Research Progress on Nanoparticles-Based CRISPR/Cas9 System for Targeted Therapy of Tumors
Abstract
:1. Introduction
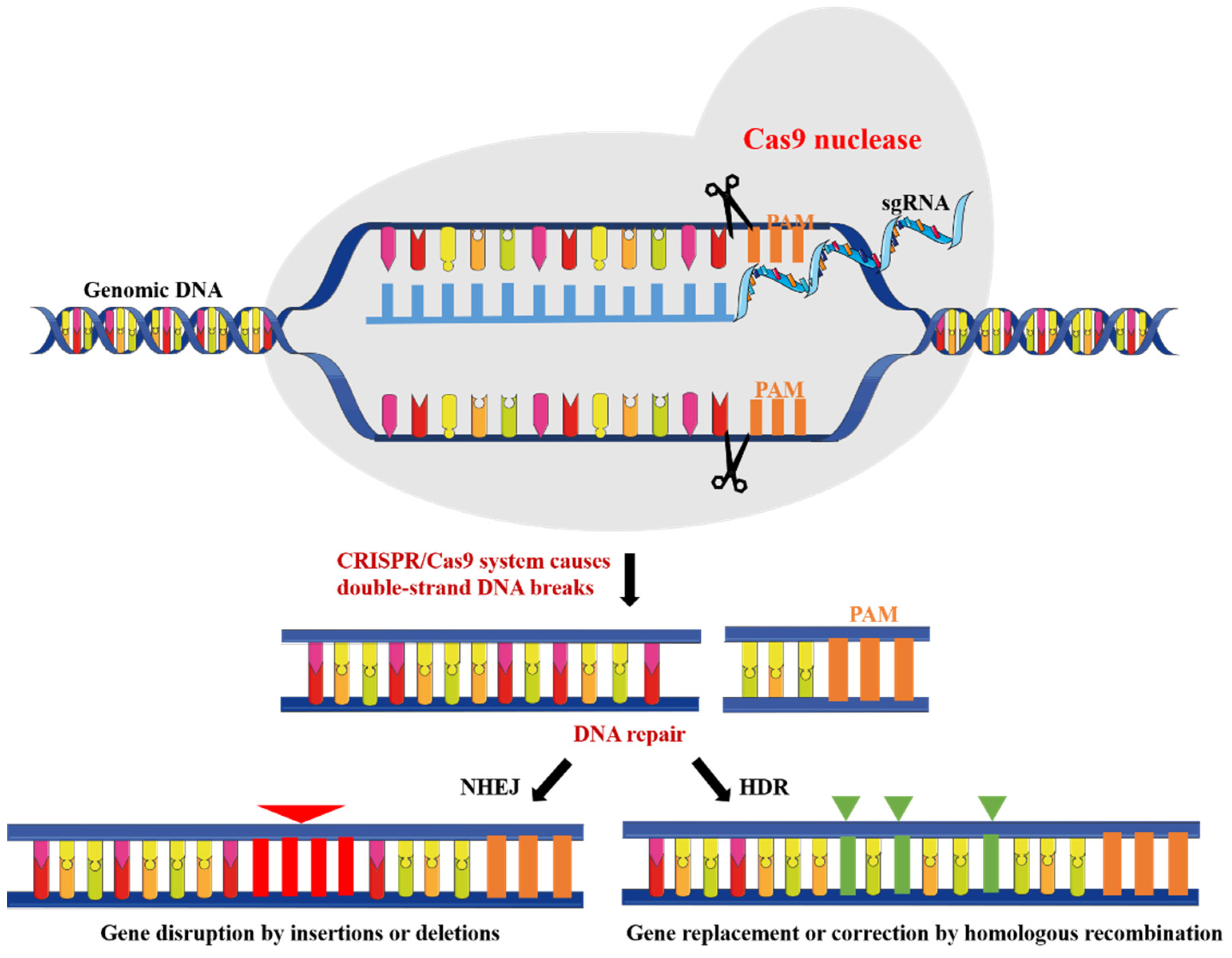
2. CRISPR/Cas9 System
2.1. Delivery of CRISPR/Cas9 System
2.2. Design of NPs-Based CRISPR/Cas9 System
3. Kinds of NPs-Based CRISPR/Cas9 System
3.1. LNPs
3.2. PNPs
3.3. INPs
3.4. NPs of Other Structures
4. Applications of NPs-Based CRISPR/Cas9 System in Cancer Therapy
4.1. Gene Therapy
4.2. Chemotherapy
4.3. Immunotherapy
4.4. Other Therapy
5. Potentials of CRISPR/Cas9 in Oncology
5.1. Clinical Trials and CAR-T Therapy
5.2. Generation of Organoid Cancer Models
6. Future Perspective and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, P.; Ahmadinejad, N.; Wang, J.; Sekulic, A.; Egan, J.B.; Asmann, Y.W.; Kumar, S.; Maley, C.; Liu, L. Somatic selection distinguishes oncogenes and tumor suppressor genes. Bioinformatics 2019, 36, 1712–1717. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-C.; Hao, J.-J.; Nagata, Y.; Xu, L.; Shang, L.; Meng, X.; Sato, Y.; Okuno, Y.; Varela, A.M.; Ding, L.-W.; et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Wardell, C.P.; Furuta, M.; Taniguchi, H.; Fujimoto, A. Cancer whole-genome sequencing: Present and future. Oncogene 2015, 34, 5943–5950. [Google Scholar] [CrossRef] [PubMed]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Holkenbrink, C.; Dam, M.I.; Kildegaard, K.R.; Beder, J.; Dahlin, J.; Belda, D.D.; Borodina, I. EasyCloneYALI: CRISPR/Cas9-Based Synthetic Toolbox for Engineering of the Yeast Yarrowia lipolytica. Biotechnol. J. 2018, 13, e1700543. [Google Scholar] [CrossRef] [Green Version]
- Vizoso, M.; van Rheenen, J. Diverse transcriptional regulation and functional effects revealed by CRISPR/Cas9-directed epigenetic editing. Oncotarget 2021, 12, 1651–1662. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, K.; Wang, C.; Zhang, Z.; Zheng, C.; Zhao, Y.; Zheng, Y.; Liu, C.; An, Y.; Shi, L.; et al. Multistage Delivery Nanoparticle Facilitates Efficient CRISPR/dCas9 Activation and Tumor Growth Suppression In Vivo. Adv. Sci. 2018, 6, 1801423. [Google Scholar] [CrossRef] [PubMed]
- Camperi, J.; Moshref, M.; Dai, L.; Lee, H.Y. Physicochemical and Functional Characterization of Differential CRISPR-Cas9 Ribonucleoprotein Complexes. Anal. Chem. 2021, 94, 1432–1440. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhou, Y.; Chen, J.; Huang, N.; Wang, Z.; Cheng, Y. Gene Therapy for Drug-Resistant Glioblastoma via Lipid-Polymer Hybrid Nanoparticles Combined with Focused Ultrasound. Int. J. Nanomed. 2021, 16, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Sarett, S.M.; Werfel, T.A.; Lee, L.; Duvall, C.L. Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. Proc. Natl. Acad. Sci. USA 2017, 114, E6490–E6497. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; Lindquist, J.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef]
- Khalaf, K.; Janowicz, K.; Dyszkiewicz-Konwińska, M.; Hutchings, G.; Dompe, C.; Moncrieff, L.; Jankowski, M.; Machnik, M.; Oleksiewicz, U.; Kocherova, I.; et al. CRISPR/Cas9 in Cancer Immunotherapy: Animal Models and Human Clinical Trials. Genes 2020, 11, 921. [Google Scholar] [CrossRef]
- Baetke, S.C.; Lammers, T.; Kiessling, F. Applications of nanoparticles for diagnosis and therapy of cancer. Br. J. Radiol. 2015, 88, 20150207. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.Y.; Rutka, J.T.; Chan, W.C. Nanomedicine. N. Engl. J. Med. 2010, 363, 2434–2443. [Google Scholar] [CrossRef]
- Nie, D.; Zhu, Y.; Guo, T.; Yue, M.; Lin, M. Research Advance in Manganese Nanoparticles in Cancer Diagnosis and Therapy. Front. Mater. 2022, 9, 857385. [Google Scholar] [CrossRef]
