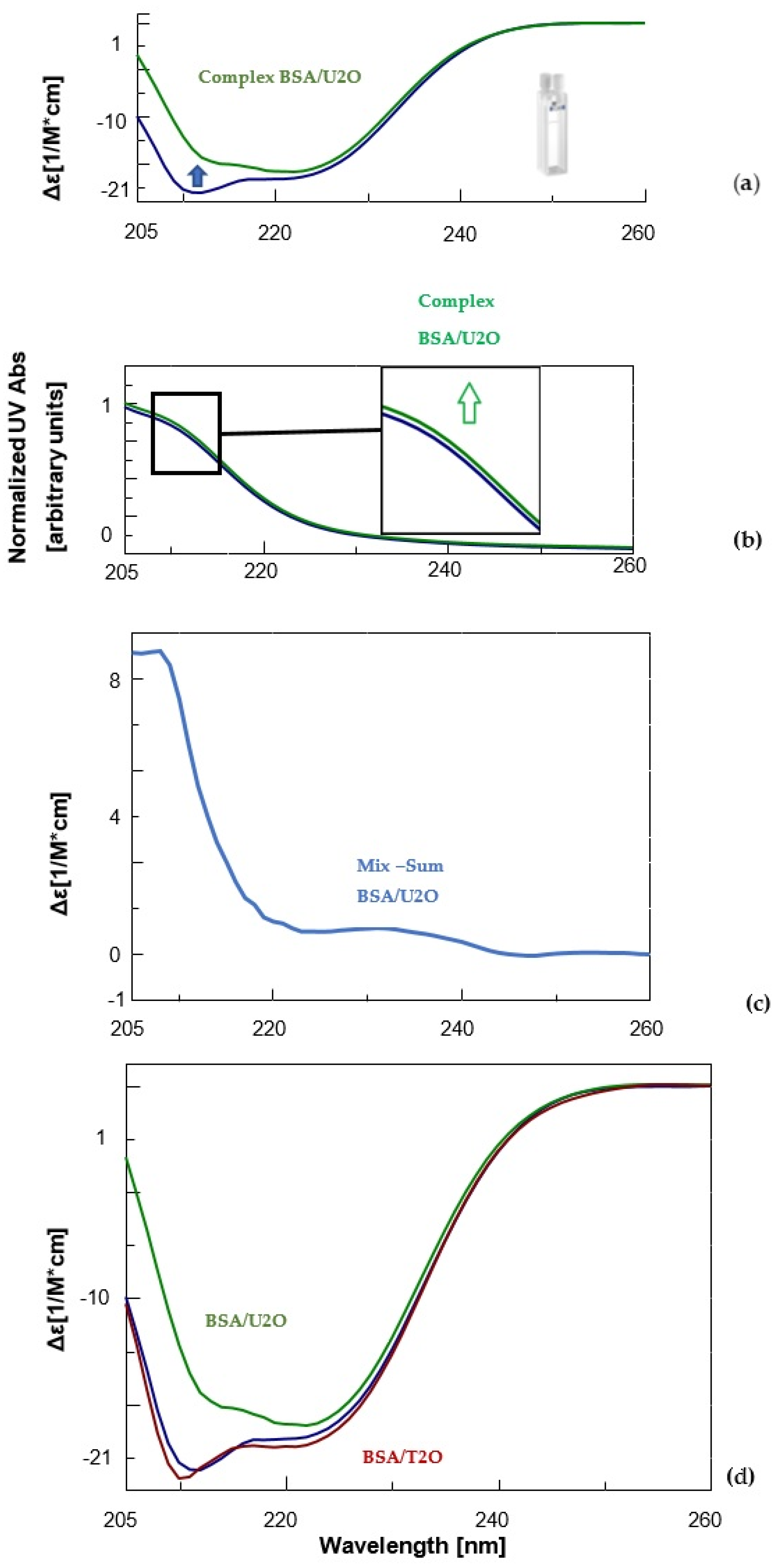
CD (a) and UV (b) spectra of BSA (1.5 μM, blue) and its complex with the 1,3-bis(1′-uracilyl)-2-propanone (6 nmol, green) in 10 mM sodium phosphate buffer (pH = 7.5) at 10 °C. Inset in Figure 2b, a zoomed-in view of UV bands between 210 and 215 nm. (c) Difference in CD spectrum of BSA in complex with U2O (CDcomplex − CDBSA). (d) Comparison of the CD spectra of BSA (blue), its complex with U2O (green) and T2O (red) under the same experimental conditions previously indicated.
3.1. Spectroscopic Binding Studies on BSA with U2O and T2O
Notoriously, the BSA structure is dominated by α-helix structures, which account for approximately 60% of its structure [86]. Moreover, about 20% of β-sheet + β-turn structures as well as ca. 20% random coil were experimentally observed for the native BSA in solution [87,88]. Accordingly, our far-UV CD experiments revealed for the unliganded serum albumin, mainly the characteristic features of the typical helical structure of proteins, i.e., two negative bands at about 208 and 222 nm (Figure 2a, blue line). After complexation with U2O, the CD band at 208 nm underwent bathochromic and hypochromic shifts (Figure 2a, green line). This evidence, together with the slight hyperchromic effect observed in the UV spectrum of the complex (Figure 2b, green line) relative to the curve of the unliganded BSA (blue line), and the non-null difference CD spectrum (Figure 2c), indicated that the BSA secondary structure underwent slight but clear modifications as a consequence of the interaction with U2O but not T2O (Figure 2d). In fact, when the CD binding experiment was performed with T2O, no substantial CD spectral difference was observed, suggesting that T2O was not able to provoke significant secondary structure perturbation in the albumin target. To achieve more quantitative information on the interaction of U2O with the protein, we performed a deconvolution of the CD spectra and reported the rates for the secondary structures’ content in the BSA and their variations in the absence and presence of an excess of U2O in Table 1.

Table 1.
Variation in the BSA secondary structures’ content (%) resulting from the addition of 1,3-bis(1′-uracilyl)-2-propanone. Please note that, besides a ~17% helical content loss detected after ligand binding, U2O determines a significant increase in β-structures.
Far-UV CD spectroscopy provides useful insights into protein conformation analysis, allowing for the monitoring of secondary structure composition and changes after molecular interactions. In fact, the deconvolution of CD spectra into secondary structure compositions allows for the quantification of a given protein α, β-sheet, β-turn and random coil contents and the exploration of the effects of biomolecular interactions on the secondary structure composition based on the resulting CD complex spectra [89,90]. According to Table 1, unliganded BSA showed a 63.0% content of helical structure (comparable to 61% found previously [87,88]) and a 21.7% of β-sheet + β-turn, which was similar to other literature reports [87,88]. On the other hand, U2O provoked significant changes in the secondary structures of BSA, with a certain loss in the helical content (−16.7%), and a concomitant increase in the β-structures (+23.6%) that are indicative of a partial protein destabilization, in agreement with the literature reports [91]. Remarkably, other nitrogen-containing heteroaromatic compounds such as alprazolam were experimentally found to be able to decrease the percentage of BSA α-helical content from 66.5 to 37.0% (Δα-helix = −29.5%) [92]. Interestingly, the interaction with U2O did not seem to provoke a significant increase in the amount of random coil structure, which indeed resulted in it being slightly decreased (−6.9%, Table 1). Moreover, the ligand-binding could determine modifications in the aromatic regions of BSA, which could explain the slight hyperchromic effect observed in the UV spectrum (Figure 2b), an aspect we investigated in silico as described below.
