Sub-Millisecond Photoinduced Dynamics of Free and EL222-Bound FMN by Stimulated Raman and Visible Absorption Spectroscopies
Abstract
:1. Introduction
2. Materials and Methods
3. Results
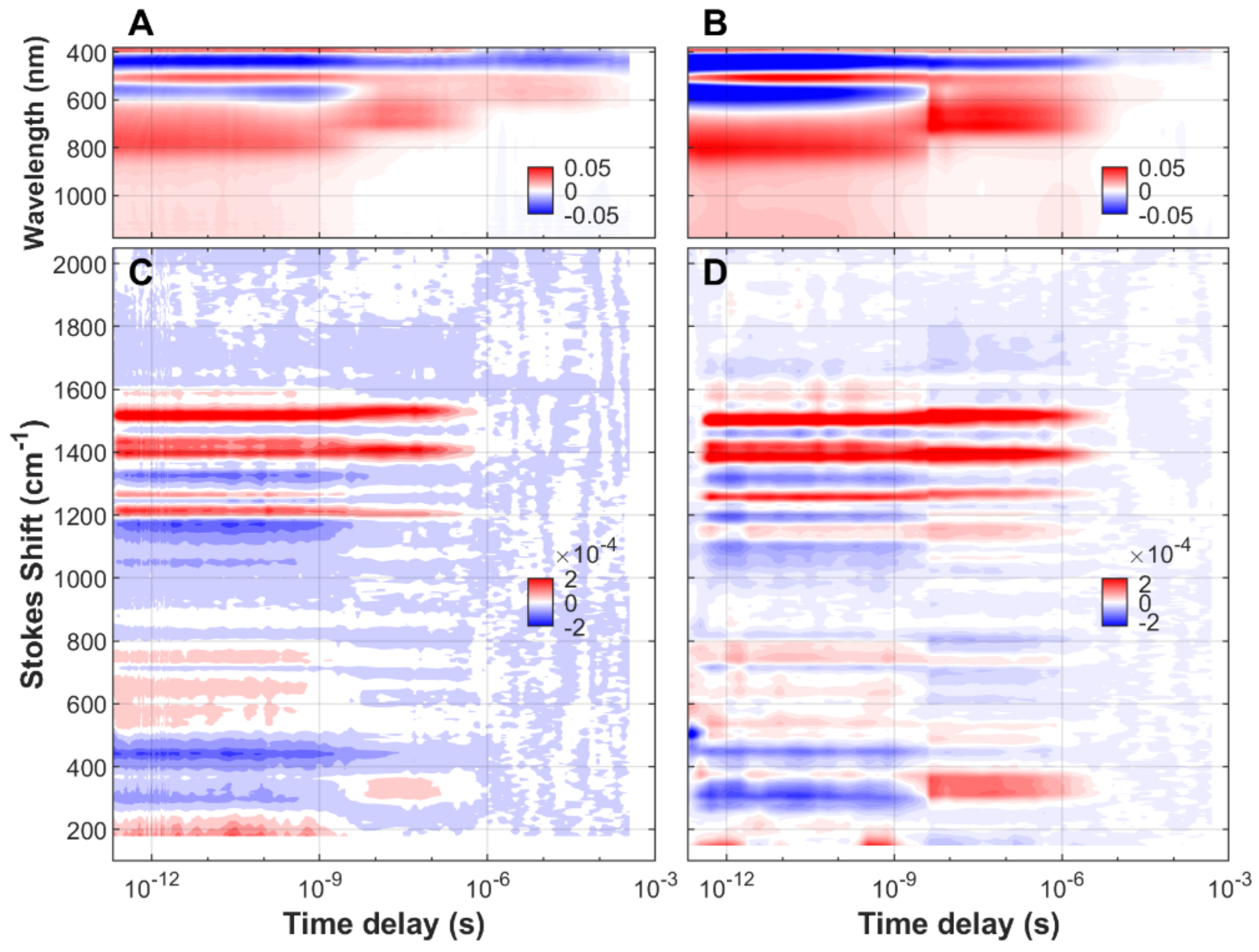
3.1. Time-Resolved FSRS/Absorption of FMN, EL222-WT and EL222-C78A
3.2. Analysis of Time-Resolved FSRS/visTA Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kottke, T.; Xie, A.; Larsen, D.S.; Hoff, W.D. Photoreceptors Take Charge: Emerging Principles for Light Sensing. Annu. Rev. Biophys. 2018, 47, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Poddar, H.; Heyes, D.J.; Schirò, G.; Weik, M.; Leys, D.; Scrutton, N.S. A guide to time-resolved structural analysis of light-activated proteins. FEBS J. 2021, 289, 576–595. [Google Scholar] [CrossRef] [PubMed]
- Herrou, J.; Crosson, S. Function, structure and mechanism of bacterial photosensory LOV proteins. Nat. Rev. Microbiol. 2011, 9, 713–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losi, A.; Gärtner, W. Solving Blue Light Riddles: New Lessons from Flavin-binding LOV Photoreceptors. Photochem. Photobiol. 2017, 93, 141–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwata, T.; Masuda, S. Photoreaction Mechanisms of Flavoprotein Photoreceptors and Their Applications. In Optogenetics; Springer: Berlin/Heidelberg, Germany, 2021; pp. 189–206. [Google Scholar] [CrossRef]
- Nash, A.I.; McNulty, R.; Shillito, M.E.; Swartz, T.E.; Bogomolni, R.A.; Luecke, H.; Gardner, K.H. Structural basis of photosensitivity in a bacterial light-oxygen-voltage/helix-turn-helix (LOV-HTH) DNA-binding protein. Proc. Natl. Acad. Sci. USA 2011, 108, 9449–9454. [Google Scholar] [CrossRef] [Green Version]
- Takakado, A.; Nakasone, Y.; Terazima, M. Sequential DNA Binding and Dimerization Processes of the Photosensory Protein EL222. Biochemistry 2018, 57, 1603–1610. [Google Scholar] [CrossRef]
- Takakado, A.; Nakasone, Y.; Terazima, M. Photoinduced dimerization of a photosensory DNA-binding protein EL222 and its LOV domain. Phys. Chem. Chem. Phys. 2017, 19, 24855–24865. [Google Scholar] [CrossRef]
- Crosson, S.; Moffat, K. Photoexcited Structure of a Plant Photoreceptor Domain Reveals a Light-Driven Molecular Switch. Plant Cell 2002, 14, 1067–1075. [Google Scholar] [CrossRef] [Green Version]
- Losi, A.; Gardner, K.H.; Möglich, A. Blue-Light Receptors for Optogenetics. Chem. Rev. 2018, 118, 10659–10709. [Google Scholar] [CrossRef]
- Seifert, S.; Brakmann, S. LOV Domains in the Design of Photoresponsive Enzymes. ACS Chem. Biol. 2018, 13, 1914–1920. [Google Scholar] [CrossRef]
- Pudasaini, A.; El-Arab, K.K.; Zoltowski, B.D. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci. 2015, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Iuliano, J.N.; French, J.B.; Tonge, P.J. Vibrational spectroscopy of flavoproteins. In New Approaches for Flavin Catalysis; Springer: Berlin/Heidelberg, Germany, 2019; pp. 189–214. [Google Scholar] [CrossRef]