- Givens, B.E.; Naguib, Y.W.; Geary, S.M.; Devor, E.J.; Salem, A.K. Nanoparticle-Based Delivery of CRISPR/Cas9 Genome-Editing Therapeutics. AAPS J. 2018, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Redman, M.; King, A.; Watson, C.; King, D. What is CRISPR/Cas9? Arch. Dis. Childhood. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2017, 13, 406–416. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Liao, C.; Beisel, C.L. The tracrRNA in CRISPR Biology and Technologies. Annu. Rev. Genet. 2021, 55, 161–181. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef]
- Tsuji, S.; Imanishi, M. Modified nucleobase-specific gene regulation using engineered transcription activator-like effectors. Adv. Drug Deliv. Rev. 2019, 147, 59–65. [Google Scholar] [CrossRef]
- Xin, Y.; Huang, M.; Guo, W.W.; Huang, Q.; Zhang, L.Z.; Jiang, G. Nano-based delivery of RNAi in cancer therapy. Mol. Cancer 2017, 16, 134. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Betge, J.; Ebert, M.P.; Boutros, M. CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 2019, 55, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 2017, 266, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Kretzmann, J.A.; Evans, C.W.; Norret, M.; Blancafort, P.; Iyer, K.S. Non-viral Methodology for Efficient Co-transfection. Methods Mol. Biol. 2018, 1767, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhan, W.; Li, H.; Xu, X.; Cao, X.; Zhu, S.; Liang, J.; Chen, X. In Vivo Dual-Modality Fluorescence and Magnetic Resonance Imaging-Guided Lymph Node Mapping with Good Biocompatibility Manganese Oxide Nanoparticles. Molecules 2017, 22, 2208. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cao, Z.; Yu, L.; Huang, Q.; Zhu, D.; Lu, C.; Lu, A.; Liu, Y. Hierarchical drug release designed Au @PDA-PEG-MTX NPs for targeted delivery to breast cancer with combined photothermal-chemotherapy. J. Nanobiotechnol. 2021, 19, 143. [Google Scholar] [CrossRef]
- Khoobchandani, M.; Katti, K.K.; Karikachery, A.R.; Thipe, V.C.; Srisrimal, D.; Mohandoss, D.K.D.; Darshakumar, R.D.; Joshi, C.M.; Katti, K.V. New Approaches in Breast Cancer Therapy Through Green Nanotechnology and Nano-Ayurvedic Medicine—Pre-Clinical and Pilot Human Clinical Investigations. Int. J. Nanomed. 2020, 15, 181–197. [Google Scholar] [CrossRef]
- Xu, S.; Olenyuk, B.Z.; Okamoto, C.T.; Hamm-Alvarez, S.F. Targeting receptor-mediated endocytotic pathways with nanoparticles: Rationale and advances. Adv. Drug Deliv. Rev. 2013, 65, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Win, K.Y.; Liu, S.; Teng, C.P.; Zheng, Y.; Han, M.-Y. Surface-functionalized nanoparticles for biosensing and imaging-guided therapeutics. Nanoscale 2013, 5, 3127–3148. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Xiang, J. Aptamer-Functionalized Nanoparticles in Targeted Delivery and Cancer Therapy. Int. J. Mol. Sci. 2020, 21, 9123. [Google Scholar] [CrossRef]
- Li, X.; Jian, M.; Sun, Y.; Zhu, Q.; Wang, Z. The Peptide Functionalized Inorganic Nanoparticles for Cancer-Related Bioanalytical and Biomedical Applications. Molecules 2021, 26, 3228. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.; Jeong, C.; Kim, W.J. Synergistic nanomedicine by combined gene and photothermal therapy. Adv. Drug Deliv. Rev. 2016, 98, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Foroozandeh, P.; Aziz, A.A. Insight into Cellular Uptake and Intracellular Trafficking of Nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Wu, Y.; Jin, Y.; Zhu, P.; Shi, W.; Li, J.; Wu, Q.; Zhang, Q.; Han, Y.; Zhao, X. Endosomal/lysosomal location of organically modified silica nanoparticles following caveolae-mediated endocytosis. RSC Adv. 2019, 9, 13855–13862. [Google Scholar] [CrossRef]
