Plasmodium falciparum Nicotinamidase as A Novel Antimalarial Target
Abstract
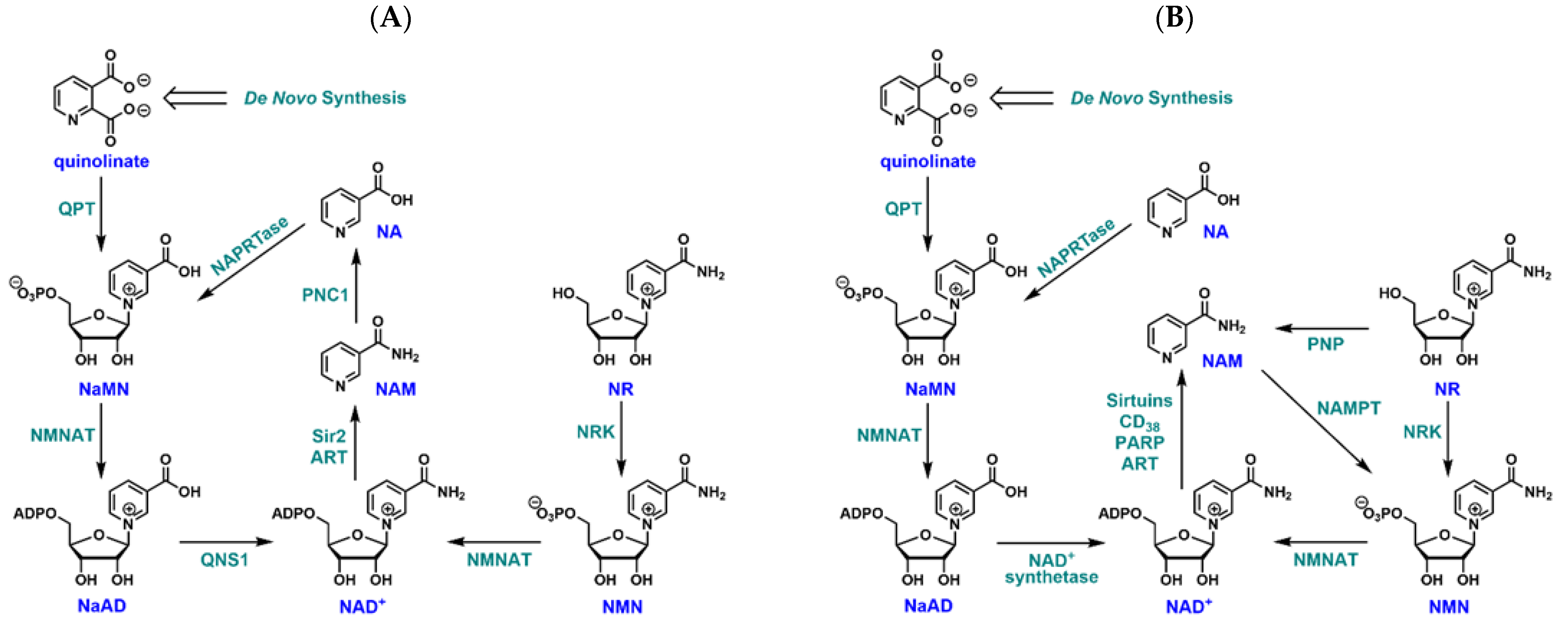
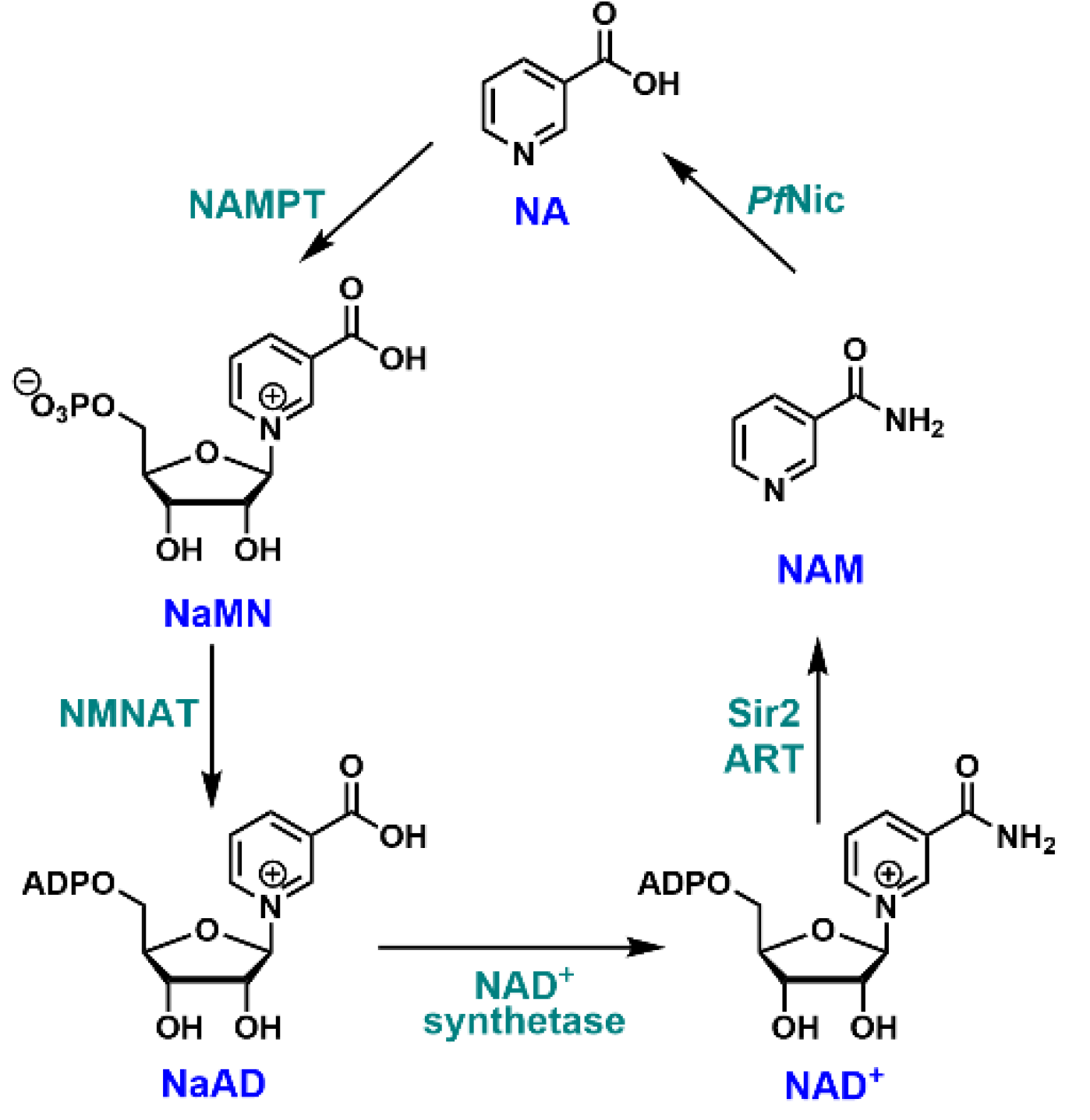
1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
2.2. Cloning and Expression of PfNic and Synthesis of Inhibitor
2.3. Parasite Culture
2.4. Inhibitor or NAD+ Precursor Treatment and NAD+ Measurements
2.5. Inhibition Study in Parasite Lysates
2.6. Luciferase Assay
2.7. Preparation of Giemsa Stained Smears
3. Results
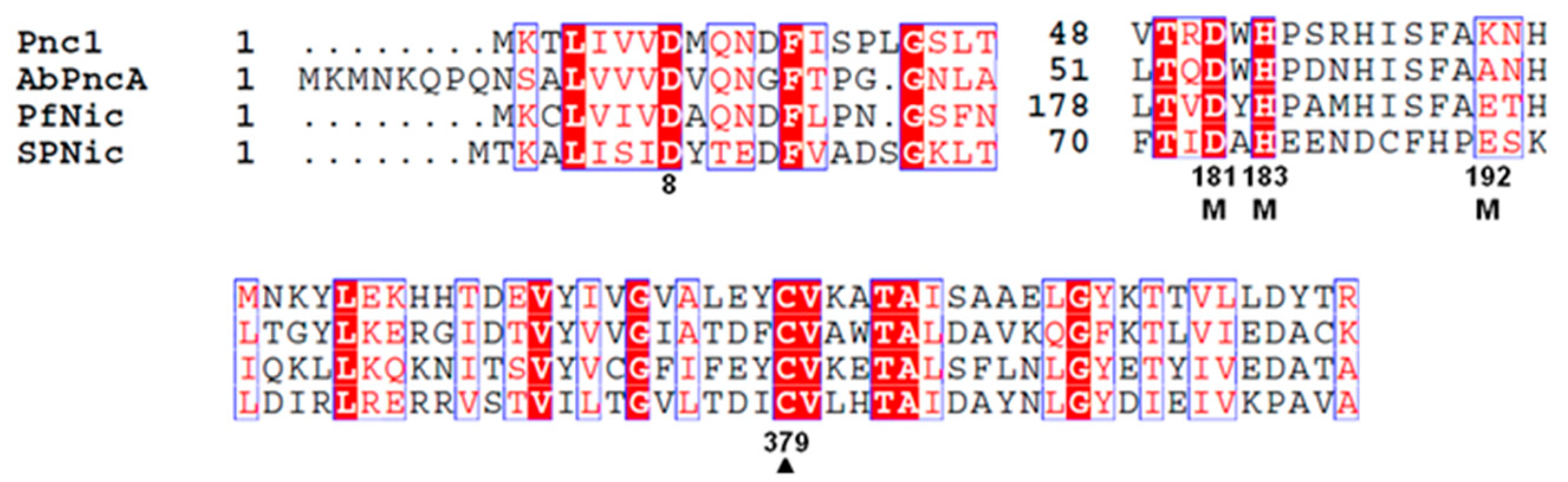
3.1. Essential Catalytic Residues Are Conserved in P. falciparum Nicotinamidase (PfNic)
3.2. Expression and Characterization of PfNic
3.3. Identification of PfNic Inhibitors
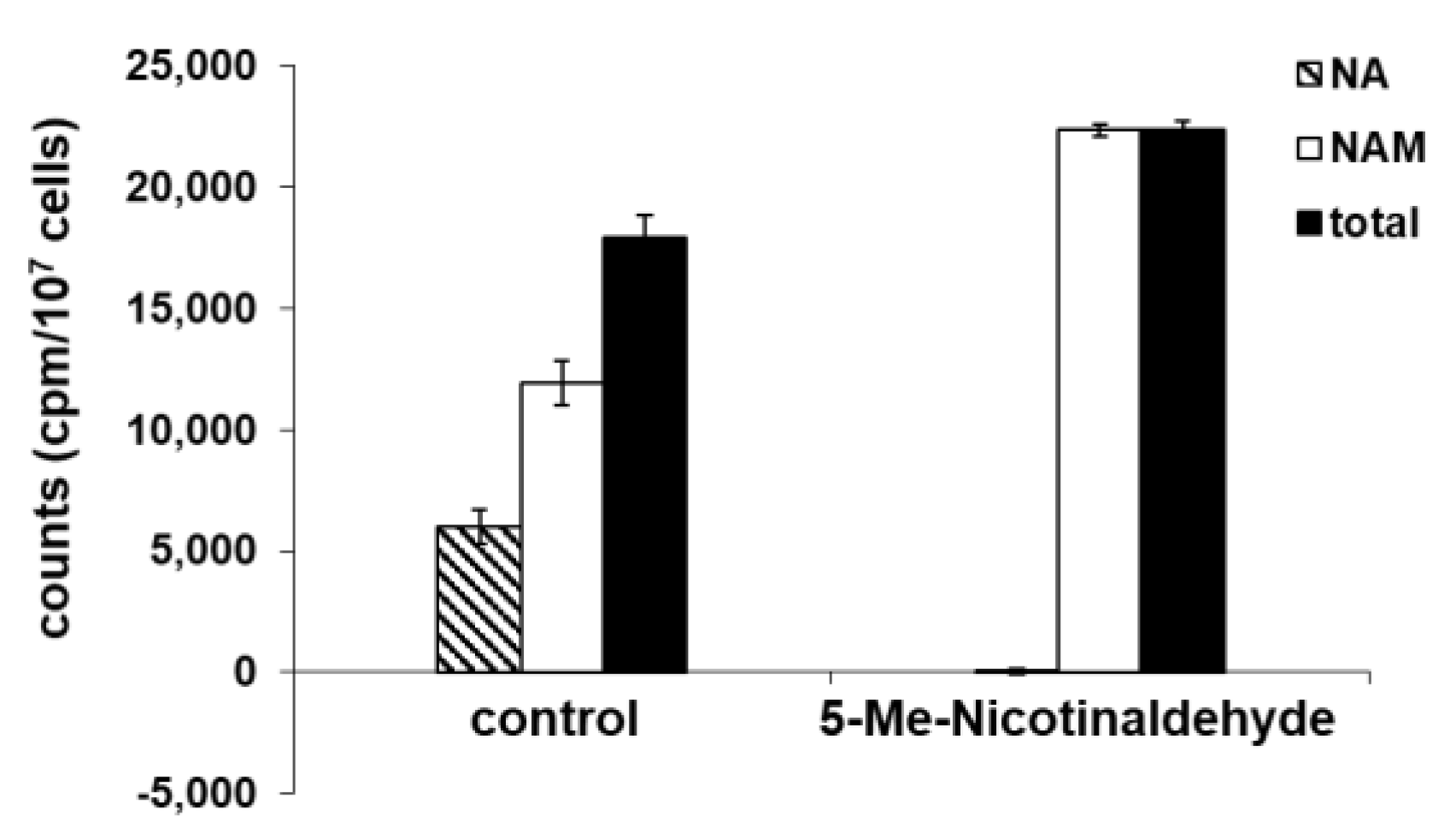
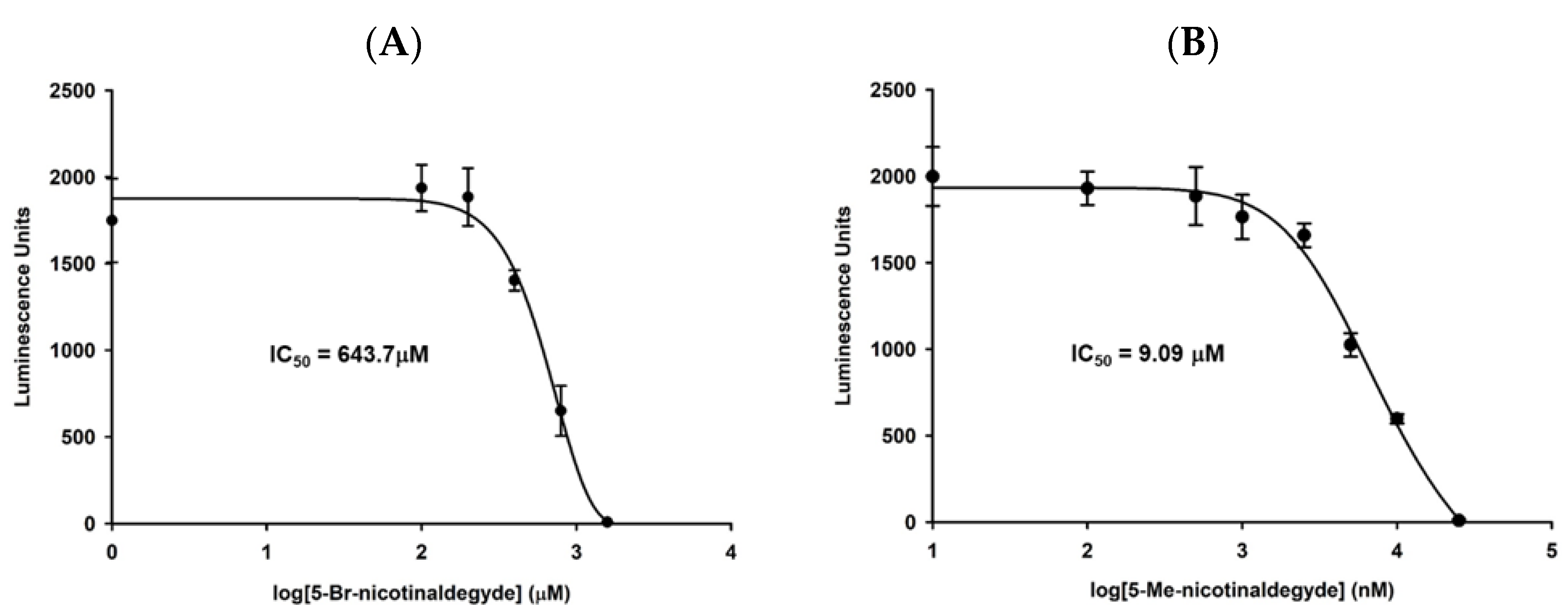
3.4. 5-Me-Nicotinaldehyde Inhibits PfNic Activity in P. falciparum Lysates
3.5. Nicotinamide Is the Preferred NAD+ Precursor in P. falciparum
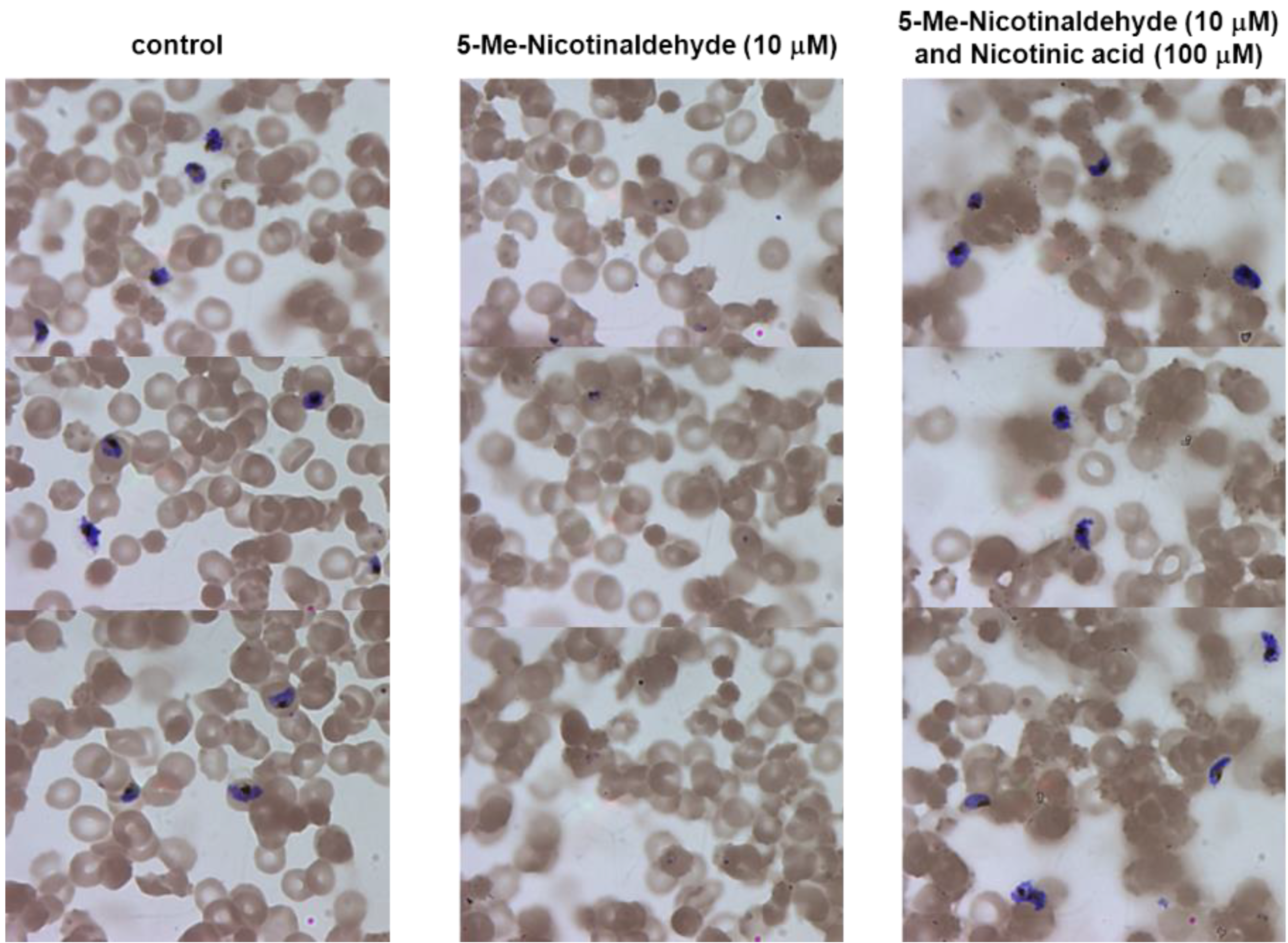
3.6. Inhibition of NAD+ Biosynthesis by Nicotinamidase Inhibitors Can Be Reversed by Nicotinic Acid
3.7. Nicotinaldehydes Inhibits P. falciparum Replication
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magni, G.; Amici, A.; Emanuelli, M.; Raffaelli, N.; Ruggieri, S. Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol. 1999, 73, 135–182. [Google Scholar]
- Shats, I.; Williams, J.G.; Liu, J.; Makarov, M.V.; Wu, X.; Lih, F.B.; Deterding, L.J.; Lim, C.; Xu, X.; Randall, T.A.; et al. Bacteria Boost Mammalian Host NAD Metabolism by Engaging the Deamidated Biosynthesis Pathway. Cell Metab. 2020, 31, 564–579.e7. [Google Scholar] [CrossRef] [PubMed]
- Sauve, A.A. NAD+ and vitamin B3: From metabolism to therapies. J. Pharmacol. Exp. Ther. 2008, 324, 883–893. [Google Scholar] [CrossRef]
- Begley, T.P.; Kinsland, C.; Mehl, R.A.; Osterman, A.; Dorrestein, P. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 2001, 61, 103–119. [Google Scholar]
