Differential Protein Content between Fresh and Freeze-Dried Plasma Rich in Growth Factors Eye Drops
Abstract
:1. Introduction
2. Materials and Methods
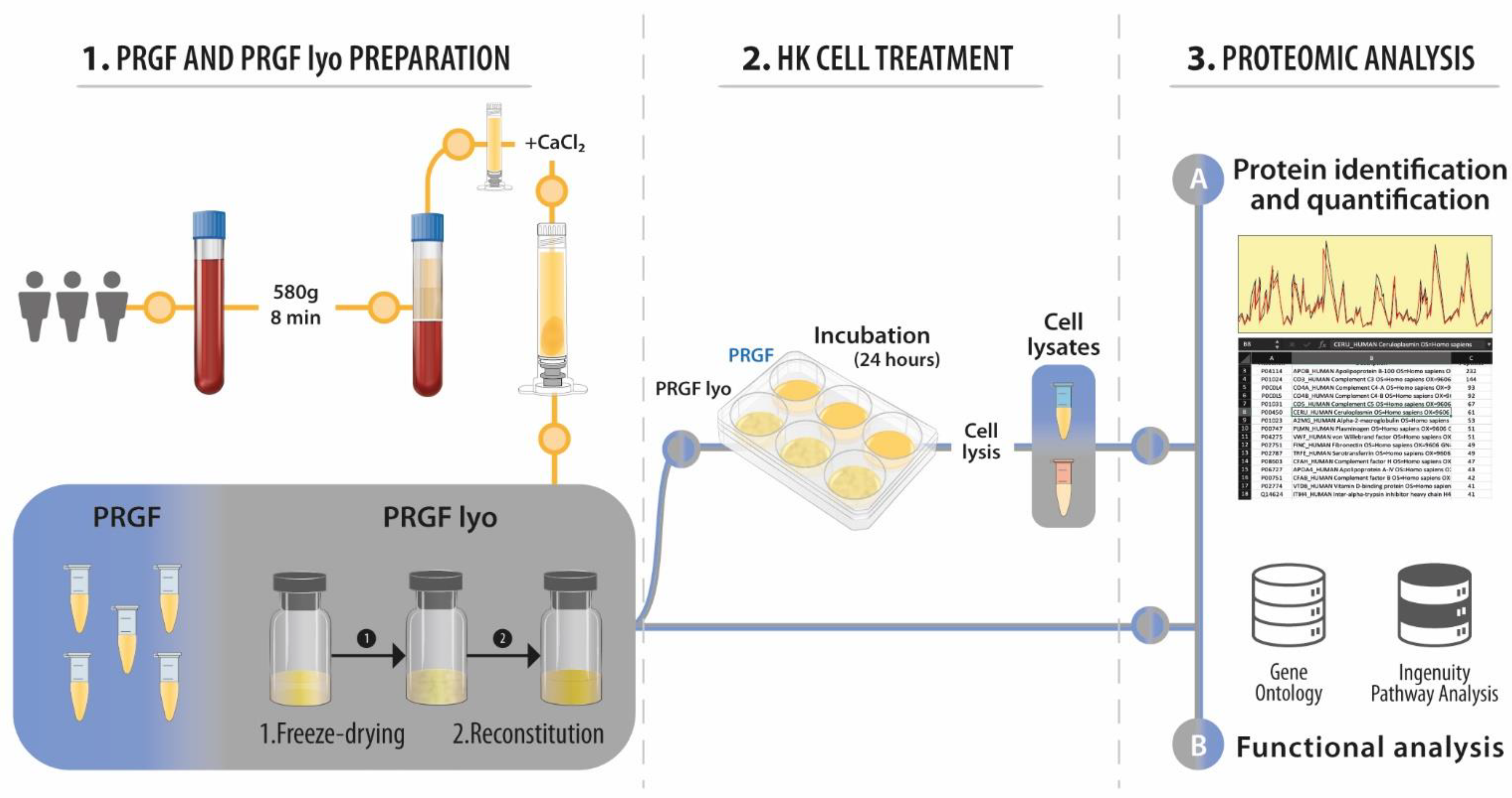
2.1. Plasma Rich in Growth Factors Preparations
2.2. Cells
2.3. Proteomic Analysis
2.4. Functional Analysis
3. Results
3.1. Proteomic Characterization of Blood-Derivative Products
3.2. Proteomic Characterization of HK Cells Treated with Blood-Derivative Products
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Solinís, M.; del Pozo-Rodríguez, A.; Apaolaza, P.S.; Rodríguez-Gascón, A. Treatment of ocular disorders by gene therapy. Eur. J. Pharm. Biopharm. 2015, 95, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.F. To engineer is to create: The link between engineering and regeneration. Trends Biotechnol. 2006, 24, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Lemp, M.A. Management of dry eye disease. Am. J. Manag. Care 2008, 14, S88–S101. [Google Scholar] [PubMed]
- Noecker, R. Effects of common ophthalmic preservatives on ocular health. Adv. Ther. 2001, 18, 205–215. [Google Scholar] [CrossRef]
- Blomquist, P.H. Ocular complications of systemic medications. Am. J. Med. Sci. 2011, 342, 62–69. [Google Scholar] [CrossRef]
- Pan, Q.; Angelina, A.; Marrone, M.; Stark, W.J.; Akpek, E.K. Autologous serum eye drops for dry eye. Cochrane Database Syst. Rev. 2017, 2, Cd009327. [Google Scholar] [CrossRef]
- Schultz, C. Safety and efficacy of cyclosporine in the treatment of chronic dry eye. Ophthalmol. Eye Dis. 2014, 6, 37–42. [Google Scholar] [CrossRef]
- Giannaccare, G.; Versura, P.; Buzzi, M.; Primavera, L.; Pellegrini, M.; Campos, E.C. Blood derived eye drops for the treatment of cornea and ocular surface diseases. Transfus. Apher. Sci. 2017, 56, 595–604. [Google Scholar] [CrossRef]
- Poon, A.C.; Geerling, G.; Dart, J.K.; Fraenkel, G.E.; Daniels, J.T. Autologous serum eyedrops for dry eyes and epithelial defects: Clinical and in vitro toxicity studies. Br. J. Ophthalmol. 2001, 85, 1188–1197. [Google Scholar] [CrossRef]
- Tsubota, K.; Goto, E.; Fujita, H.; Ono, M.; Inoue, H.; Saito, I.; Shimmura, S. Treatment of dry eye by autologous serum application in Sjogren's syndrome. Br. J. Ophthalmol. 1999, 83, 390–395. [Google Scholar] [CrossRef]
- Anitua, E.; Muruzabal, F.; Tayebba, A.; Riestra, A.; Perez, V.L.; Merayo-Lloves, J.; Orive, G. Autologous serum and plasma rich in growth factors in ophthalmology: Preclinical and clinical studies. Acta Ophthalmol. 2015, 93, e605–e614. [Google Scholar] [CrossRef] [PubMed]
- Riestra, A.C.; Alonso-Herreros, J.M.; Merayo-Lloves, J. Plasma rico en plaquetas en superficie ocular. Arch. De La Soc. Esp. De Oftalmol. 2016, 91, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; Muruzabal, F.; Pino, A.; Prado, R.; Azkargorta, M.; Elortza, F.; Merayo-Lloves, J. Proteomic Characterization of Plasma Rich in Growth Factors and Undiluted Autologous Serum. Int. J. Mol. Sci. 2021, 22, 12176. [Google Scholar] [CrossRef] [PubMed]