- Sahni, A.; Qian, Z.; Pei, D. Cell-Penetrating Peptides Escape the Endosome by Inducing Vesicle Budding and Collapse. ACS Chem. Biol. 2020, 15, 2485–2492. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.J.; Billingsley, M.M.; Haley, R.M.; Wechsler, M.E.; Peppas, N.A.; Langer, R. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 2020, 20, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Thuy, L.T.; Mallick, S.; Choi, J.S. Polyamidoamine (PAMAM) dendrimers modified with short oligopeptides for early endosomal escape and enhanced gene delivery. Int. J. Pharm. 2015, 492, 233–243. [Google Scholar] [CrossRef]
- Pei, M.; Xu, R.; Zhang, C.; Wang, X.; Li, C.; Hu, Y. Mannose-functionalized antigen nanoparticles for targeted dendritic cells, accelerated endosomal escape and enhanced MHC-I antigen presentation. Colloids Surf. B Biointerfaces 2021, 197, 111378. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [PubMed]
- Sung, Y.H.; Kim, J.M.; Kim, H.-T.; Lee, J.; Jeon, J.; Jin, Y.; Choi, J.-H.; Ban, Y.H.; Ha, S.-J.; Kim, C.-H.; et al. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 2013, 24, 125–131. [Google Scholar] [CrossRef]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef]
- Laughery, M.F.; Wyrick, J.J. Simple CRISPR-Cas9 Genome Editing in Saccharomyces cerevisiae. Curr. Protoc. Mol. Biol. 2019, 129, e110. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Varanasi, M.; Mendes, S.; Yamagata, H.M.; Wilson, D.R.; Green, J.J. Poly(Beta-Amino Ester) Nanoparticles Enable Nonviral Delivery of CRISPR-Cas9 Plasmids for Gene Knockout and Gene Deletion. Mol. Ther.-Nucleic Acids 2020, 20, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Nishikawa, A.; Kume, S.; Chayama, K.; Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014, 4, 5400. [Google Scholar] [CrossRef] [PubMed]
- Hiranniramol, K.; Chen, Y.; Liu, W.; Wang, X. Generalizable sgRNA design for improved CRISPR/Cas9 editing efficiency. Bioinformatics 2020, 36, 2684–2689. [Google Scholar] [CrossRef] [PubMed]
- Hennig, S.L.; Owen, J.R.; Lin, J.C.; Young, A.E.; Ross, P.J.; Van Eenennaam, A.L.; Murray, J.D. Evaluation of mutation rates, mosaicism and off target mutations when injecting Cas9 mRNA or protein for genome editing of bovine embryos. Sci. Rep. 2020, 10, 22309. [Google Scholar] [CrossRef] [PubMed]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massouridès, E.; Sohier, T.J.M.; Corbin, A.; et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.M.; Ikeda, K.; Cromer, M.K.; Uchida, N.; Nishimura, T.; Romano, R.; Tong, A.J.; Lemgart, V.T.; Camarena, J.; Pavel-Dinu, M.; et al. Highly Efficient and Marker-free Genome Editing of Human Pluripotent Stem Cells by CRISPR-Cas9 RNP and AAV6 Donor-Mediated Homologous Recombination. Cell Stem Cell 2019, 24, 821–828.e5. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, J.; Roscoe, B.P.; Liu, P.; Yao, Q.; Lazzarotto, C.R.; Clement, M.K.; Cole, M.; Luk, K.; Baricordi, C.; et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019, 25, 776–783. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.-Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2014, 33, 73–80. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew. Chem. Int. Ed. 2017, 57, 1491–1496. [Google Scholar] [CrossRef]
- Pandey, V.; Kohli, S. Lipids and Surfactants: The Inside Story of Lipid-Based Drug Delivery Systems. Crit. Rev. Ther. Drug Carr. Syst. 2018, 35, 99–155. [Google Scholar] [CrossRef]