- Kurnasov, O.; Goral, V.; Colabroy, K.; Gerdes, S.; Anantha, S.; Osterman, A.; Begley, T.P. NAD biosynthesis: Identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol. 2003, 10, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, F.; Stebbins, R.; Chang, S.Z.; McPeek, M.A.; Brenner, C. Microbial NAD metabolism: Lessons from comparative genomics. Microbiol. Mol. Biol. Rev. 2009, 73, 529–541. [Google Scholar] [CrossRef]
- Belenky, P.; Bogan, K.L.; Brenner, C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007, 32, 12–19. [Google Scholar] [CrossRef]
- Bogan, K.L.; Brenner, C. Nicotinic acid, nicotinamide, and nicotinamide riboside: A molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu. Rev. Nutr. 2008, 28, 115–130. [Google Scholar] [CrossRef]
- Chauhan, N.; Poddar, R. In silico pharmacophore modeling and simulation studies for searching potent antileishmanials targeted against Leishmania donovani nicotinamidase. Comput. Biol. Chem. 2019, 83, 107150. [Google Scholar] [CrossRef] [PubMed]
- Shang, F.; Chen, J.; Wang, L.; Jin, L.; Zou, L.; Bu, T.; Dong, Y.; Ha, N.C.; Nam, K.H.; Quan, C.; et al. Crystal structure of the nicotinamidase/pyrazinamidase PncA from Bacillus subtilis. Biochem. Biophys. Res. Commun. 2018, 503, 2906–2911. [Google Scholar] [CrossRef] [PubMed]
- Curry, A.; White, D.; Cen, Y. Small Molecule Regulators Targeting NAD(+) Biosynthetic Enzymes. Curr. Med. Chem. 2022, 29, 1718–1738. [Google Scholar] [CrossRef] [PubMed]
- Osterman, A.L.; Rodionova, I.; Li, X.; Sergienko, E.; Ma, C.T.; Catanzaro, A.; Pettigrove, M.E.; Reed, R.W.; Gupta, R.; Rohde, K.H.; et al. Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase. ACS Chem. Biol. 2019, 14, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Rodionova, I.A.; Zuccola, H.J.; Sorci, L.; Aleshin, A.E.; Kazanov, M.D.; Ma, C.T.; Sergienko, E.; Rubin, E.J.; Locher, C.P.; Osterman, A.L. Mycobacterial nicotinate mononucleotide adenylyltransferase: Structure, mechanism, and implications for drug discovery. J. Biol. Chem. 2015, 290, 7693–7706. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ahn, Y.M.; Lentscher, A.G.; Lister, J.S.; Brothers, R.C.; Kneen, M.M.; Gerratana, B.; Boshoff, H.I.; Dowd, C.S. Design, synthesis, and evaluation of substituted nicotinamide adenine dinucleotide (NAD(+)) synthetase inhibitors as potential antitubercular agents. Bioorg. Med. Chem. Lett. 2017, 27, 4426–4430. [Google Scholar] [CrossRef]