3.2. Computational Studies
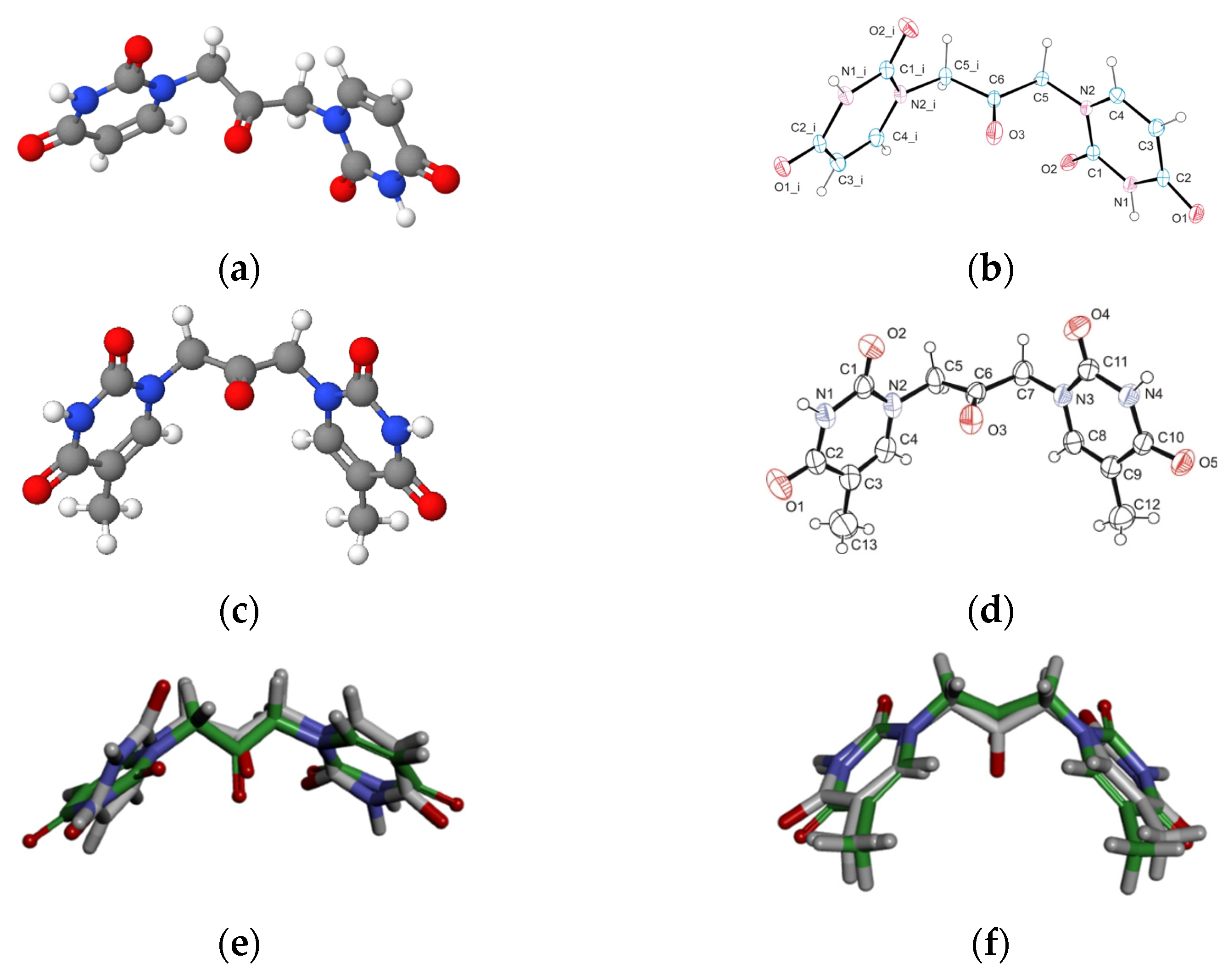
Aiming to give a tentative interpretation of some of the binding evidence from our experiments described in the previous sections, we performed a molecular docking study. Within this scope, the HDOCK server was used for blind molecular docking between BSA and U2O. First, we obtained the energy-minimized 3D structure model of U2O by MOLVIEW (http://molview.org, accessed on 30 May 2022), and the 3D structure (Figure 3a) was compared to that (Figure 3b) previously described by some of us through X-ray crystallography that was in good agreement with the predicted structure (Figure 3e) [41]. Similarly, the T2O computational 3D model (Figure 3c) obtained following the same procedure seems to resemble the experimental structure very closely, in terms of the orientation of both heteroaromatic rings as well as C=O, and NH moieties with respect to the central carbonyl moiety (Figure 3d,f). Afterward, the 3D structure of U2O was uploaded as a ligand into the HDOCK server, and the top-10 solutions resulting from the blind rigid docking were analyzed. The results of the docking of U2O with BSA are presented in Table 2 and Figure 4.
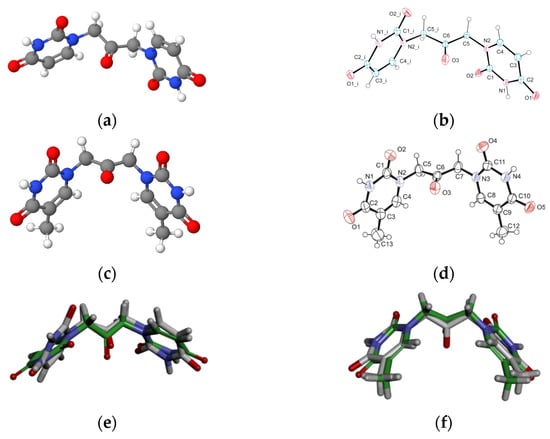
Figure 3.
The energy-minimized 3D structure models of U2O (a) and T2O (c) obtained by MolView software (TU Delft, Deft, Netherlands, v2.4) compared to the X-ray solved structures (b,d, respectively). Superimposed U2O (e) and T2O (f) 3D computational models (green) and their X-ray solved structures. The original files for the two X-ray structures can be freely downloaded from https://www.ccdc.cam.ac.uk/ (accessed on 14 July 2022) CCDC 932,679 (U2O) and 821,514 (T2O) [39,41].

Table 2.
HDOCK docking results for the best pose and mean values from the top-1–3 and top-1–10 poses of U2O, T2O, and reference compound (HA 14−1, [73]) complexed with BSA. The interface residues within 5.0 Å from the ligand in the top-1 complexes are reported in the last column.
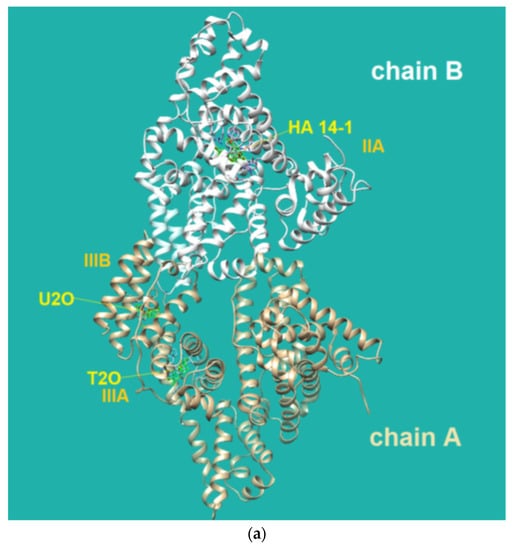
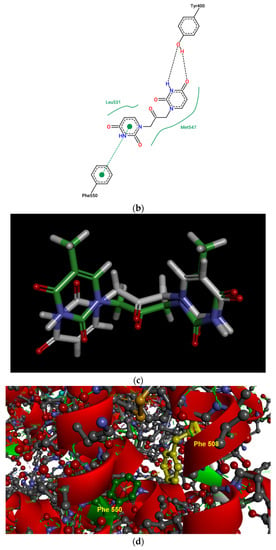
Figure 4.