- Bargui, R.; Solgadi, A.; Prost, B.; Chester, M.; Ferreiro, A.; Piquereau, J.; Moulin, M. Phospholipids: Identification and Implication in Muscle Pathophysiology. Int. J. Mol. Sci. 2021, 22, 8176. [Google Scholar] [CrossRef]
- Li, Y.; Gao, L.; Tan, X.; Li, F.; Zhao, M.; Peng, S. Lipid rafts-mediated endocytosis and physiology-based cell membrane traffic models of doxorubicin liposomes. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 1801–1811. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Hersch, N.; Gerlach, S.; Dreissen, G.; Springer, R.; Merkel, R.; Csiszár, A.; Hoffmann, B. Complex Size and Surface Charge Determine Nucleic Acid Transfer by Fusogenic Liposomes. Int. J. Mol. Sci. 2020, 21, 2244. [Google Scholar] [CrossRef]
- Zhen, S.; Li, X. Liposomal delivery of CRISPR/Cas9. Cancer Gene Ther. 2020, 27, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xie, H.; Liu, Y.; Xia, C.; Cun, X.; Long, Y.; Chen, X.; Deng, M.; Guo, R.; Zhang, Z.; et al. Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer. J. Control. Release 2019, 304, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Rosenblum, D.; Gutkin, A.; Kedmi, R.; Ramishetti, S.; Veiga, N.; Jacobi, A.M.; Schubert, M.S.; Friedmann-Morvinski, D.; Cohen, Z.R.; Behlke, M.A.; et al. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 2020, 6, eabc9450. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cheng, Q.; Wei, T.; Yu, X.; Johnson, L.T.; Farbiak, L.; Siegwart, D.J. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 2021, 20, 701–710. [Google Scholar] [CrossRef]
- Li, C.; Li, Z.; Gong, X.; Liu, J.; Zheng, T.; Wang, F.; Wu, J.; Zhang, B. Acidic tumor microenvironment-sensitive liposomes enhance colorectal cancer therapy by acting on both tumor cells and cancer-associated fibroblasts. Nanoscale 2021, 13, 10509–10525. [Google Scholar] [CrossRef]
- Tu, K.; Deng, H.; Kong, L.; Wang, Y.; Yang, T.; Hu, Q.; Hu, M.; Yang, C.; Zhang, Z. Reshaping Tumor Immune Microenvironment through Acidity-Responsive Nanoparticles Featured with CRISPR/Cas9-Mediated Programmed Death-Ligand 1 Attenuation and Chemotherapeutics-Induced Immunogenic Cell Death. ACS Appl. Mater. Interfaces 2020, 12, 16018–16030. [Google Scholar] [CrossRef]
- Zhen, S.; Liu, Y.; Lu, J.; Tuo, X.; Yang, X.; Chen, H.; Chen, W.; Li, X. Human Papillomavirus Oncogene Manipulation Using Clustered Regularly Interspersed Short Palindromic Repeats/Cas9 Delivered by pH-Sensitive Cationic Liposomes. Hum. Gene Ther. 2020, 31, 309–324. [Google Scholar] [CrossRef]
- Yin, H.; Sun, L.; Pu, Y.; Yu, J.; Feng, W.; Dong, C.; Zhou, B.; Du, D.; Zhang, Y.; Chen, Y.; et al. Ultrasound-Controlled CRISPR/Cas9 System Augments Sonodynamic Therapy of Hepatocellular Carcinoma. ACS Central Sci. 2021, 7, 2049–2062. [Google Scholar] [CrossRef]
- Zhang, Z.; Wan, T.; Chen, Y.; Chen, Y.; Sun, H.; Cao, T.; Songyang, Z.; Tang, G.; Wu, C.; Ping, Y.; et al. Cationic Polymer-Mediated CRISPR/Cas9 Plasmid Delivery for Genome Editing. Macromol. Rapid Commun. 2018, 40, e1800068. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Ringel-Scaia, V.M.; McDaniel, D.K.; Thomas, C.A.; Zhang, R.; Riffle, J.S.; Allen, I.C.; Davis, R.M. Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR–Cas9 plasmid. J. Nanobiotechnol. 2020, 18, 16. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.-C.; Chou, Y.-H.; Hung, Y.-N.; Hu, S.-H.; Chiang, W.-H. Hybrid polymeric nanoparticles with high zoledronic acid payload and proton sponge-triggered rapid drug release for anticancer applications. Mater. Sci. Eng. C 2020, 116, 111277. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-C.; Wu, P.-Y.; Zou, J.-J.; Jiang, J.-L.; Zhao, R.-R.; Luo, B.-Y.; Liao, Y.-Q.; Shao, J.-W. Efficient CRISPR/Cas9 gene-chemo synergistic cancer therapy via a stimuli-responsive chitosan-based nanocomplex elicits anti-tumorigenic pathway effect. Chem. Eng. J. 2020, 393, 124688. [Google Scholar] [CrossRef]