- Teixeira, C.S.S.; Cerqueira, N.; Sousa, S.F. Multifunctional Enzymes as Targets for the Treatment of Tuberculosis: Paving the Way for New Anti-TB Drugs. Curr. Med. Chem. 2021, 28, 5847–5882. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, X.; Bheda, P.; Revollo, J.R.; Imai, S.; Wolberger, C. Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme. Nat. Struct. Mol. Biol. 2006, 13, 661–662. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.S.; Fuller, L.; Yero, I.L.; Martinez, L. Nicotinamide mononucleotide pyrophosphorylase activity in animal tissues. J. Biol. Chem. 1966, 241, 188–191. [Google Scholar] [CrossRef]
- Preiss, J.; Handler, P. Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem. 1958, 233, 488–492. [Google Scholar] [CrossRef]
- Xu, P.; Sauve, A.A. Vitamin B3, the nicotinamide adenine dinucleotides and aging. Mech. Ageing Dev. 2010, 131, 287–298. [Google Scholar] [CrossRef]
- Geleta, G.; Ketema, T.; Severe, M. Associated with Plasmodium falciparum and P. vivax among Children in Pawe Hospital, Northwest Ethiopia. Malar. Res. Treat. 2016, 2016, 1240962. [Google Scholar] [CrossRef]
- Florini, F.; Visone, J.E.; Deitsch, K.W. Shared Mechanisms for Mutually Exclusive Expression and Antigenic Variation by Protozoan Parasites. Front. Cell Dev. Biol. 2022, 10, 852239. [Google Scholar] [CrossRef]
- Sakoguchi, A.; Arase, H. Mechanisms for Host Immune Evasion Mediated by Plasmodium falciparum-Infected Erythrocyte Surface Antigens. Front. Immunol. 2022, 13, 901864. [Google Scholar] [CrossRef]
- Tchekounou, C.; Zida, A.; Zongo, C.; Soulama, I.; Sawadogo, P.M.; Guiguemde, K.T.; Sangare, I.; Guiguemde, R.T.; Traore, Y. Antimalarial drugs resistance genes of Plasmodium falciparum: A review. Ann. Parasitol. 2022, 68, 215–225. [Google Scholar]
- Gil, J.P.; Fancony, C. Plasmodium falciparum Multidrug Resistance Proteins (pfMRPs). Front. Pharmacol. 2021, 12, 759422. [Google Scholar] [CrossRef]