- Zayner, J.P.; Sosnick, T.R. Factors That Control the Chemistry of the LOV Domain Photocycle. PLoS ONE 2014, 9, e87074. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Tang, L. Mapping Structural Dynamics of Proteins with Femtosecond Stimulated Raman Spectroscopy. Annu. Rev. Phys. Chem. 2020, 71, 239–265. [Google Scholar] [CrossRef] [Green Version]
- Dietze, D.R.; Mathies, R.A. Femtosecond Stimulated Raman Spectroscopy. ChemPhysChem 2016, 17, 1224–1251. [Google Scholar] [CrossRef]
- Kukura, P.; McCamant, D.W.; Mathies, R.A. Femtosecond Stimulated Raman Spectroscopy. Annu. Rev. Phys. Chem. 2007, 58, 461–488. [Google Scholar] [CrossRef]
- Kottke, T.; Lórenz-Fonfría, V.A.; Heberle, J. The Grateful Infrared: Sequential Protein Structural Changes Resolved by Infrared Difference Spectroscopy. J. Phys. Chem. B 2016, 121, 335–350. [Google Scholar] [CrossRef]
- Schubert, L.; Langner, P.; Ehrenberg, D.; Lorenz-Fonfria, V.A.; Heberle, J. Protein conformational changes and protonation dynamics probed by a single shot using quantum-cascade-laser-based IR spectroscopy. J. Chem. Phys. 2022, 156, 204201. [Google Scholar] [CrossRef]
- Greetham, G.M.; Sole, D.; Clark, I.P.; Parker, A.W.; Pollard, M.R.; Towrie, M. Time-resolved multiple probe spectroscopy. Rev. Sci. Instrum. 2012, 83, 103107. [Google Scholar] [CrossRef]
- Weigel, A.; Dobryakov, A.; Klaumünzer, B.; Sajadi, M.; Saalfrank, P.; Ernsting, N.P. Femtosecond Stimulated Raman Spectroscopy of Flavin after Optical Excitation. J. Phys. Chem. B 2011, 115, 3656–3680. [Google Scholar] [CrossRef]
- Andrikopoulos, P.C.; Liu, Y.; Picchiotti, A.; Lenngren, N.; Kloz, M.; Chaudhari, A.S.; Precek, M.; Rebarz, M.; Andreasson, J.; Hajdu, J.; et al. Femtosecond-to-nanosecond dynamics of flavin mononucleotide monitored by stimulated Raman spectroscopy and simulations. Phys. Chem. Chem. Phys. 2020, 22, 6538–6552. [Google Scholar] [CrossRef]
- Iuliano, J.N.; Hall, C.R.; Green, D.; Jones, G.A.; Lukacs, A.; Illarionov, B.; Bacher, A.; Fischer, M.; French, J.B.; Tonge, P.J.; et al. Excited State Vibrations of Isotopically Labeled FMN Free and Bound to a Light–Oxygen–Voltage (LOV) Protein. J. Phys. Chem. B 2020, 124, 7152–7165. [Google Scholar] [CrossRef] [PubMed]
- Iuliano, J.N.; Gil, A.A.; Laptenok, S.P.; Hall, C.R.; Tolentino Collado, J.; Lukacs, A.; Hag Ahmed, S.A.; Abyad, J.; Daryaee, T.; Greetham, G.M.; et al. Variation in LOV Photoreceptor Activation Dynamics Probed by Time-Resolved Infrared Spectroscopy. Biochemistry 2018, 57, 620–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrante, C.; Batignani, G.; Pontecorvo, E.; Montemiglio, L.C.; Vos, M.H.; Scopigno, T. Ultrafast Dynamics and Vibrational Relaxation in Six-Coordinate Heme Proteins Revealed by Femtosecond Stimulated Raman Spectroscopy. J. Am. Chem. Soc. 2020, 142, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Frontiera, R.R.; Tran, R.; Mathies, R.A. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature 2009, 462, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Kukura, P.; McCamant, D.W.; Yoon, S.; Wandschneider, D.B.; Mathies, R.A. Structural Observation of the Primary Isomerization in Vision with Femtosecond-Stimulated Raman. Science 2005, 310, 1006–1009. [Google Scholar] [CrossRef] [Green Version]