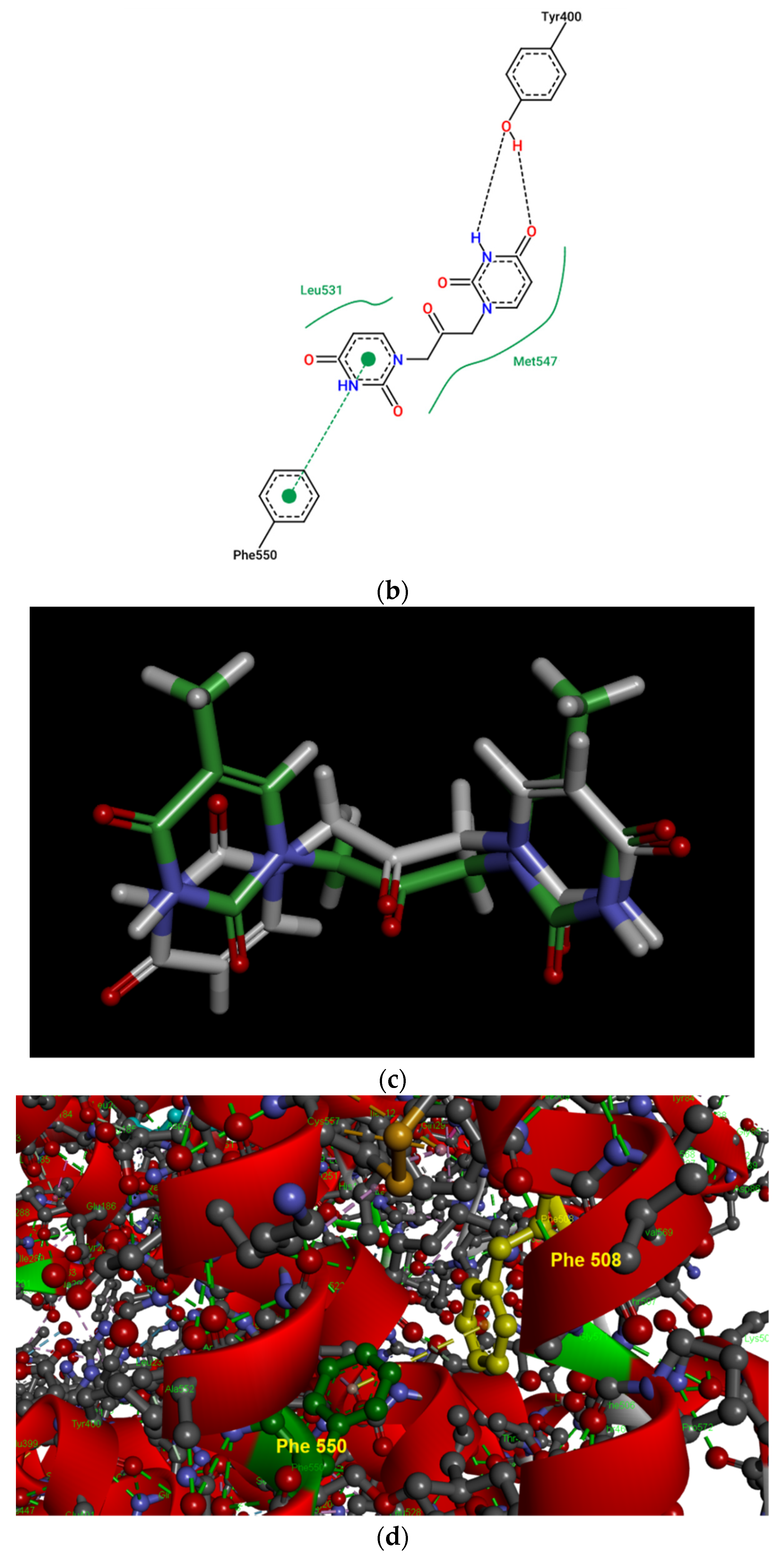
(a) Pose view for the complex of U2O with BSA as obtained by blind rigid docking with the HDOCK server and visualized in UCSF Chimera. (b) 2D protein–ligand interaction diagram obtained by ProteinPlus with BSA and U2O. H-bond length for C=O—H-O (Tyr 400): 2.44 Å. (c) Superimposed 3D models for U2O and T2O. (d) Detail of the 3D structure of BSA (PDB ID: 4f5s) as visualized by the software Discovery Studio (Dassault Systèmes Corporate, Waltham, MA, USA, v.2021) showing a π–π stacking interaction between Phe-550 and Phe-508.
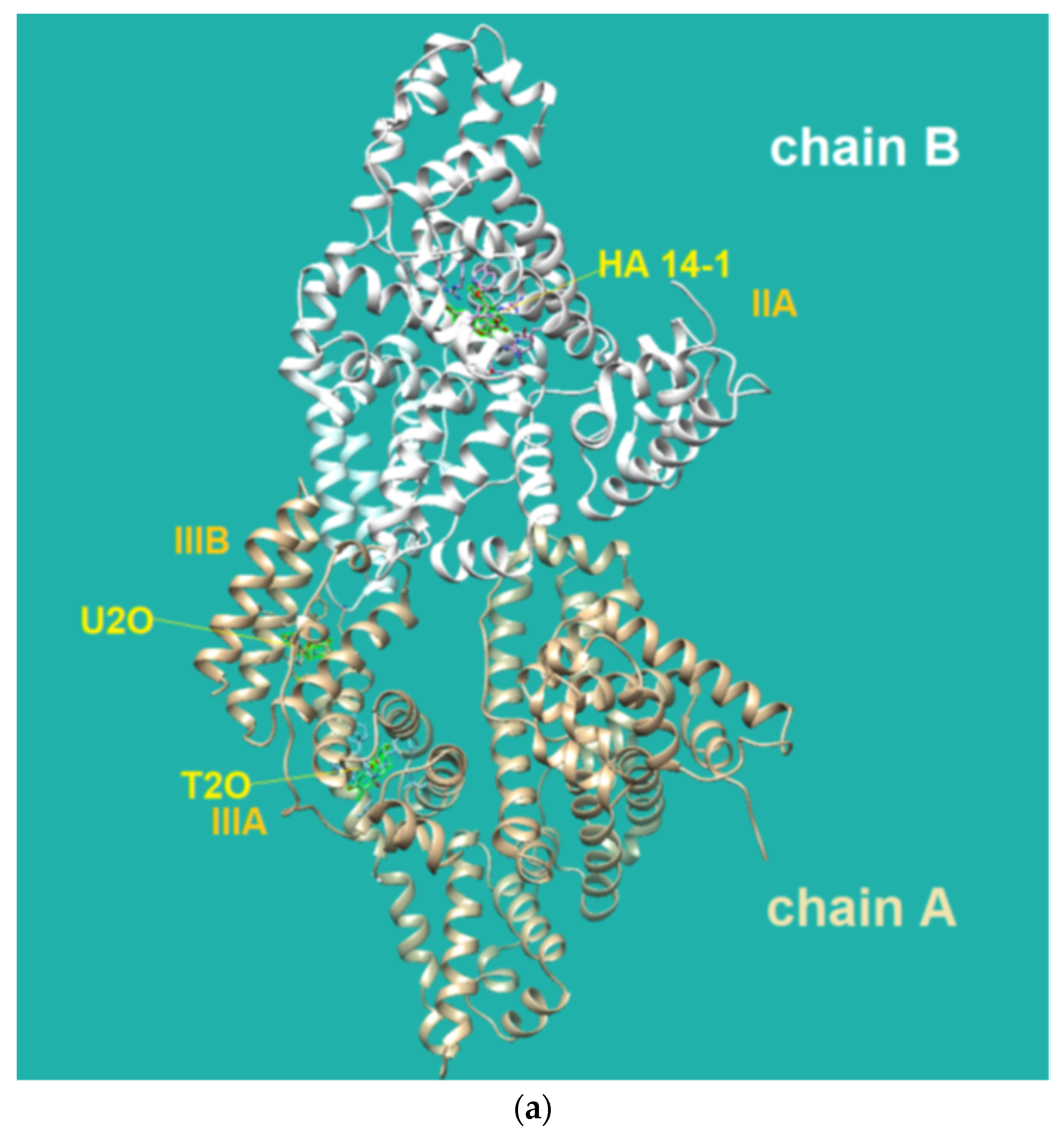
The HDOCK scores listed in Table 2 are dimensionless and are helpful only in achieving comparisons between ligands of the same target. Therefore, we also used the same procedure with T2O, which showed a lower affinity for the same protein as revealed by the HDOCK scores and AutoDock Vina binding energies (Table 2). Similarly, we also repeated the blind docking with the reference literature BSA ligand HA 14−1 [73]. Our docking seems to indicate that U2O could bind BSA with higher affinity than HA 14−1, as suggested by the larger negative scores for U2O observed for the top-1, top 1–3, and top 1–10 poses (−158.56 vs. −137.72, −157.11 ± 1.38 vs. −129.80 ± 6.92, −155.18 ± 1.60 vs. −124.72 ± 5.15, respectively, Table 2). Accordingly, the binding energy was more favorable in the case of U2O with respect to T2O and HA 14−1 (−7.6 vs. −7.0 and −5.9 kcal/mol, respectively). Moreover, the amino acid residues involved in the three predicted interactions are different, suggesting that while HA 14−1 binds the chain B of BSA at the subdomain IIA (Figure 4a) as reported in the literature [73], T2O and U2O more likely recognize the albumin (Figure 4a) at the level of chain A, with T2O binding the subdomain IIIA while U2O interacts with Tyr-400 (with predicted H-bonding, C=O—H-O H-bond length: 2.44 Å) and residues such as Phe-550 (π-stacking), Leu-531 and Met-547 (hydrophobic interaction, Figure 4b) of the subdomain IIIB. The differences in BSA binding observed experimentally could be due to the higher steric hindrance determined by the two methyl groups in T2O as well as the different orientation of the carbonyl and NH moieties in this latter compound with respect to U2O (Figure 4c), not allowing the simultaneous H-bond, hydrophobic and aromatic interactions stabilizing the BSA/U2O complex. Our studies suggest that U2O interacts with the protein backbone via hydrogen bonding, and interactions with hydrophobic residues interfere with the mutual attraction of BSA nonpolar groups, as well as π–π stacking interactions leading to the observed UV absorbance differences (e.g., interference with π–π Phe 550–Phe 508 stacking, Figure 4d).