- Rasmussen, C.; Alonso, P.; Ringwald, P. Current and emerging strategies to combat antimalarial resistance. Expert Rev. Anti. Infect. Ther. 2022, 20, 353–372. [Google Scholar] [CrossRef]
- Guidelines for the Treatment of Malaria, 3rd ed.; World Health Organization: Geneva, Switzerland, 2015.
- Tisnerat, C.; Dassonville-Klimpt, A.; Gosselet, F.; Sonnet, P. Antimalarial Drug Discovery: From Quinine to the Most Recent Promising Clinical Drug Candidates. Curr. Med. Chem. 2022, 29, 3326–3365. [Google Scholar] [CrossRef]
- Miller, L.H.; Baruch, D.I.; Marsh, K.; Doumbo, O.K. The pathogenic basis of malaria. Nature 2002, 415, 673–679. [Google Scholar] [CrossRef]
- Buffet, P.A.; Safeukui, I.; Deplaine, G.; Brousse, V.; Prendki, V.; Thellier, M.; Turner, G.D.; Mercereau-Puijalon, O. The pathogenesis of Plasmodium falciparum malaria in humans: Insights from splenic physiology. Blood 2011, 117, 381–392. [Google Scholar] [CrossRef]
- Plata, G.; Hsiao, T.L.; Olszewski, K.L.; Llinas, M.; Vitkup, D. Reconstruction and flux-balance analysis of the Plasmodium falciparum metabolic network. Mol. Syst. Biol. 2010, 6, 408. [Google Scholar] [CrossRef]
- Fulco, M.; Cen, Y.; Zhao, P.; Hoffman, E.P.; McBurney, M.W.; Sauve, A.A.; Sartorelli, V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell 2008, 14, 661–673. [Google Scholar] [CrossRef]
- Trammell, S.A.; Schmidt, M.S.; Weidemann, B.J.; Redpath, P.; Jaksch, F.; Dellinger, R.W.; Li, Z.; Abel, E.D.; Migaud, M.E.; Brenner, C. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun. 2016, 7, 12948. [Google Scholar] [CrossRef] [PubMed]
- Zerez, C.R.; Roth, E.F., Jr.; Schulman, S.; Tanaka, K.R. Increased nicotinamide adenine dinucleotide content and synthesis in Plasmodium falciparum-infected human erythrocytes. Blood 1990, 75, 1705–1710. [Google Scholar] [CrossRef] [PubMed]
- French, J.B.; Cen, Y.; Vrablik, T.L.; Xu, P.; Allen, E.; Hanna-Rose, W.; Sauve, A.A. Characterization of nicotinamidases: Steady state kinetic parameters, classwide inhibition by nicotinaldehydes, and catalytic mechanism. Biochemistry 2010, 49, 10421–10439. [Google Scholar] [CrossRef] [PubMed]