- Nguyen, D.N.; Roth, T.; Li, P.J.; Chen, P.A.; Apathy, R.; Mamedov, M.R.; Vo, L.T.; Tobin, V.; Goodman, D.; Shifrut, E.; et al. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2019, 38, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Bayda, S.; Hadla, M.; Palazzolo, S.; Riello, P.; Corona, G.; Toffoli, G.; Rizzolio, F. Inorganic Nanoparticles for Cancer Therapy: A Transition from Lab to Clinic. Curr. Med. Chem. 2018, 25, 4269–4303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-C.; Luo, B.-Y.; Zou, J.-J.; Wu, P.-Y.; Jiang, J.-L.; Le, J.-Q.; Zhao, R.-R.; Chen, L.; Shao, J.-W. Co-delivery of Sorafenib and CRISPR/Cas9 Based on Targeted Core–Shell Hollow Mesoporous Organosilica Nanoparticles for Synergistic HCC Therapy. ACS Appl. Mater. Interfaces 2020, 12, 57362–57372. [Google Scholar] [CrossRef]
- Singh, P.; Pandit, S.; Mokkapati, V.; Garg, A.; Ravikumar, V.; Mijakovic, I. Gold Nanoparticles in Diagnostics and Therapeutics for Human Cancer. Int. J. Mol. Sci. 2018, 19, 1979. [Google Scholar] [CrossRef]
- Rohiwal, S.S.; Dvorakova, N.; Klima, J.; Vaskovicova, M.; Senigl, F.; Slouf, M.; Pavlova, E.; Stepanek, P.; Babuka, D.; Benes, H.; et al. Polyethylenimine based magnetic nanoparticles mediated non-viral CRISPR/Cas9 system for genome editing. Sci. Rep. 2020, 10, 4619. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Gao, L.; Fan, T.; Zhang, C.; Zhang, B.; Al-Hartomy, O.A.; Al-Ghamdi, A.; Wageh, S.; Qiu, M.; Zhang, H. Strategic Design of Intelligent-Responsive Nanogel Carriers for Cancer Therapy. ACS Appl. Mater. Interfaces 2021, 13, 54621–54647. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liu, F.; Chen, Y.; Liu, J.; Wang, X.; Chen, A.T.; Deng, G.; Zhang, H.; Liu, J.; Hong, Z.; et al. Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles. Adv. Funct. Mater. 2017, 27, 1703036. [Google Scholar] [CrossRef] [PubMed]
- Doucey, M.A.; Carrara, S. Nanowire Sensors in Cancer. Trends Biotechnol. 2019, 37, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. Engl. 2015, 54, 12029–12033. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sunami, Y.; Hashimoto, H. Mini Review: Nanosheet Technology towards Biomedical Application. Nanomaterials 2017, 7, 246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Cui, H.; Ying, L.; Yu, X.-F. Enhanced Cytosolic Delivery and Release of CRISPR/Cas9 by Black Phosphorus Nanosheets for Genome Editing. Angew. Chem. Int. Ed. Engl. 2018, 57, 10268–10272. [Google Scholar] [CrossRef]
- Sun, W.; Shi, Q.; Zhang, H.; Yang, K.; Ke, Y.; Wang, Y.; Qiao, L. Advances in the techniques and methodologies of cancer gene therapy. Discov. Med. 2019, 27, 45–55. [Google Scholar]
- van den Bulk, J.; Verdegaal, E.M.; de Miranda, N.F. Cancer immunotherapy: Broadening the scope of targetable tumours. Open Biol. 2018, 8, 180037. [Google Scholar] [CrossRef]
- Li, X.; Lovell, J.F.; Yoon, J.; Chen, X. Clinical development and potential of photothermal and photodynamic therapies for cancer. Nat. Rev. Clin. Oncol. 2020, 17, 657–674. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Wang, X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef]
- Liu, Y.; Hu, X.; Han, C.; Wang, L.; Zhang, X.; He, X.; Lu, X. Targeting tumor suppressor genes for cancer therapy. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015, 37, 1277–1286. [Google Scholar] [CrossRef]