The three-dimensional structure of the serum albumin comprises three helical domains (I, II, and III), each of which is divided into two subdomains (A and B). Several molecules, including warfarin, indomethacin, and phenylbutazone (PB), bind at the binding site IIA, which is also indicated as drug site 1, while others such as diflunisal, diazepam, and iophenoxic acid bind to the site IIIA (also called drug site 2) [73].
Flexible docking was also performed on U2O in complex with BSA using the program FireDock [79]. This study confirmed the tendency of U2O to bind the albumin in the subdomain IIIB (Figure 5a) involving again Phe-550 in aromatic interactions and Leu-531 (hydrophobic interaction) together with Phe-506 (aromatic interaction), Gln 579 (H-bonding), and Ala-527 (hydrophobic interaction, Figure 5b). Overall, all BSA residues involved in the interaction with U2O predicted after both the rigid and flexible dockings belong to α-helical regions (Figure 5c) of the albumin, suggesting that this molecular recognition can alter the α-helical content of BSA. Finally, in predicting some pharmacokinetics for U2O, we found that the compound, not likely to permeate the blood–brain barrier (BBB, Table 3), is slightly more soluble in water than in organic solvents (cLogP = −0.83, Table 3). Nonetheless, it shows a favorable drug-likeness profile (0 violations of Lipinski’s rule of five, Table 3) [93]. In this prediction, U2O lacks any unspecific biomolecular interaction tendency (PAINS score: 0, Table 3) [94]. On the other side, T2O shows similar predicted pharmacokinetic properties but is endowed with higher hydrophobicity (cLogP = −0.15, Table 3) because of the two methyl moieties of thymine bases.
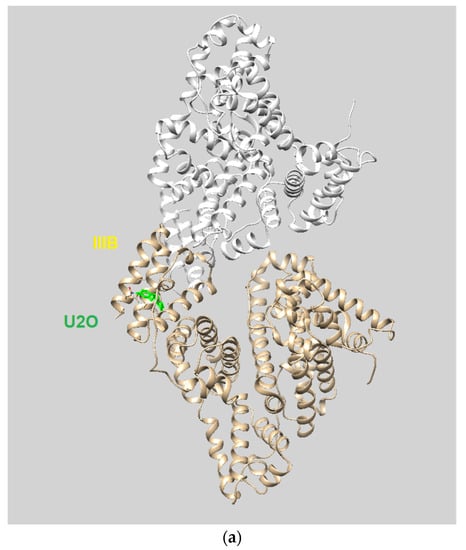
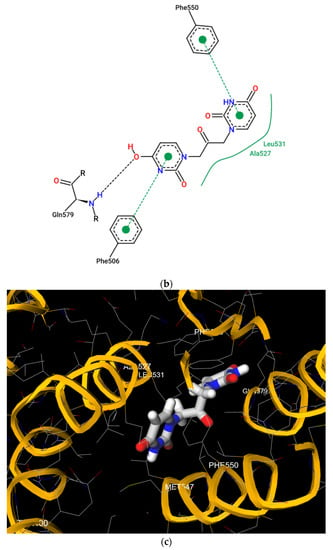
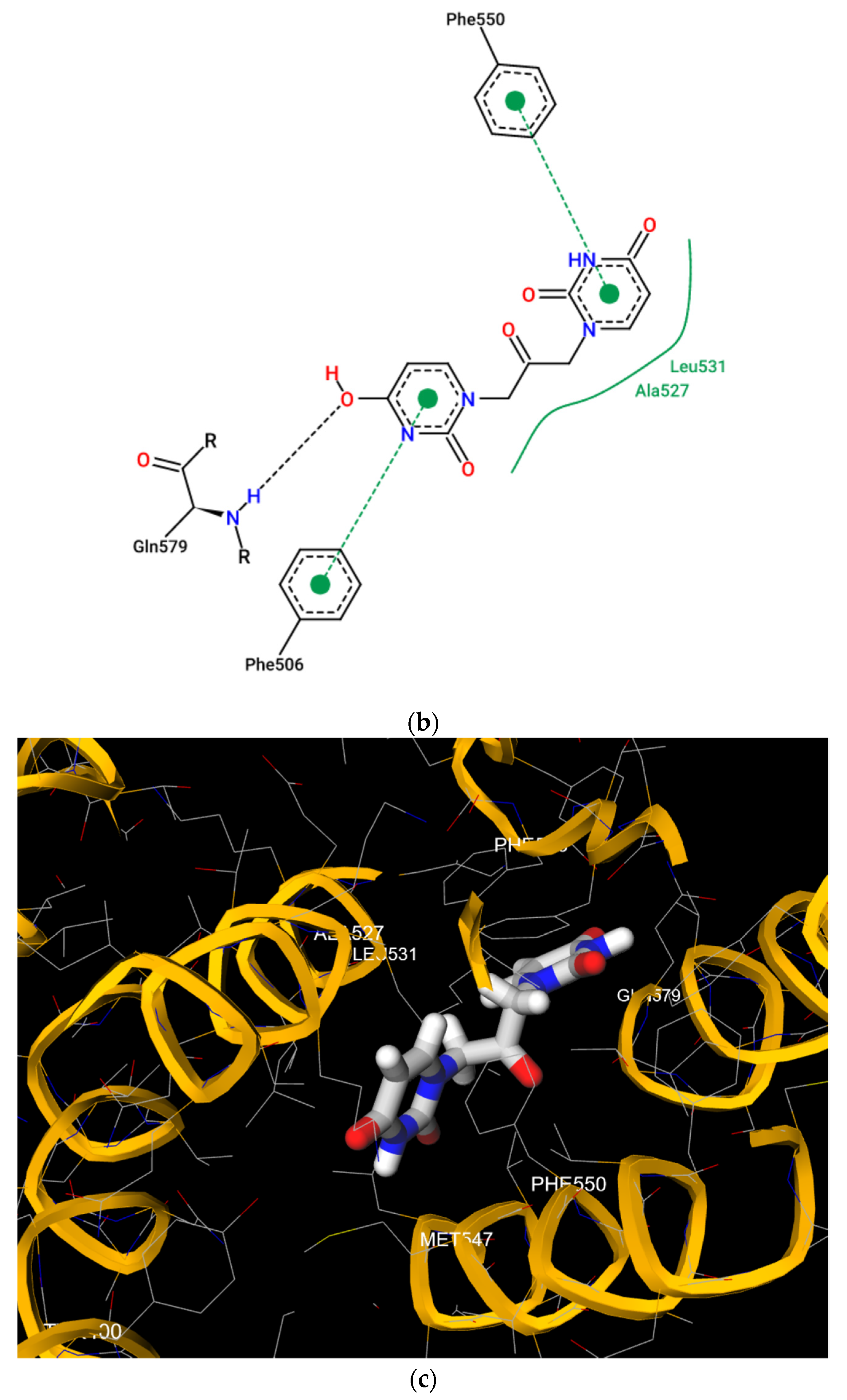
Figure 5.