- Konold, P.E.; Mathes, T.; Weiβenborn, J.; Groot, M.L.; Hegemann, P.; Kennis, J.T.M. Unfolding of the C-Terminal Jα Helix in the LOV2 Photoreceptor Domain Observed by Time-Resolved Vibrational Spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3472–3476. [Google Scholar] [CrossRef] [Green Version]
- Nakasone, Y.; Eitoku, T.; Matsuoka, D.; Tokutomi, S.; Terazima, M. Dynamics of Conformational Changes of Arabidopsis Phototropin 1 LOV2 with the Linker Domain. J. Mol. Biol. 2007, 367, 432–442. [Google Scholar] [CrossRef]
- Hontani, Y.; Broser, M.; Luck, M.; Weißenborn, J.; Kloz, M.; Hegemann, P.; Kennis, J.T.M. Dual Photoisomerization on Distinct Potential Energy Surfaces in a UV-Absorbing Rhodopsin. J. Am. Chem. Soc. 2020, 142, 11464–11473. [Google Scholar] [CrossRef]
- Takaya, T.; Anan, M.; Iwata, K. Vibrational relaxation dynamics of β-carotene and its derivatives with substituents on terminal rings in electronically excited states as studied by femtosecond time-resolved stimulated Raman spectroscopy in the near-IR region. Phys. Chem. Chem. Phys. 2018, 20, 3320–3327. [Google Scholar] [CrossRef]
- Challa, J.R.; Du, Y.; McCamant, D.W. Femtosecond Stimulated Raman Spectroscopy Using a Scanning Multichannel Technique. Appl. Spectrosc. 2012, 66, 227–232. [Google Scholar] [CrossRef]
- Lipkin, M.; Nixdorf, J.; Gilch, P. Optimized amplitude modulation in femtosecond stimulated Raman microscopy. Opt. Lett. 2020, 45, 4204–4207. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Chen, C.; Zhu, R.; Jiang, J.; Weng, T.-C.; Ji, Q.; Huang, Y.; Fang, C.; Liu, W. Mapping the Complete Photocycle that Powers a Large Stokes Shift Red Fluorescent Protein. Angew. Chem. Int. Ed. 2022, e202212209. [Google Scholar] [CrossRef]
- Andrikopoulos, P.C.; Chaudhari, A.S.; Liu, Y.; Konold, P.E.; Kennis, J.T.M.; Schneider, B.; Fuertes, G. QM calculations predict the energetics and infrared spectra of transient glutamine isomers in LOV photoreceptors. Phys. Chem. Chem. Phys. 2021, 23, 13934–13950. [Google Scholar] [CrossRef]
- Green, D.; Roy, P.; Hall, C.R.; Iuliano, J.N.; Jones, G.A.; Lukacs, A.; Tonge, P.J.; Meech, S.R. Excited State Resonance Raman of Flavin Mononucleotide: Comparison of Theory and Experiment. J. Phys. Chem. A 2021, 125, 6171–6179. [Google Scholar] [CrossRef]