- Swamy, L.; Amulic, B.; Deitsch, K.W. Plasmodium falciparum var gene silencing is determined by cis DNA elements that form stable and heritable interactions. Eukaryot. Cell 2011, 10, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Radfar, A.; Mendez, D.; Moneriz, C.; Linares, M.; Marin-Garcia, P.; Puyet, A.; Diez, A.; Bautista, J.M. Synchronous culture of Plasmodium falciparum at high parasitemia levels. Nat. Protoc. 2009, 4, 1899–1915. [Google Scholar] [CrossRef]
- Bai, P.; Canto, C.; Brunyanszki, A.; Huber, A.; Szanto, M.; Cen, Y.; Yamamoto, H.; Houten, S.M.; Kiss, B.; Oudart, H.; et al. PARP-2 regulates SIRT1 expression and whole-body energy expenditure. Cell Metab. 2011, 13, 450–460. [Google Scholar] [CrossRef]
- North, B.J.; Rosenberg, M.A.; Jeganathan, K.B.; Hafner, A.V.; Michan, S.; Dai, J.; Baker, D.J.; Cen, Y.; Wu, L.E.; Sauve, A.A.; et al. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. EMBO J. 2014, 33, 1438–1453. [Google Scholar] [CrossRef]
- Yang, Y.; Sauve, A.A. Assays for Determination of Cellular and Mitochondrial NAD(+) and NADH Content. Methods Mol. Biol. 2021, 2310, 271–285. [Google Scholar]
- Epp, C.; Raskolnikov, D.; Deitsch, K.W. A regulatable transgene expression system for cultured Plasmodium falciparum parasites. Malar. J. 2008, 7, 86. [Google Scholar] [CrossRef]
- French, J.B.; Cen, Y.; Sauve, A.A.; Ealick, S.E. High-resolution crystal structures of Streptococcus pneumoniae nicotinamidase with trapped intermediates provide insights into the catalytic mechanism and inhibition by aldehydes. Biochemistry 2010, 49, 8803–8812. [Google Scholar] [CrossRef]
- Fyfe, P.K.; Rao, V.A.; Zemla, A.; Cameron, S.; Hunter, W.N. Specificity and mechanism of Acinetobacter baumanii nicotinamidase: Implications for activation of the front-line tuberculosis drug pyrazinamide. Angew. Chem. Int. Ed. Engl. 2009, 48, 9176–9179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, J.Y.; Bi, L.J.; Zhou, Y.F.; Zhang, Z.P.; Zhang, C.G.; Zhang, Y.; Zhang, X.E. Characterization of Mycobacterium tuberculosis nicotinamidase/pyrazinamidase. FEBS J. 2008, 275, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Taylor, A.B.; McAlister-Henn, L.; Hart, P.J. Crystal structure of the yeast nicotinamidase Pnc1p. Arch. Biochem. Biophys. 