(a) Pose view for the complex of U2O with BSA as obtained by blind flexible docking with the FireDock server and visualized in UCSF Chimera. (b) 2D protein–ligand interaction diagram obtained by ProteinPlus with BSA/U2O complex after FireDock docking. (c) Pose view for the complex BSA/U2O showing the residues involved in the interactions predicted after both the rigid and flexible dockings.

Table 3.
Drug-likeness and pharmacokinetic properties predicted for U2O and T2O by the SwissADME software (Swiss Institute of Bioinformatics, Geneva, Switzerland).
4. Conclusions
We demonstrated that 1,3-bis(1′-uracilyl)-2-propanone (U2O) could bind to serum albumins, which is particularly important in drug delivery applications where these proteins can act as carriers of these bioactive heterocyclic molecules. Our combined experimental (CD- and UV-based) and computational (molecular docking) investigation led us to conclude that the U2O structure can interact with BSA at the level of the subdomain IIIB of the serum albumin in the vicinity of residues such as Phe-550 (π–π stacking), and Leu-531 (hydrophobic interaction) involved in complex formation. In our predictions, the affinity of U2O for BSA was higher than both T2O and the literature ligand HA 14−1 (binding energies: −7.6 vs. −7.0 and −5.9 kcal/mol, respectively). The experimentally found changes in albumin secondary structure content induced by U2O (Δβ-turn + Δβ-sheet = +23.6%, Δα = −16.7%) and the slight hyperchromic effect observed by UV spectroscopy could be explained by the partial protein destabilization and the binding with residues such as Phe-550 that could determine higher exposure of aromatic rings previously involved in π–π stacking with other BSA residues to the solvent, thus increasing the UV absorbance. Overall, all BSA residues involved in the interaction with U2O predicted after both rigid and flexible dockings belong to α-helical regions (Figure 5c) of the albumin, suggesting that this molecular recognition can alter the α-helical content of BSA.
In conclusion, U2O, a building block for the realization of biomedical nanostructures, binds BSA and could likely be efficiently transported in human serum by albumins, thus being effective in the modulation of protein-driven processes.
Author Contributions
Conceptualization, G.O., G.N.R. and N.B.; data curation, A.P.F., M.T. and G.N.R.; funding acquisition, G.P. and N.B.; investigation, F.G., A.P.F., C.D. and G.N.R.; methodology, M.T., G.P., G.O. and G.N.R.; supervision, G.N.R.; writing—original draft, G.N.R.; writing—review and editing, G.N.R. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by Ministero dell’Università e della Ricerca FOE 2017-ISBE-IT Joint Research Unit (G.P., N.B.) and by the Department of Pharmacy-University of Naples Federico II grant “Sostegno allo Sviluppo della Ricerca Dipartimentale” (N.B.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We thank Antonietta Gargiulo for her technical assistance and help in the literature search. We are grateful to Gergely Prikler and Gergely Takács (https://mcule.com, accessed on 14 July 2022, Mcule team, Hungary) for their kind support with the 1-Click Mcule online platform. BSA and HSA structures were superimposed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Majorek, K.A.; Porebski, P.J.; Dayal, A.; Zimmerman, M.D.; Jablonska, K.; Stewart, A.J.; Chruszcz, M.; Minor, W. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol. 2012, 52, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Kosa, T.; Maruyama, T.; Otagiri, M. Species differences of serum albumins: I. Drug binding sites. Pharm. Res. 1997, 14, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho Bertozo, L.; Kogut, M.; Maszota-Zieleniak, M.; Samsonov, S.A.; Ximenes, V.F. Induced circular dichroism as a tool to monitor the displacement of ligands between albumins. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 278, 121374. [Google Scholar] [CrossRef] [PubMed]
- Peters, T., Jr. Serum albumin. Adv. Protein Chem. 1985, 37, 161–245. [Google Scholar]
- Carter, D.C.; Ho, J.X. Structure of serum albumin. Adv. Protein Chem. 1994, 45, 153–203. [Google Scholar]
- Wani, T.A.; Alanazi, M.M.; Alsaif, N.A.; Bakheit, A.H.; Zargar, S.; Alsalami, O.M.; Khan, A.A. Interaction Characterization of a Tyrosine Kinase Inhibitor Erlotinib with a Model Transport Protein in the Presence of Quercetin: A Drug–Protein and Drug–Drug Interaction Investigation Using Multi-Spectroscopic and Computational Approaches. Molecules 2022, 27, 1265. [Google Scholar] [CrossRef]
- Roviello, V.; Musumeci, D.; Mokhir, A.; Roviello, G.N. Evidence of Protein Binding by a Nucleopeptide Based on a Thyminedecorated L-Diaminopropanoic Acid through CD and In Silico Studies. Curr. Med. Chem. 2021, 28, 5004–5015. [Google Scholar] [CrossRef]
- Polat, H.; Eren, M.C.; Polat, M. The effect of protein BSA on the stability of lipophilic drug (docetaxel)-loaded polymeric micelles. Colloids Surf. A Physicochem. Eng. Asp. 2021, 631, 127712. [Google Scholar] [CrossRef]
- Akdogan, Y.; Emrullahoglu, M.; Tatlidil, D.; Ucuncu, M.; Cakan-Akdogan, G. EPR studies of intermolecular interactions and competitive binding of drugs in a drug–BSA binding model. Phys. Chem. Chem. Phys. 2016, 18, 22531–22539. [Google Scholar] [CrossRef]
- Fik-Jaskółka, M.A.; Mkrtchyan, A.F.; Saghyan, A.S.; Palumbo, R.; Belter, A.; Hayriyan, L.A.; Simonyan, H.; Roviello, V.; Roviello, G.N. Spectroscopic and SEM evidences for G4-DNA binding by a synthetic alkyne-containing amino acid with anticancer activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 229, 117884. [Google Scholar] [CrossRef]
- Fik-Jaskółka, M.A.; Mkrtchyan, A.F.; Saghyan, A.S.; Palumbo, R.; Belter, A.; Hayriyan, L.A.; Simonyan, H.; Roviello, V.; Roviello, G.N. Biological macromolecule binding and anticancer activity of synthetic alkyne-containing l-phenylalanine derivatives. Amino Acids 2020, 52, 755–769. [Google Scholar] [CrossRef]
- Liu, E.-H.; Qi, L.-W.; Li, P. Structural relationship and binding mechanisms of five flavonoids with bovine serum albumin. Molecules 2010, 15, 9092–9103. [Google Scholar] [CrossRef]
- Szymańska, M.; Pospieszna-Markiewicz, I.; Mańka, M.; Insińska-Rak, M.; Dutkiewicz, G.; Patroniak, V.; Fik-Jaskółka, M.A. Synthesis and Spectroscopic Investigations of Schiff Base Ligand and Its Bimetallic Ag (I) Complex as DNA and BSA Binders. Biomolecules 2021, 11, 1449. [Google Scholar] [CrossRef]