- Freire, V.; Andollo, N.; Etxebarria, J.; Duran, J.A.; Morales, M.C. In vitro effects of three blood derivatives on human corneal epithelial cells. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5571–5578. [Google Scholar] [CrossRef]
- Anitua, E.; de la Fuente, M.; Muruzabal, F.; Riestra, A.; Merayo-Lloves, J.; Orive, G. Plasma rich in growth factors (PRGF) eye drops stimulates scarless regeneration compared to autologous serum in the ocular surface stromal fibroblasts. Exp. Eye Res. 2015, 135, 118–126. [Google Scholar] [CrossRef]
- Anitua, E.; Muruzabal, F.; de la Fuente, M.; Riestra, A.; Merayo-Lloves, J.; Orive, G. PRGF exerts more potent proliferative and anti-inflammatory effects than autologous serum on a cell culture inflammatory model. Exp. Eye Res. 2016, 151, 115–121. [Google Scholar] [CrossRef]
- Merayo-Lloves, J.; Sanchez-Avila, R.M.; Riestra, A.C.; Anitua, E.; Begona, L.; Orive, G.; Fernandez-Vega, L. Safety and Efficacy of Autologous Plasma Rich in Growth Factors Eye Drops for the Treatment of Evaporative Dry Eye. Ophthalmic Res. 2016, 56, 68–73. [Google Scholar] [CrossRef]
- Sanchez-Avila, R.M.; Merayo-Lloves, J.; Muruzabal, F.; Orive, G.; Anitua, E. Plasma rich in growth factors for the treatment of dry eye from patients with graft versus host diseases. Eur. J. Ophthalmol. 2020, 30, 94–103. [Google Scholar] [CrossRef]
- Sanchez-Avila, R.M.; Merayo-Lloves, J.; Riestra, A.C.; Fernandez-Vega Cueto, L.; Anitua, E.; Begoña, L.; Muruzabal, F.; Orive, G. Treatment of patients with neurotrophic keratitis stages 2 and 3 with plasma rich in growth factors (PRGF-Endoret) eye-drops. Int. Ophthalmol. 2017, 38, 1193–1204. [Google Scholar] [CrossRef]
- Anitua, E. Plasma rich in growth factors: Preliminary results of use in the preparation of future sites for implants. Int. J. Oral Maxillofac Implant. 1999, 14, 529–535. [Google Scholar]
- Anitua, E.; de la Sen-Corcuera, B.; Orive, G.; Sánchez-Ávila, R.M.; Heredia, P.; Muruzabal, F.; Merayo-Lloves, J. Progress in the use of plasma rich in growth factors in ophthalmology: From ocular surface to ocular fundus. Exp. Opin. Biol. Ther. 2022, 22, 31–45. [Google Scholar] [CrossRef]
- Anitua, E.; de la Fuente, M.; Muruzábal, F.; Merayo-Lloves, J. Short- and Long-Term Stability of Plasma Rich in Growth Factors Eye Drops. Cornea 2021, 40, 107–112. [Google Scholar] [CrossRef]
- Anitua, E.; de la Fuente, M.; Riestra, A.; Merayo-Lloves, J.; Muruzabal, F.; Orive, G. Preservation of Biological Activity of Plasma and Platelet-Derived Eye Drops After Their Different Time and Temperature Conditions of Storage. Cornea 2015, 34, 1144–1148. [Google Scholar] [CrossRef]