- Ju, E.; Li, T.; da Silva, S.R.; Gao, S.-J. Gold Nanocluster-Mediated Efficient Delivery of Cas9 Protein through pH-Induced Assembly-Disassembly for Inactivation of Virus Oncogenes. ACS Appl. Mater. Interfaces 2019, 11, 34717–34724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qin, C.; An, C.; Zheng, X.; Wen, S.; Chen, W.; Liu, X.; Lv, Z.; Yang, P.; Xu, W.; et al. Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Mol. Cancer 2021, 20, 126. [Google Scholar] [CrossRef] [PubMed]
- Robert, J. Epirubicin. Clinical pharmacology and dose-effect relationship. Drugs 1993, 45 (Suppl. 2), 20–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yao, Z.; Wang, L.; Ding, H.; Shao, J.; Chen, A.; Zhang, F.; Zheng, S. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy 2018, 14, 2083–2103. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-S.; Chang, C.-H.; Tzeng, T.-Y.; Lin, A.M.-Y.; Lo, Y.-L. Gene-editing by CRISPR-Cas9 in combination with anthracycline therapy via tumor microenvironment-switchable, EGFR-targeted, and nucleus-directed nanoparticles for head and neck cancer suppression. Nanoscale Horiz. 2021, 6, 729–743. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, M.; Hou, B.; Zheng, B.; Wang, Z.; Huang, M.; Xu, Y.; Chang, J.; Wang, T. CRISPR/Cas9 nanoeditor of double knockout large fragments of E6 and E7 oncogenes for reversing drugs resistance in cervical cancer. J. Nanobiotechnol. 2021, 19, 231. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Zhang, Q.; Feng, S.; Li, C.; Wang, L.; Zhao, X.; Yang, Z.; Li, Z.; Luo, H.; Liu, R.; et al. Safety and Efficacy of PD-1/PD-L1 inhibitors combined with radiotherapy in patients with non-small-cell lung cancer: A systematic review and meta-analysis. Cancer Med. 2021, 10, 1222–1239. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.Y.; Jiang, X.M.; Wang, B.L.; Sun, Y.; Lu, J.-J. Combination therapy with PD-1/PD-L1 blockade in non-small cell lung cancer: Strategies and mechanisms. Pharmacol. Ther. 2021, 219, 107694. [Google Scholar] [CrossRef]
- Deng, H.; Tan, S.; Gao, X.; Zou, C.; Xu, C.; Tu, K.; Song, Q.; Fan, F.; Huang, W.; Zhang, Z. Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm. Sin. B 2020, 10, 358–373. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Sun, H.; Feng, Y.; Xu, L.; Chan, A.; Tong, J.H.; Wong, J.; Chong, C.; Lai, P.B.S.; et al. Hepatoma-intrinsic CCRK inhibition diminishes myeloid-derived suppressor cell immunosuppression and enhances immune-checkpoint blockade efficacy. Gut 2018, 67, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Song, N.; Dong, Y.; Li, S.; Li, L.; Liu, Y.; Li, Z.; Yang, D. A Proton-Activatable DNA-Based Nanosystem Enables Co-Delivery of CRISPR/Cas9 and DNAzyme for Combined Gene Therapy. Angew. Chem. Int. Ed. Engl. 2022, 61, e202116569. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. CRISPR gene-editing tested in a person for the first time. Nature 2016, 539, 479. [Google Scholar] [CrossRef]
- Hong, M.; Clubb, J.D.; Chen, Y.Y. Engineering CAR-T Cells for Next-Generation Cancer Therapy. Cancer Cell 2020, 38, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Larson, R.C.; Maus, M.V. Recent advances and discoveries in the mechanisms and functions of CAR T cells. Nat. Rev. Cancer 2021, 21, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Sterner, R.C.; Sterner, R.M. CAR-T cell therapy: Current limitations and potential strategies. Blood Cancer J. 2021, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, Z.; Liu, Y.; Han, W. New development in CAR-T cell therapy. J. Hematol. Oncol. 2017, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wu, Z.; Jia, H.; Tong, C.; Guo, Y.; Ti, D.; Han, X.; Liu, Y.; Zhang, W.; Wang, C.; et al. Bispecific CAR-T cells targeting both CD19 and CD22 for therapy of adults with relapsed or refractory B cell acute lymphoblastic leukemia. J. Hematol. Oncol. 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Ying, Z.; Huang, X.F.; Xiang, X.; Liu, Y.; Kang, X.; Song, Y.; Guo, X.; Liu, H.; Ding, N.; Zhang, T.; et al. A safe and potent anti-CD19 CAR T cell therapy. Nat. Med. 2019, 25, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef] [Green Version]