- Izawa, H.; Kinai, M.; Ifuku, S.; Morimoto, M.; Saimoto, H. Guanidinylation of Chitooligosaccharides involving internal cyclization of the Guanidino group on the reducing end and effect of Guanidinylation on protein binding ability. Biomolecules 2019, 9, 259. [Google Scholar] [CrossRef]
- Losytskyy, M.; Chornenka, N.; Vakarov, S.; Meier-Menches, S.M.; Gerner, C.; Potocki, S.; Arion, V.B.; Gumienna-Kontecka, E.; Voloshin, Y.; Kovalska, V. Sensing of Proteins by ICD Response of Iron (II) Clathrochelates Functionalized by Carboxyalkylsulfide Groups. Biomolecules 2020, 10, 1602. [Google Scholar] [CrossRef]
- Gelamo, E.; Silva, C.; Imasato, H.; Tabak, M. Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: Spectroscopy and modelling. Biochim. et Biophys. Acta (BBA)-Protein Struct. Mol. Enzymol. 2002, 1594, 84–99. [Google Scholar] [CrossRef]
- Ghosh, S.; Jana, S.; Guchhait, N. Domain specific association of small fluorescent probe trans-3-(4-monomethylaminophenyl)-acrylonitrile (MMAPA) with bovine serum albumin (BSA) and its dissociation from protein binding sites by Ag nanoparticles: Spectroscopic and molecular docking study. J. Phys. Chem. B 2012, 116, 1155–1163. [Google Scholar] [CrossRef]
- Simard, J.R.; Zunszain, P.A.; Hamilton, J.A.; Curry, S. Location of high and low affinity fatty acid binding sites on human serum albumin revealed by NMR drug-competition analysis. J. Mol. Biol. 2006, 361, 336–351. [Google Scholar] [CrossRef]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef]
- Loureiro, A.; Azoia, N.G.; Gomes, A.C.; Cavaco-Paulo, A. Albumin-based nanodevices as drug carriers. Curr. Pharm. Des. 2016, 22, 1371–1390. [Google Scholar] [CrossRef]
- Parodi, A.; Miao, J.; Soond, S.M.; Rudzińska, M.; Zamyatnin, A.A. Albumin nanovectors in cancer therapy and imaging. Biomolecules 2019, 9, 218. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Camacho, M.; Encinar, J.A.; Martínez-Tomé, M.J.; Esquembre, R.; Mateo, C.R. The Interaction of Temozolomide with Blood Components Suggests the Potential Use of Human Serum Albumin as a Biomimetic Carrier for the Drug. Biomolecules 2020, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Yadav, A.B. Preparation and characterization of BSA as a model protein loaded chitosan nanoparticles for the development of protein-/peptide-based drug delivery system. Future J. Pharm. Sci. 2021, 7, 200. [Google Scholar] [CrossRef]
- D’Errico, S.; Borbone, N.; Catalanotti, B.; Secondo, A.; Petrozziello, T.; Piccialli, I.; Pannaccione, A.; Costantino, V.; Mayol, L.; Piccialli, G.; et al. Synthesis and Biological Evaluation of a New Structural Simplified Analogue of cADPR, a Calcium-Mobilizing Secondary Messenger Firstly Isolated from Sea Urchin Eggs. Mar. Drugs 2018, 16, 89. [Google Scholar] [CrossRef]
- D’Errico, S.; Oliviero, G.; Amato, J.; Borbone, N.; Cerullo, V.; Hemminki, A.; Piccialli, V.; Zaccaria, S.; Mayol, L.; Piccialli, G. Synthesis and biological evaluation of unprecedented ring-expanded nucleosides (RENs) containing the imidazo [4,5-d][1,2,6]oxadiazepine ring system. Chem. Commun. 2012, 48, 9310. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Piccialli, G.; Mayol, L. Synthesis of N-1 and ribose modified inosine analogues on solid support. Tetrahedron Lett. 2007, 48, 397–400. [Google Scholar] [CrossRef]
- Feng, Z.; Mohapatra, S.; Klimko, P.G.; Hellberg, M.R.; May, J.A.; Kelly, C.; Williams, G.; McLaughlin, M.A.; Sharif, N.A. Novel benzodifuran analogs as potent 5-HT2A receptor agonists with ocular hypotensive activity. Bioorganic Med. Chem. Lett. 2007, 17, 2998–3002. [Google Scholar] [CrossRef]
- Alper-Hayta, S.; Arisoy, M.; Temiz-Arpaci, Ö.; Yildiz, I.; Aki, E.; Özkan, S.; Kaynak, F. Synthesis, antimicrobial activity, pharmacophore analysis of some new 2-(substitutedphenyl/benzyl)-5-[(2-benzofuryl)carboxamido]benzoxazoles. Eur. J. Med. Chem. 2008, 43, 2568–2578. [Google Scholar] [CrossRef]
- Soni, J.N.; Soman, S.S. Synthesis and antimicrobial evaluation of amide derivatives of benzodifuran-2-carboxylic acid. Eur. J. Med. Chem. 2014, 75, 77–81. [Google Scholar] [CrossRef]
- Ashok, D.; Sudershan, K.; Khalilullah, M. Solvent-free microwave-assisted synthesis of E-(1)-(6-benzoyl-3,5-dimethylfuro[3′,2′:4,5]benzo[b]furan-2-yl)-3-(aryl)-2-propen-1-ones and their antibacterial activity. Green Chem. Lett. Rev. 2012, 5, 121–125. [Google Scholar] [CrossRef]
- Hayakawa, I.; Shioya, R.; Agatsuma, T.; Furukawa, H.; Naruto, S.; Sugano, Y. 4-Hydroxy-3-methyl-6-phenylbenzofuran-2-carboxylic acid ethyl ester derivatives as potent anti-tumor agents. Bioorganic Med. Chem. Lett. 2004, 14, 455–458. [Google Scholar] [CrossRef]
- Xie, F.; Zhu, H.; Zhang, H.; Lang, Q.; Tang, L.; Huang, Q.; Yu, L. In vitro and in vivo characterization of a benzofuran derivative, a potential anticancer agent, as a novel Aurora B kinase inhibitor. Eur. J. Med. Chem. 2015, 89, 310–319. [Google Scholar] [CrossRef]
- Musumeci, D.; Roviello, G.N.; Rigione, G.; Capasso, D.; Di Gaetano, S.; Riccardi, C.; Roviello, V.; Montesarchio, D. Benzodifuran Derivatives as Potential Antiproliferative Agents: Possible Correlation between Their Bioactivity and Aggregation Properties. ChemPlusChem 2017, 82, 251–260. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Liu, M.-F.; Hou, W.-Z.; Xu, R.-M.; Gao, J.; Lu, A.-Q.; Xie, M.-P.; Li, L.; Zhang, J.-J.; Peng, Y.; et al. Bioactive Benzofuran Derivatives from Cortex Mori Radicis, and Their Neuroprotective and Analgesic Activities Mediated by mGluR1. Molecules 2017, 22, 236. [Google Scholar] [CrossRef]
- Thévenin, M.; Thoret, S.; Grellier, P.; Dubois, J. Synthesis of polysubstituted benzofuran derivatives as novel inhibitors of parasitic growth. Bioorganic Med. Chem. 2013, 21, 4885–4892. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, V.; Musumeci, D.; Pedone, C. Synthesis of a novel benzodifuran derivative and its molecular recognition of poly rA RNA. Biol. Chem. 2013, 394, 1235–1239. [Google Scholar] [CrossRef]
- Carella, A.; Roviello, V.; Iannitti, R.; Palumbo, R.; La Manna, S.; Marasco, D.; Trifuoggi, M.; Diana, R.; Roviello, G.N. Evaluating the biological properties of synthetic 4-nitrophenyl functionalized benzofuran derivatives with telomeric DNA binding and antiproliferative activities. Int. J. Biol. Macromol. 2019, 121, 77–88. [Google Scholar] [CrossRef]