2007, 461, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, W.; Kim, R.; Yakota, H.; Nguyen, H.; Kim, S.H. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry 2001, 40, 14166–14172. [Google Scholar] [CrossRef]
- Seiner, D.R.; Hegde, S.S.; Blanchard, J.S. Kinetics and inhibition of nicotinamidase from Mycobacterium tuberculosis. Biochemistry 2010, 49, 9613–9619. [Google Scholar] [CrossRef][Green Version]
- Yan, C.; Sloan, D.L. Purification and characterization of nicotinamide deamidase from yeast. J. Biol. Chem. 1987, 262, 9082–9087. [Google Scholar] [CrossRef]
- Sato, S. Plasmodium-a brief introduction to the parasites causing human malaria and their basic biology. J. Physiol. Anthropol. 2021, 40, 1. [Google Scholar] [CrossRef]
- Tran, A.; Yokose, R.; Cen, Y. Chemo-enzymatic synthesis of isotopically labeled nicotinamide riboside. Org. Biomol. Chem. 2018, 16, 3662–3671. [Google Scholar] [CrossRef]
- Curry, A.M.; Barton, E.; Kang, W.; Mongeluzi, D.V.; Cen, Y. Development of Second Generation Activity-Based Chemical Probes for Sirtuins. Molecules 2020, 26, 11. [Google Scholar] [CrossRef]
- Rising, K.A.; Schramm, V.L. Enzymatic-Synthesis of Nad(+) with the Specific Incorporation of Atomic Labels. J. Am. Chem. Soc. 1994, 116, 6531–6536. [Google Scholar] [CrossRef]
- Cen, Y.; Sauve, A.A. Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches. J. Am. Chem. Soc. 2010, 132, 12286–12298. [Google Scholar] [CrossRef][Green Version]
- Moore, G.E.; Gerner, R.E.; Franklin, H.A. Culture of normal human leukocytes. JAMA 1967, 199, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Merckx, A.; Nivez, M.P.; Bouyer, G.; Alano, P.; Langsley, G.; Deitsch, K.; Thomas, S.; Doerig, C.; Egee, S. Plasmodium falciparum regulatory subunit of cAMP-dependent PKA and anion channel conductance. PLoS Pathog. 2008, 4, e19. [Google Scholar] [CrossRef] [PubMed]