- Hall, C.R.; Heisler, I.A.; Jones, G.A.; Frost, J.E.; Gil, A.A.; Tonge, P.J.; Meech, S.R. Femtosecond stimulated Raman study of the photoactive flavoprotein AppABLUF. Chem. Phys. Lett. 2017, 683, 365–369. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, A.S.; Chatterjee, A.; Domingos, C.A.O.; Andrikopoulos, P.C.; Liu, Y.; Andersson, I.; Schneider, B.; Lórenz-Fonfría, V.A.; Fuertes, G. Genetically encoded non-canonical amino acids reveal asynchronous dark reversion of chromophore, backbone and side-chains in EL222. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kloz, M.; Weißenborn, J.; Polívka, T.; Frank, H.A.; Kennis, J.T.M. Spectral watermarking in femtosecond stimulated Raman spectroscopy: Resolving the nature of the carotenoid S* state. Phys. Chem. Chem. Phys. 2016, 18, 14619–14628. [Google Scholar] [CrossRef] [Green Version]
- Pimenta, F.M.; Jensen, R.L.; Breitenbach, T.; Etzerodt, M.; Ogilby, P.R. Oxygen-Dependent Photochemistry and Photophysics of “MiniSOG”, a Protein-Encased Flavin. Photochem. Photobiol. 2013, 89, 1116–1126. [Google Scholar] [CrossRef]
- McIntosh, J.R.; Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A Genetically Encoded Tag for Correlated Light and Electron Microscopy of Intact Cells, Tissues, and Organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef] [Green Version]
- Lórenz-Fonfría, V.A.; Kandori, H. Bayesian Maximum Entropy (Two-Dimensional) Lifetime Distribution Reconstruction from Time-Resolved Spectroscopic Data. Appl. Spectrosc. 2016, 61, 428–443. [Google Scholar] [CrossRef]
- Lórenz-Fonfría, V.A.; Kandori, H. Practical Aspects of the Maximum Entropy Inversion of the Laplace Transform for the Quantitative Analysis of Multi-Exponential Data. Appl. Spectrosc. 2016, 61, 74–84. [Google Scholar] [CrossRef]
- Lórenz-Fonfría, V.A.; Kandori, H. Transformation of Time-Resolved Spectra to Lifetime-Resolved Spectra by Maximum Entropy Inversion of the Laplace Transform. Appl. Spectrosc. 2016, 60, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Bozovic, O.; Zanobini, C.; Gulzar, A.; Jankovic, B.; Buhrke, D.; Post, M.; Wolf, S.; Stock, G.; Hamm, P. Real-time observation of ligand-induced allosteric transitions in a PDZ domain. Proc. Natl. Acad. Sci. USA 2020, 117, 26031–26039. [Google Scholar] [CrossRef] [PubMed]
- Stock, G.; Hamm, P. A non-equilibrium approach to allosteric communication. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Stokkum, I.H.M.; Larsen, D.S.; van Grondelle, R. Global and target analysis of time-resolved spectra. Biochim. Biophys. Acta (BBA) Bioenerg. 2004, 1657, 82–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snellenburg, J.J.; Laptenok, S.P.; Seger, R.; Mullen, K.M.; Stokkum, I.H.M.v. Glotaran: AJava-Based Graphical User Interface for theRPackageTIMP. J. Stat. Softw. 2012, 49, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Beckwith, J.S.; Rumble, C.A.; Vauthey, E. Data analysis in transient electronic spectroscopy—An experimentalist’s view. Int. Rev. Phys. Chem. 2020, 39, 135–216. [Google Scholar] [CrossRef]
- Slavov, C.; Hartmann, H.; Wachtveitl, J. Implementation and Evaluation of Data Analysis Strategies for Time-Resolved Optical Spectroscopy. Anal. Chem. 2015, 87, 2328–2336. [Google Scholar] [CrossRef]
- Sun, M.; Moore, T.A.; Song, P.-S. Molecular luminescence studies of flavines. I. Excited states of flavines. J. Am. Chem. Soc. 2002, 94, 1730–1740. [Google Scholar] [CrossRef]