- Anitua, E.; Muruzabal, F.; Pino, A.; Merayo-Lloves, J.; Orive, G. Biological Stability of Plasma Rich in Growth Factors Eye Drops After Storage of 3 Months. Cornea 2013, 32, 1380–1386. [Google Scholar] [CrossRef] [PubMed]
- Samarkanova, D.; Martin, S.; Bisbe, L.; Puig, J.; Calatayud-Pinuaga, M.; Rodriguez, L.; Azqueta, C.; Coll, R.; Casaroli-Marano, R.; Madrigal, A.; et al. Clinical evaluation of allogeneic eye drops from cord blood platelet lysate. Blood Transfus. 2021, 19, 347–356. [Google Scholar]
- Anitua, E.; de la Fuente, M.; Alcalde, I.; Sanchez, C.; Merayo-Lloves, J.; Muruzabal, F. Development and Optimization of Freeze-Dried Eye Drops Derived From Plasma Rich in Growth Factors Technology. Transl. Vis. Sci. Technol. 2020, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Anitua, E.; de la Fuente, M.; Muruzabal, F.; Merayo-Lloves, J. Stability of freeze-dried plasma rich in growth factors eye drops stored for 3 months at different temperature conditions. Eur. J. Ophthalmol. 2020, 31, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, T.; Prestrelski, S.J.; Kenney, W.C.; Carpenter, J.F. Factors affecting short-term and long-term stabilities of proteins. Adv. Drug Deliv. Rev. 2001, 46, 307–326. [Google Scholar] [CrossRef]
- Izutsu, K.I. Applications of Freezing and Freeze-Drying in Pharmaceutical Formulations. Adv. Exp. Med. Biol 2018, 1081, 371–383. [Google Scholar]
- Anitua, E.; Sanchez, M.; Merayo-Lloves, J.; De la Fuente, M.; Muruzabal, F.; Orive, G. Plasma rich in growth factors (PRGF-Endoret) stimulates proliferation and migration of primary keratocytes and conjunctival fibroblasts and inhibits and reverts TGF-beta1-Induced myodifferentiation. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6066–6073. [Google Scholar] [CrossRef]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Pulcini, S.; Merolle, L.; Marraccini, C.; Quartieri, E.; Mori, D.; Schiroli, D.; Berni, P.; Iotti, B.; Di Bartolomeo, E.; Baricchi, R.; et al. Apheresis Platelet Rich-Plasma for Regenerative Medicine: An In Vitro Study on Osteogenic Potential. Int. J. Mol. Sci. 2021, 22, 8764. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. The human plasma proteome: History, character, and diagnostic prospects. Mol. Cell Proteom. 2002, 1, 845–867. [Google Scholar] [CrossRef]
- Anderson, N.L.; Polanski, M.; Pieper, R.; Gatlin, T.; Tirumalai, R.S.; Conrads, T.P.; Veenstra, T.D.; Adkins, J.N.; Pounds, J.G.; Fagan, R.; et al. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol. Cell Proteom. 2004, 3, 311–326. [Google Scholar] [CrossRef]
- Quartieri, E.; Marraccini, C.; Merolle, L.; Pulcini, S.; Buzzi, M.; Guardi, M.; Schiroli, D.; Baricchi, R.; Pertinhez, T.A. Metabolomics comparison of cord and peripheral blood-derived serum eye drops for the treatment of dry eye disease. Transfus. Apher. Sci. 2021, 60, 103155. [Google Scholar] [CrossRef]