- Norelli, M.; Camisa, B.; Barbiera, G.; Falcone, L.; Purevdorj, A.; Genua, M.; Sanvito, F.; Ponzoni, M.; Doglioni, C.; Cristofori, P.; et al. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 2018, 24, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Paszkiewicz, P.J.; Fräßle, S.P.; Srivastava, S.; Sommermeyer, D.; Hudecek, M.; Drexler, I.; Sadelain, M.; Liu, L.; Jensen, M.C.; Riddell, S.R. Targeted antibody-mediated depletion of murine CD19 CAR T cells permanently reverses B cell aplasia. J. Clin. Investig. 2016, 126, 4262–4272. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Fang, Y.; Chen, X.; Wang, Z.; Liang, X.; Zhang, T.; Liu, M.; Zhou, N.; Lv, J.; Tang, K.; et al. Gasdermin E–mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. Sci. Immunol. 2020, 5, eaax7969. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhou, Y.; Zhang, M.; Ge, W.; Li, Y.; Yang, L.; Wei, G.; Han, L.; Wang, H.; Yu, S.; et al. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2021, 27, 2764–2772. [Google Scholar] [CrossRef]
- Shi, L.; Meng, T.; Zhao, Z.; Han, J.; Zhang, W.; Gao, F.; Cai, J. CRISPR knock out CTLA-4 enhances the anti-tumor activity of cytotoxic T lymphocytes. Gene 2017, 636, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, X.; Liu, X.; Fang, C.; Jiang, S.; June, C.H.; Zhao, Y. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget 2017, 8, 17002–17011. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Wang, X. Stem cells in tissues, organoids, and cancers, Cellular and molecular life sciences. Cell. Mol. Life Sci. 2019, 76, 4043–4070. [Google Scholar] [CrossRef] [PubMed]
- Schwank, G.; Koo, B.-K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional Repair of CFTR by CRISPR/Cas9 in Intestinal Stem Cell Organoids of Cystic Fibrosis Patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; Van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; de Ligt, J.; Behjati, S.; Grolleman, J.E.; van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, S.; Zhao, G.; Xu, C.-F.; Zhang, H.-B.; Luo, Y.-L.; Cao, Z.-T.; Shi, J.; Zhao, Z.-B.; Lian, Z.-X.; et al. In situ repurposing of dendritic cells with CRISPR/Cas9-based nanomedicine to induce transplant tolerance. Biomaterials 2019, 217, 119302. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Watanabe, S.; Ando, N.; Ishihara, M.; Sato, M. Transplacental Gene Delivery (TPGD) as a Noninvasive Tool for Fetal Gene Manipulation in Mice. Int. J. Mol. Sci. 2019, 20, 5926. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-Y.; Liu, Z.-H.; Weng, W.-H.; Chang, C.-W. Magnetic nanocomplexes for gene delivery applications. J. Mater. Chem. B 2021, 9, 4267–4286. [Google Scholar] [CrossRef] [PubMed]



| Delivery Approaches | NPs Formulation | CRISPR/Cas9 Cargo | Efficiency | Application | Reference |
|---|---|---|---|---|---|
| LNPs | Cationic liposomes | Cas9 sgRNA complexes | 80% | in vitro and in vivo | [66] |
| Cationic liposomes modified with R8-DGR | sgRNA | -- | in vitro and in vivo | [73] | |