- Van Stokkum, I.H.M.; Gauden, M.; Crosson, S.; van Grondelle, R.; Moffat, K.; Kennis, J.T.M. The Primary Photophysics of the Avena sativa Phototropin 1 LOV2 Domain Observed with Time-resolved Emission Spectroscopy†. Photochem. Photobiol. 2011, 87, 534–541. [Google Scholar] [CrossRef]
- Kutta, R.J.; Magerl, K.; Kensy, U.; Dick, B. A search for radical intermediates in the photocycle of LOV domains. Photochem. Photobiol. Sci. 2015, 14, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Yee, E.F.; Diensthuber, R.P.; Vaidya, A.T.; Borbat, P.P.; Engelhard, C.; Freed, J.H.; Bittl, R.; Möglich, A.; Crane, B.R. Signal transduction in light–oxygen–voltage receptors lacking the adduct-forming cysteine residue. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Dietler, J.; Gelfert, R.; Kaiser, J.; Borin, V.; Renzl, C.; Pilsl, S.; Ranzani, A.T.; García de Fuentes, A.; Gleichmann, T.; Diensthuber, R.P.; et al. Signal transduction in light-oxygen-voltage receptors lacking the active-site glutamine. Nat. Commun. 2022, 13, 10079. [Google Scholar] [CrossRef]
- Kopka, B.; Magerl, K.; Savitsky, A.; Davari, M.D.; Röllen, K.; Bocola, M.; Dick, B.; Schwaneberg, U.; Jaeger, K.-E.; Krauss, U. Electron transfer pathways in a light, oxygen, voltage (LOV) protein devoid of the photoactive cysteine. Sci. Rep. 2017, 7, 13346. [Google Scholar] [CrossRef] [Green Version]
- Atzrodt, J.; Derdau, V.; Kerr, W.J.; Reid, M. Deuterium- and Tritium-Labelled Compounds: Applications in the Life Sciences. Angew. Chem. Int. Ed. 2018, 57, 1758–1784. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, M.T.A.; Domratcheva, T.; Bonetti, C.; van Wilderen, L.J.G.W.; van Grondelle, R.; Groot, M.-L.; Hellingwerf, K.J.; Kennis, J.T.M. Primary Reactions of the LOV2 Domain of Phototropin Studied with Ultrafast Mid-Infrared Spectroscopy and Quantum Chemistry. Biophys. J. 2009, 97, 227–237. [Google Scholar] [CrossRef] [Green Version]
- Alexandre, M.T.A.; Purcell, E.B.; van Grondelle, R.; Robert, B.; Kennis, J.T.M.; Crosson, S. Electronic and Protein Structural Dynamics of a Photosensory Histidine Kinase. Biochemistry 2010, 49, 4752–4759. [Google Scholar] [CrossRef] [Green Version]
- Corchnoy, S.B.; Swartz, T.E.; Lewis, J.W.; Szundi, I.; Briggs, W.R.; Bogomolni, R.A. Intramolecular Proton Transfers and Structural Changes during the Photocycle of the LOV2 Domain of Phototropin 1. J. Biol. Chem. 2003, 278, 724–731. [Google Scholar] [CrossRef]
- Sakai, M.; Takahashi, H. One-electron photoreduction of flavin mononucleotide: Time-resolved resonance Raman and absorption study. J. Mol. Struct. 1996, 379, 9–18. [Google Scholar] [CrossRef]
- Li, G.; Glusac, K.D. Light-Triggered Proton and Electron Transfer in Flavin Cofactors. J. Phys. Chem. A 2008, 112, 4573–4583. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Byrdin, M.; Aubert, C.; Eker, A.P.M.; Brettel, K.; Vos, M.H. Excited-State Properties of Flavin Radicals in Flavoproteins: Femtosecond Spectroscopy of DNA Photolyase, Glucose Oxidase, and Flavodoxin. J. Phys. Chem. B 2004, 108, 10160–10167. [Google Scholar] [CrossRef]