- Gygli, S.M.; Borrell, S.; Trauner, A.; Gagneux, S. Antimicrobial resistance in Mycobacterium tuberculosis: Mechanistic and evolutionary perspectives. FEMS Microbiol. Rev. 2017, 41, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Gorvel, J.P. Brucella: A Mr “Hide” converted into Dr Jekyll. Microbes Infect. 2008, 10, 1010–1013. [Google Scholar] [CrossRef]
- Zapata-Perez, R.; Garcia-Saura, A.G.; Jebbar, M.; Golyshin, P.N.; Sanchez-Ferrer, A. Combined Whole-Cell High-Throughput Functional Screening for Identification of New Nicotinamidases/Pyrazinamidases in Metagenomic/Polygenomic Libraries. Front. Microbiol. 2016, 7, 1915. [Google Scholar] [CrossRef][Green Version]
- Pizarro-Cerda, J.; Meresse, S.; Parton, R.G.; van der Goot, G.; Sola-Landa, A.; Lopez-Goni, I.; Moreno, E.; Gorvel, J.P. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 1998, 66, 5711–5724. [Google Scholar] [CrossRef]
- Kim, S.; Kurokawa, D.; Watanabe, K.; Makino, S.; Shirahata, T.; Watarai, M. Brucella abortus nicotinamidase (PncA) contributes to its intracellular replication and infectivity in mice. FEMS Microbiol. Lett. 2004, 234, 289–295. [Google Scholar] [CrossRef]
- Garcia, C.K.; Brown, M.S.; Pathak, R.K.; Goldstein, J.L. cDNA cloning of MCT2, a second monocarboxylate transporter expressed in different cells than MCT1. J. Biol. Chem. 1995, 270, 1843–1849. [Google Scholar] [CrossRef]
- Spector, R. Niacin and niacinamide transport in the central nervous system. In vivo studies. J. Neurochem. 1979, 33, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, J.B. Niacin status impacts chromatin structure. J. Nutr. 2009, 139, 2397–2401. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kirkland, J.B. Niacin status, NAD distribution and ADP-ribose metabolism. Curr. Pharm. Des. 2009, 15, 3–11. [Google Scholar] [CrossRef]
- Yamada, K.; Hara, N.; Shibata, T.; Osago, H.; Tsuchiya, M. The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal. Biochem. 2006, 352, 282–285. [Google Scholar] [CrossRef]