- Brogna, R.; Oldenhof, H.; Sieme, H.; Figueiredo, C.; Kerrinnes, T.; Wolkers, W.F. Increasing storage stability of freeze-dried plasma using trehalose. PLoS ONE 2020, 15, e0234502. [Google Scholar] [CrossRef]
- Chen, L.W.; Huang, C.J.; Tu, W.H.; Lu, C.J.; Sun, Y.C.; Lin, S.Y.; Chen, W.L. The corneal epitheliotrophic abilities of lyophilized powder form human platelet lysates. PLoS ONE 2018, 13, e0194345. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Protein Accession Number | Gene Name | Protein Description | Fold Change | p-Value |
|---|---|---|---|---|
| P22352 | GPX3 | Glutathione peroxidase 3 | 0.7 | 0.0077 |
| P08603 | CFAH | Complement factor H | 0.9 | 0.0142 |
| P04196 | HRG | Histidine-rich glycoprotein | 1.1 | 0.0245 |
| Q9BYE9 | CDHR2 | Cadherin-related family member 2 | 0.6 | 0.0252 |
| P02749 | APOH | Beta-2-glycoprotein 1 | 0.8 | 0.0336 |
| P37802 | TAGL2 | Transgelin-2 | 0.5 | 0.0436 |
| P02649 | APOE | Apolipoprotein E | 0.6 | 0.0464 |
| Q9UHG3 | PCYOX | Prenylcysteine oxidase 1 | 0.8 | 0.0488 |
| GO Term | GO Definition | Genes | % | p-Value |
|---|---|---|---|---|
| 0051918 | Negative regulation of fibrinolysis | P02749, P04196 | 25 | 0.0042 |
| 0030195 | Negative regulation of blood coagulation | P02749, P02649 | 25 | 0.0050 |
| 0043537 | Negative regulation of blood vessel endothelial cell migration | P02649, P04196 | 25 | 0.0058 |
| 0001937 | Negative regulation of endothelial cell proliferation | P02749, P02649 | 25 | 0.0120 |
| 0006641 | Triglyceride metabolic process | P02749, P02649 | 25 | 0.0145 |
| 0000302 | Response to reactive oxygen species | P02649, P22352 | 25 | 0.0161 |
| 0016525 | Negative regulation of angiogenesis | P02749, P04196 | 25 | 0.0256 |
| 0098869 | Cellular oxidant detoxification | P02649, P22352 | 25 | 0.0288 |
| 0030855 | Epithelial cell differentiation | Q9BYE9, P37802 | 25 | 0.0288 |
| 0010468 | Regulation of gene expression | P02649, P04196 | 25 | 0.0409 |
| 0002576 | Platelet degranulation | P02749, P04196 | 25 | 0.0421 |
| 0006979 | Response to oxidative stress | P02649, P22352 | 25 | 0.0449 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anitua, E.; Pino, A.; Azkargorta, M.; Elortza, F.; Merayo-Lloves, J.; Muruzabal, F. Differential Protein Content between Fresh and Freeze-Dried Plasma Rich in Growth Factors Eye Drops. Biomolecules 2022, 12, 1215. https://doi.org/10.3390/biom12091215
Anitua E, Pino A, Azkargorta M, Elortza F, Merayo-Lloves J, Muruzabal F. Differential Protein Content between Fresh and Freeze-Dried Plasma Rich in Growth Factors Eye Drops. Biomolecules. 2022; 12(9):1215. https://doi.org/10.3390/biom12091215
Chicago/Turabian StyleAnitua, Eduardo, Ander Pino, Mikel Azkargorta, Felix Elortza, Jesús Merayo-Lloves, and Francisco Muruzabal. 2022. "Differential Protein Content between Fresh and Freeze-Dried Plasma Rich in Growth Factors Eye Drops" Biomolecules 12, no. 9: 1215. https://doi.org/10.3390/biom12091215