- Glantz, S.T.; Carpenter, E.J.; Melkonian, M.; Gardner, K.H.; Boyden, E.S.; Wong, G.K.-S.; Chow, B.Y. Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. USA 2016, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, G.; Weeks, C.L.; Ibrahim, M.; Soldatova, A.V.; Spiro, T.G. Protein dynamics from time resolved UV Raman spectroscopy. Curr. Opin. Struct. Biol. 2008, 18, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Halavaty, A.S.; Moffat, K. N- and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena sativa. Biochemistry 2007, 46, 14001–14009. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]



| FMN | EL222-WT | EL222-C78A | Sample | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2O | D2O | H2O | D2O | H2O | D2O | Buffer | |||||||||||||
| RH | RL | TA | RH | RL | TA | RH | RL | TA | RH | RL | TA | RH | RL | TA | RH | RL | TA | Scale | Interpretation |
115 (α) | 1.7 (ε) 212 (ζ) | 1.3 (γ) 152 (δ) | ps | Solvent [22] Ribityl [22] | |||||||||||||||
2.9 | 3.3 | 3.8 | 3.2 | 1.9 (η) 3.1(θ) | 2.9 | 2.5 | 2.7 | 2.6 | 2.5 | 3.7 | 2.6 | 3.9 | 4.4 | 3.2 | 3.1 | 3.4 | 2.7 | ns | ? ① ISC ① |
| 0.32 | 0.40 | 0.32 | 1.7 | 1.5 | 3.0 | ?② | |||||||||||||
| 0.55 | 0.66 | 0.63 | 4.2 | 7.6 | 4.9 | µs | Adduct② | ||||||||||||
| 91 (β) | 87 | 208 | 109 | 269 | Phospho- rescence or FMNH•? ② | ||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chaudhari, A.S.; Chatterjee, A.; Andrikopoulos, P.C.; Picchiotti, A.; Rebarz, M.; Kloz, M.; Lorenz-Fonfria, V.A.; Schneider, B.; Fuertes, G. Sub-Millisecond Photoinduced Dynamics of Free and EL222-Bound FMN by Stimulated Raman and Visible Absorption Spectroscopies. Biomolecules 2023, 13, 161. https://doi.org/10.3390/biom13010161
Liu Y, Chaudhari AS, Chatterjee A, Andrikopoulos PC, Picchiotti A, Rebarz M, Kloz M, Lorenz-Fonfria VA, Schneider B, Fuertes G. Sub-Millisecond Photoinduced Dynamics of Free and EL222-Bound FMN by Stimulated Raman and Visible Absorption Spectroscopies. Biomolecules. 2023; 13(1):161. https://doi.org/10.3390/biom13010161
Chicago/Turabian StyleLiu, Yingliang, Aditya S. Chaudhari, Aditi Chatterjee, Prokopis C. Andrikopoulos, Alessandra Picchiotti, Mateusz Rebarz, Miroslav Kloz, Victor A. Lorenz-Fonfria, Bohdan Schneider, and Gustavo Fuertes. 2023. "Sub-Millisecond Photoinduced Dynamics of Free and EL222-Bound FMN by Stimulated Raman and Visible Absorption Spectroscopies" Biomolecules 13, no. 1: 161. https://doi.org/10.3390/biom13010161
APA StyleLiu, Y., Chaudhari, A. S., Chatterjee, A., Andrikopoulos, P. C., Picchiotti, A., Rebarz, M., Kloz, M., Lorenz-Fonfria, V. A., Schneider, B., & Fuertes, G. (2023). Sub-Millisecond Photoinduced Dynamics of Free and EL222-Bound FMN by Stimulated Raman and Visible Absorption Spectroscopies. Biomolecules, 13(1), 161. https://doi.org/10.3390/biom13010161