- Inaba, S.; Rieke, R.D. Metallic nickel-mediated synthesis of ketones by the reaction of benzylic, allylic, vinylic, and pentafluorophenyl halides with acid halides. J. Org. Chem. 2002, 50, 1373–1381. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, G.; Musumeci, D.; Bucci, E.M.; Pedone, C. Dakin–West reaction on 1-thyminyl acetic acid for the synthesis of 1,3-bis(1-thyminyl)-2-propanone, a heteroaromatic compound with nucleopeptide-binding properties. Amino Acids 2012, 43, 1615–1623. [Google Scholar] [CrossRef]
- Srinivas, A. Synthesis and Antimicrobial Activity of Bis-[4-methoxy-3-(6-aryl-7H-[1,2,4]triazolo[3,4-b][1,3,4]-thiadiazin-3-yl)phenyl]methanes and Bis-[(triazolo[3,4-b]thiadiazipin-3-yl)phenyl]methanes. Acta Chim. Slov. 2016, 63, 173–179. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, G.; Musumeci, D.; Capasso, D.; Di Gaetano, S.; Costanzo, M.; Pedone, C. Synthesis and supramolecular assembly of 1,3-bis(1′-uracilyl)-2-propanone. RSC Advances 2014, 4, 28691. [Google Scholar] [CrossRef]
- Rother, A.; Bobbitt, J.; Schwarting, A. Structure and synthesis of the alkaloid anaferine. In Soc Chemical Industry 14 Belgrave Square, London sw1x 8ps; Chemistry & Industry: London, UK, 1962; pp. 654–655. [Google Scholar]
- Kaiser, C.; Audia, V.H.; Carter, J.P.; McPherson, D.W.; Waid, P.P.; Lowe, V.C.; Noronha-Blob, L. Synthesis and antimuscarinic activity of some 1-cycloalkyl-1-hydroxy-1-phenyl-3-(4-substituted piperazinyl)-2-propanones and related compounds. J. Med. Chem. 1993, 36, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Brindisi, M. Urea Derivatives in Modern Drug Discovery and Medicinal Chemistry. J. Med. Chem. 2019, 63, 2751–2788. [Google Scholar] [CrossRef] [PubMed]
- Dakin, H.; West, R. A general reaction of amino acids. J. Biol. Chem. 1928, 78, 91–104. [Google Scholar] [CrossRef]
- Curran, T.T. Implementation of the Dakin-West reaction for the preparation of an α-amino-pentafluoroethyl ketone. J. Fluor. Chem. 1995, 74, 107–112. [Google Scholar] [CrossRef]
- Tran, K.-V.; Bickar, D. Dakin—West Synthesis of β-Aryl Ketones. J. Org. Chem. 2006, 71, 6640–6643. [Google Scholar] [CrossRef]
- Kawase, M.; Hirabayashi, M.; Kumakura, H.; Saito, S.; Yamamoto, K. The Dakin-West reaction of N-alkoxycarbonyl-N-alkyl-α-amino acids employing trifluoroacetic anhydride. Chem. Pharm. Bull. 2000, 48, 114–119. [Google Scholar] [CrossRef][Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Musumeci, D.; Mokhir, A.; Roviello, G.N. Synthesis and nucleic acid binding evaluation of a thyminyl l-diaminobutanoic acid-based nucleopeptide. Bioorganic Chem. 2020, 100, 103862. [Google Scholar] [CrossRef]
- D’Atri, V.; Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Mayol, L.; Piccialli, V.; Haider, S.; Hoorelbeke, B.; Balzarini, J.; et al. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: Effect of the base sequence on anti-HIV activity. Chem. Commun. 2012, 48, 9516. [Google Scholar] [CrossRef]
- Amato, J.; Oliviero, G.; De Pauw, E.; Gabelica, V. Hybridization of short complementary PNAs to G-quadruplex forming oligonucleotides: An electrospray mass spectrometry study. Biopolymers 2009, 91, 244–255. [Google Scholar] [CrossRef]
- Esposito, V.; Virgilio, A.; Pepe, A.; Oliviero, G.; Mayol, L.; Galeone, A. Effects of the introduction of inversion of polarity sites in the quadruplex forming oligonucleotide TGGGT. Bioorganic Med. Chem. 2009, 17, 1997–2001. [Google Scholar] [CrossRef]
- Scuotto, M.; Persico, M.; Bucci, M.; Vellecco, V.; Borbone, N.; Morelli, E.; Oliviero, G.; Novellino, E.; Piccialli, G.; Cirino, G.; et al. Outstanding effects on antithrombin activity of modified TBA diastereomers containing an optically pure acyclic nucleotide analogue. Org. Biomol. Chem. 2014, 12, 5235–5242. [Google Scholar] [CrossRef]
- Scognamiglio, P.L.; Vicidomini, C.; Fontanella, F.; De Stefano, C.; Palumbo, R.; Roviello, G.N. Protein Binding of Benzofuran Derivatives: A CD Spectroscopic and In Silico Comparative Study of the Effects of 4-Nitrophenyl Functionalized Benzofurans and Benzodifurans on BSA Protein Structure. Biomolecules 2022, 12, 262. [Google Scholar] [CrossRef]
- Greco, F.; Musumeci, D.; Borbone, N.; Falanga, A.P.; D’Errico, S.; Terracciano, M.; Piccialli, I.; Roviello, G.N.; Oliviero, G. Exploring the Parallel G-Quadruplex Nucleic Acid World: A Spectroscopic and Computational Investigation on the Binding of the c-myc Oncogene NHE III1 Region by the Phytochemical Polydatin. Molecules 2022, 27, 2997. [Google Scholar] [CrossRef]
- Roviello, G.N.; Musumeci, D.; Castiglione, M.; Bucci, E.M.; Pedone, C.; Benedetti, E. Solid phase synthesis and RNA-binding studies of a serum-resistant nucleo-ε-peptide. J. Pept. Sci. 2009, 15, 155–160. [Google Scholar] [CrossRef]
- Roviello, G.N.; Roviello, V.; Autiero, I.; Saviano, M. Solid phase synthesis of TyrT, a thymine–tyrosine conjugate with poly(A) RNA-binding ability. RSC Adv. 2016, 6, 27607–27613. [Google Scholar] [CrossRef]
- Roviello, G.N.; Moccia, M.; Sapio, R.; Valente, M.; Bucci, E.M.; Castiglione, M.; Pedone, C.; Perretta, G.; Benedetti, E.; Musumeci, D. Synthesis, characterization and hybridization studies of new nucleo-gamma-peptides based on diaminobutyric acid. J. Pept. Sci. 2006, 12, 829–835. [Google Scholar] [CrossRef]
- Genta-Jouve, G.; Weinberg, L.; Cocandeau, V.; Maestro, Y.; Thomas, O.P.; Holderith, S. Revising the Absolute Configurations of Coatlines via Density Functional Theory Calculations of Electronic Circular Dichroism Spectra. Chirality 2013, 25, 180–184. [Google Scholar] [CrossRef]
- Castiglioni, E.; Abbate, S.; Lebon, F.; Longhi, G. Ultraviolet, Circular Dichroism, Fluorescence, and Circularly Polarized Luminescence Spectra of Regioregular Poly-[3-((S)-2-Methylbutyl)-Thiophene] in Solution. Chirality 2012, 24, 725–730. [Google Scholar] [CrossRef]
- Abriata, L.A. Online interactive fitting and simulation of protein circular dichroism spectra for use in education and for preliminary spectral analysis. arXiv 2020, arXiv:2006.06275. [Google Scholar]
- Abriata, L.A. A simple spreadsheet program to simulate and analyze the far-UV circular dichroism spectra of proteins. J. Chem. Educ. 2011, 88, 1268–1273. [Google Scholar] [CrossRef]
- Musumeci, D.; Ullah, S.; Ikram, A.; Roviello, G.N. Novel insights on nucleopeptide binding: A spectroscopic and in silico investigation on the interaction of a thymine-bearing tetrapeptide with a homoadenine DNA. J. Mol. Liq. 2022, 347, 117975. [Google Scholar] [CrossRef]