- Sullivan, D.J., Jr.; Gluzman, I.Y.; Russell, D.G.; Goldberg, D.E. On the molecular mechanism of chloroquine’s antimalarial action. Proc. Natl. Acad. Sci. USA 1996, 93, 11865–11870. [Google Scholar] [CrossRef]
- Gardner, M.J.; Hall, N.; Fung, E.; White, O.; Berriman, M.; Hyman, R.W.; Carlton, J.M.; Pain, A.; Nelson, K.E.; Bowman, S.; et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002, 419, 498–511. [Google Scholar] [CrossRef]
- Duraisingh, M.T.; Voss, T.S.; Marty, A.J.; Duffy, M.F.; Good, R.T.; Thompson, J.K.; Freitas-Junior, L.H.; Scherf, A.; Crabb, B.S.; Cowman, A.F. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell 2005, 121, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Tonkin, C.J.; Carret, C.K.; Duraisingh, M.T.; Voss, T.S.; Ralph, S.A.; Hommel, M.; Duffy, M.F.; Silva, L.M.; Scherf, A.; Ivens, A.; et al. Sir2 paralogues cooperate to regulate virulence genes and antigenic variation in Plasmodium falciparum. PLoS Biol. 2009, 7, e84. [Google Scholar] [CrossRef] [PubMed]
- Dzikowski, R.; Deitsch, K.W. Genetics of antigenic variation in Plasmodium falciparum. Curr. Genet. 2009, 55, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Rivero, F.D.; Saura, A.; Prucca, C.G.; Carranza, P.G.; Torri, A.; Lujan, H.D. Disruption of antigenic variation is crucial for effective parasite vaccine. Nat. Med. 2010, 16, 551–557. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donu, D.; Sharma, C.; Cen, Y. Plasmodium falciparum Nicotinamidase as A Novel Antimalarial Target. Biomolecules 2022, 12, 1109. https://doi.org/10.3390/biom12081109
Donu D, Sharma C, Cen Y. Plasmodium falciparum Nicotinamidase as A Novel Antimalarial Target. Biomolecules. 2022; 12(8):1109. https://doi.org/10.3390/biom12081109
Chicago/Turabian StyleDonu, Dickson, Chiranjeev Sharma, and Yana Cen. 2022. "Plasmodium falciparum Nicotinamidase as A Novel Antimalarial Target" Biomolecules 12, no. 8: 1109. https://doi.org/10.3390/biom12081109
APA StyleDonu, D., Sharma, C., & Cen, Y. (2022). Plasmodium falciparum Nicotinamidase as A Novel Antimalarial Target. Biomolecules, 12(8), 1109. https://doi.org/10.3390/biom12081109