| Amino-ionizable lipid NPs | Cas9 mRNA and sgRNA | ~70% (aggressive orthotopic glioblastoma) ~80% (disseminated ovarian tumors) | in vitro and in vivo | [74] | |
| SORT | Cas9 mRNA and sgRNA | 40% (epithelial cells) 65% (endothelial cells) 12% (B cells) 10% (T cells) 93% (hepatocytes) 20% (liver) 50% (lung) 30% (spleen) | in vitro and in vivo | [75] | |
| iPhos | Cas9 mRNA and sgRNA | ~91% (hepatocytes) ~34% (lung endothelial cells) ~20% (lung epithelial cells) ~13% (lung immune cells) ~30% (splenic macrophages) 6% (Splenic B cells) | in vivo | [76] | |
| pH-sensitive cationic liposomes | Cas9 mRNA and sgRNA | -- | in vitro and in vivo | [79] | |
| HMME@Lip-Cas9 | RNP | 17.28% (HMME@Lip-Cas9) 58.77% (HMME@Lip-Cas9 + ultrasound) | in vitro and in vivo | [80] | |
| PNPs | PEI-β-cyclodextrin cationic polymers | pDNA | 19.1% (HBB locus) 7% (RHBDF1 locus) | in vitro | [81] |
| Poly lactic-co-glycolic acid (PLGA) NPs | pDNA | 95% (murine bone marrow derived macrophages) | in vitro | [83] | |
| MDNP | pDNA | -- | in vitro and in vivo | [12] | |
| Polyglutamic acid-modified NPs | Cas9 RNPs | ~2 fold increase | in vitro | [86] | |
| INPs | SEHPA NPs | RNP | >60% (EGFR editing efficiency) | in vitro and in vivo | [88] |
| LACP | RNP | 68% | in vitro and in vivo | [67] | |
| PEI-coated magnetic Fe3O4 NPs | pDNA | 13% (with magnetic field) 10% (without magnetic field) | in vitro | [90] | |
| NPs of other structures | LHNPs | Cas9 protein and minicircle gRNA | 1.3 times more efficiently than Lip2k | in vitro and in vivo | [92] |
| DNA nanowires | Cas9 protein and sgRNA | 36% | in vitro and in vivo | [94] | |
| Cas9 N3BPs | Cas9 sgRNA complexes | 26.7% (Target 1) 32.1% (GRIN2B) | in vitro and in vivo | [96] |
| Identifier | Target Gene | Phase | Condition |
|---|---|---|---|
| NCT03057912 | HPV E6/E7 | I | Human Papillomavirus-Related Malignant Neoplasm |
| NCT03164135 | CISH | I/II | Gastrointestinal Epithelial Cancer, Gastrointestinal Neoplasms |
| NCT04976218 | TGF-β | I | Solid Tumor |
| NCT04767308 | CD5 | I | Relapsed/Refractory Hematopoietic Malignancies |
| NCT03545815 | PD-1, TCR | I | Solid Tumor, Adult |
| NCT05066165 | WT1 | I/II | Acute Myeloid Leukemia |
| NCT05309733 | CD33 | Leukemia, Myeloid, Acute | |
| NCT03747965 | PD-1 | I | Solid Tumor, Adult |
| NCT04035434 | CD19 | I | B-cell Malignancy Non-Hodgkin Lymphoma B-cell Lymphoma Adult B Cell ALL |
| NCT04502446 | CD70 | I | T Cell Lymphoma |
| NCT03081715 | PD-1 | -- | Esophageal Cancer |
| NCT05037669 | CD19 | I | Acute Lymphoblastic Leukemia, Chronic Lymphocytic Leukemia, Non Hodgkin Lymphoma |
| NCT04244656 | BCMA | I | Multiple Myeloma |
| NCT04438083 | CD70 | I | Renal Cell Carcinoma |
| NCT03166878 | CD19 | I/II | B Cell Leukemia, B Cell Lymphoma |
| NCT03398967 | CD19/CD20/CD22 | I/II | B Cell Leukemia, B Cell Lymphoma |
| NCT04557436 | CD52, TRAC | I | B Acute Lymphoblastic Leukemia |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, D.; Guo, T.; Yue, M.; Li, W.; Zong, X.; Zhu, Y.; Huang, J.; Lin, M. Research Progress on Nanoparticles-Based CRISPR/Cas9 System for Targeted Therapy of Tumors. Biomolecules 2022, 12, 1239. https://doi.org/10.3390/biom12091239
Nie D, Guo T, Yue M, Li W, Zong X, Zhu Y, Huang J, Lin M. Research Progress on Nanoparticles-Based CRISPR/Cas9 System for Targeted Therapy of Tumors. Biomolecules. 2022; 12(9):1239. https://doi.org/10.3390/biom12091239
Chicago/Turabian StyleNie, Dengyun, Ting Guo, Miao Yue, Wenya Li, Xinyu Zong, Yinxing Zhu, Junxing Huang, and Mei Lin. 2022. "Research Progress on Nanoparticles-Based CRISPR/Cas9 System for Targeted Therapy of Tumors" Biomolecules 12, no. 9: 1239. https://doi.org/10.3390/biom12091239