- Roviello, V.; Scognamiglio, P.L.; Caruso, U.; Vicidomini, C.; Roviello, G.N. Evaluating In Silico the Potential Health and Environmental Benefits of Houseplant Volatile Organic Compounds for an Emerging ‘Indoor Forest Bathing’ Approach. Int. J. Environ. Res. Public Health 2021, 19, 273. [Google Scholar] [CrossRef] [PubMed]
- Roviello, V.; Roviello, G.N. Less COVID-19 deaths in southern and insular Italy explained by forest bathing, Mediterranean environment, and antiviral plant volatile organic compounds. Environ. Chem. Lett. 2021, 20, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Vicidomini, C.; Roviello, V.; Roviello, G.N. In Silico Investigation on the Interaction of Chiral Phytochemicals from Opuntia ficus-indica with SARS-CoV-2 Mpro. Symmetry 2021, 13, 1041. [Google Scholar] [CrossRef]
- Roviello, V.; Gilhen-Baker, M.; Vicidomini, C.; Roviello, G.N. The Healing Power of Clean Rivers: In Silico Evaluation of the Antipsoriatic Potential of Apiin and Hyperoside Plant Metabolites Contained in River Waters. Int. J. Environ. Res. Public Health 2022, 19, 2502. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Majumder, A.; Mondal, S.K.; Mukhoty, S.; Bag, S.; Mondal, A.; Begum, Y.; Sharma, K.; Banik, A. Virtual Screening and Docking Analysis of Novel Ligands for Selective Enhancement of Tea (Camellia sinensis) Flavonoids. Food Chem. X 2022, 13, 100212. [Google Scholar] [CrossRef]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef]
- Zhao, R.; Xie, Y.; Tan, Y.; Tan, C.; Jiang, Y. Binding of a bcl-2 Family Inhibitor to Bovine Serum Albumin: Fluorescence Quenching and Molecular Docking Study. Protein Pept. Lett. 2012, 19, 949–954. [Google Scholar] [CrossRef]
- Belinskaia, D.A.; Voronina, P.A.; Shmurak, V.I.; Jenkins, R.O.; Goncharov, N.V. Serum Albumin in Health and Disease: Esterase, Antioxidant, Transporting and Signaling Properties. Int. J. Mol. Sci. 2021, 22, 10318. [Google Scholar] [CrossRef]
- Subramanyam, R.; Nusrat, S.; Siddiqi, M.K.; Zaman, M.; Zaidi, N.; Ajmal, M.R.; Alam, P.; Qadeer, A.; Abdelhameed, A.S.; Khan, R.H. A Comprehensive Spectroscopic and Computational Investigation to Probe the Interaction of Antineoplastic Drug Nordihydroguaiaretic Acid with Serum Albumins. PLoS ONE 2016, 11, e0158833. [Google Scholar]
- Ràfols, C.; Amézqueta, S.; Fuguet, E.; Bosch, E. Molecular interactions between warfarin and human (HSA) or bovine (BSA) serum albumin evaluated by isothermal titration calorimetry (ITC), fluorescence spectrometry (FS) and frontal analysis capillary electrophoresis (FA/CE). J. Pharm. Biomed. Anal. 2018, 150, 452–459. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Zou, X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins Struct. Funct. Bioinform. 2008, 72, 557–579. [Google Scholar] [CrossRef]
- Rarey, M.; Steinegger, R.; Nittinger, E.; Meyder, A.; Flachsenberg, F.; Fährrolfes, R.; Diedrich, K.; Schöning-Stierand, K. ProteinsPlus: Interactive analysis of protein–ligand binding interfaces. Nucleic Acids Res. 2020, 48, W48–W53. [Google Scholar]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins: Structure, Funct. Bioinform. 2007, 69, 139–159. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33 (Suppl. S2), W363–W367. [Google Scholar] [CrossRef]
- Potemkin, V.; Potemkin, A.; Grishina, M. Internet Resources for Drug Discovery and Design. Curr. Top. Med. Chem. 2019, 18, 1955–1975. [Google Scholar] [CrossRef]
- Kiss, R.; Sandor, M.; Szalai, F.A. http://Mcule.com: A public web service for drug discovery. J. Cheminformatics 2012, 4, P17. [Google Scholar] [CrossRef]
- Karst, J.C.; Sotomayor Pérez, A.C.; Guijarro, J.I.; Raynal, B.; Chenal, A.; Ladant, D. Calmodulin-Induced Conformational and Hydrodynamic Changes in the Catalytic Domain of Bordetella pertussis Adenylate Cyclase Toxin. Biochemistry 2009, 49, 318–328. [Google Scholar] [CrossRef]
- Ostrowski, M.; Porowinska, D.; Prochnicki, T.; Prevost, M.; Raynal, B.; Baron, B.; Sauguet, L.; Corringer, P.-J.; Faure, G. Neurotoxic phospholipase A2 from rattlesnake as a new ligand and new regulator of prokaryotic receptor GLIC (proton-gated ion channel from G. violaceus). Toxicon 2016, 116, 63–71. [Google Scholar] [CrossRef]
- Gospodarska, E.; Kupniewska-Kozak, A.; Goch, G.; Dadlez, M. Binding studies of truncated variants of the Aβ peptide to the V-domain of the RAGE receptor reveal Aβ residues responsible for binding. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2011, 1814, 592–609. [Google Scholar] [CrossRef]
- Ye, F.; An, Y.; Qin, D.; Yang, L.; She, L.; Xing, R. Spectroscopic study on the effect of crystallization of the hydroxyapatite on the secondary structure of bovine serum albumin. Guang Pu Xue Yu Guang Pu Fen Xi = Guang Pu 2007, 27, 321–324. [Google Scholar]
- Gülseren, İ.; Güzey, D.; Bruce, B.D.; Weiss, J. Structural and functional changes in ultrasonicated bovine serum albumin solutions. Ultrason. Sonochemistry 2007, 14, 173–183. [Google Scholar] [CrossRef]
- Guzey, D.; Gulseren, I.; Bruce, B.; Weiss, J. Interfacial properties and structural conformation of thermosonicated bovine serum albumin. Food Hydrocoll. 2006, 20, 669–677. [Google Scholar] [CrossRef]
- Fasman, G.D. The measurement of transmembrane helices by the deconvolution of CD spectra of membrane proteins: A review. Biopolym. Orig. Res. Biomol. 1995, 37, 339–362. [Google Scholar] [CrossRef]
- Perczel, A.; Park, K.; Fasman, G.D. Deconvolution of the circular dichroism spectra of proteins: The circular dichroism spectra of the antiparallel β-sheet in proteins. Proteins Struct. Funct. Bioinform. 1992, 13, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Dubeau, S.; Bourassa, P.; Thomas, T.J.; Tajmir-Riahi, H.A. Biogenic and Synthetic Polyamines Bind Bovine Serum Albumin. Biomacromolecules 2010, 11, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Paul, S.S.; Mukherjea, K.K. Interaction of bovine serum albumin with a psychotropic drug alprazolam: Physicochemical, photophysical and molecular docking studies. J. Lumin. 2013, 142, 220–230. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Baell, J.B.; Nissink, J.W.M. Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017 Utility and Limitations. ACS Chem. Biol. 2018, 13, 36–44. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).