Identification of Bottle Gourd (Lagenaria siceraria) OVATE Family Genes and Functional Characterization of LsOVATE1
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Phylogenetic Analysis of the OVATE Gene Family
2.3. Sequence Alignment, Chromosomal Location, and Three-Dimensional Protein Structure Prediction
2.4. RNA Extraction and RT-qPCR Analysis
2.5. Transcriptional Profiling
2.6. In Situ Hybridization
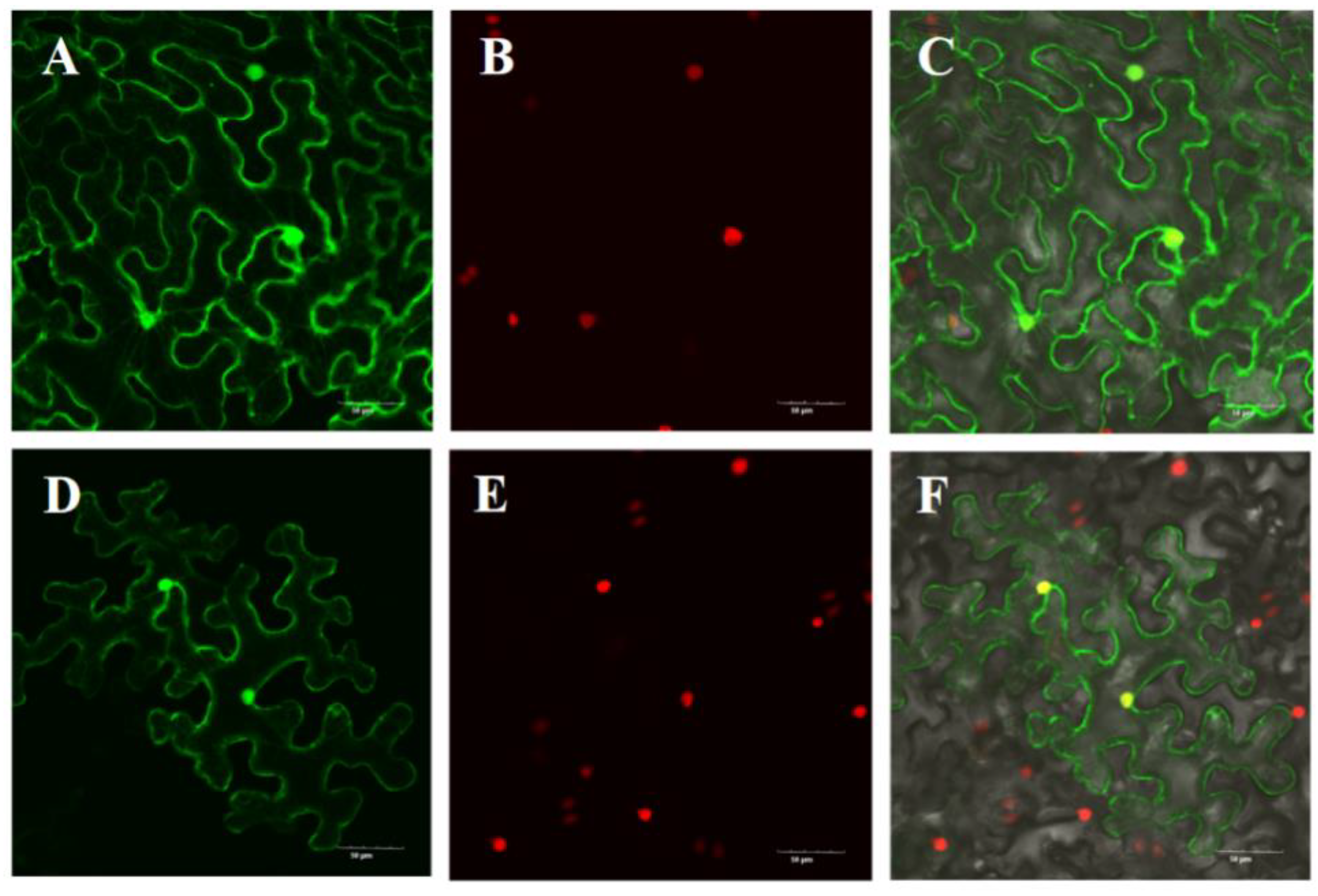
2.7. Subcellular Localization
2.8. Construction of Overexpression Plasmid and Phenotypic Evaluation of Transgenic Plants
3. Results
3.1. Identification of OVATE Family Genes in Bottle Gourd
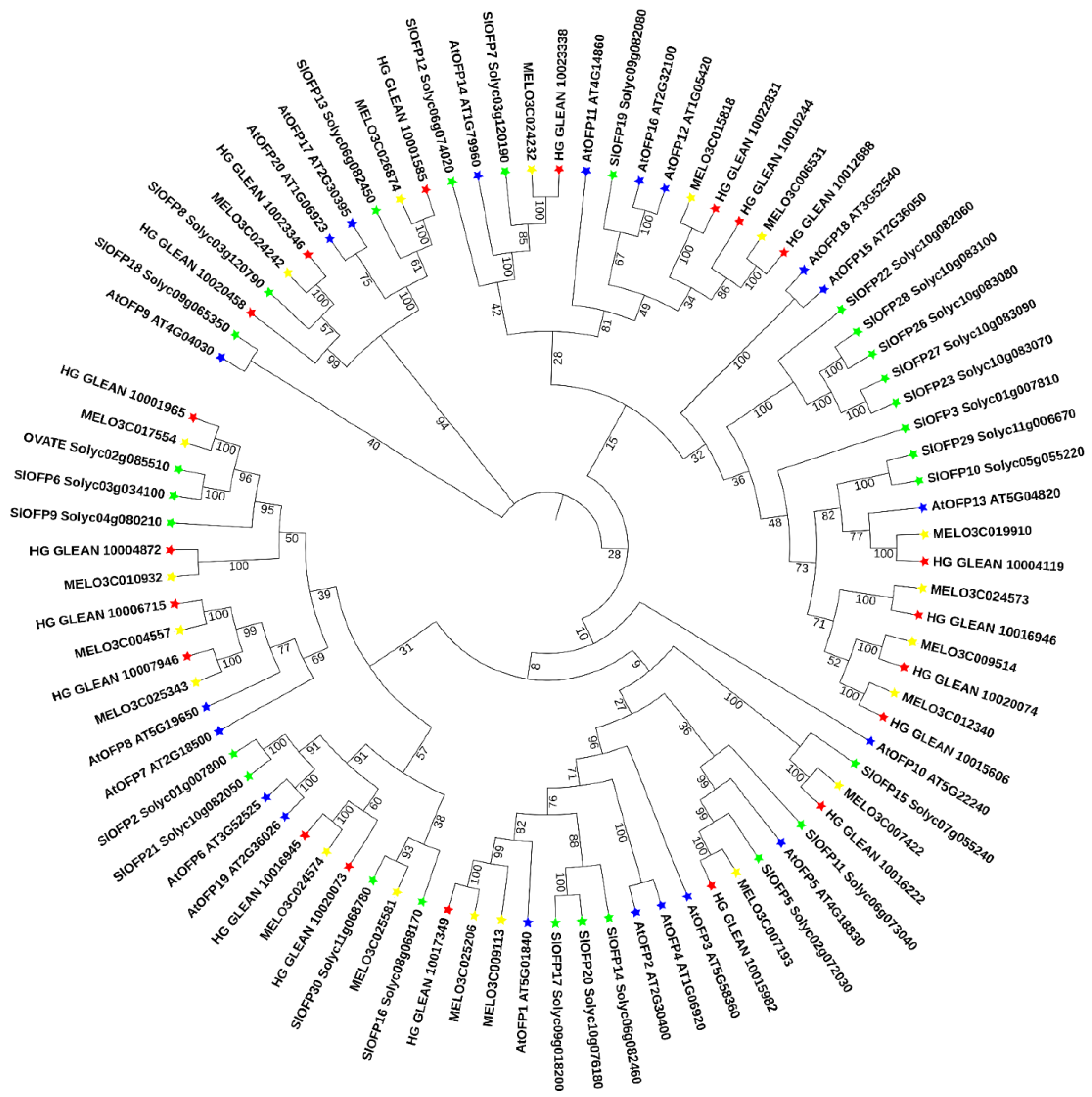
3.2. Phylogenetic Relationship of the OVATE Family Genes in Bottle Gourd
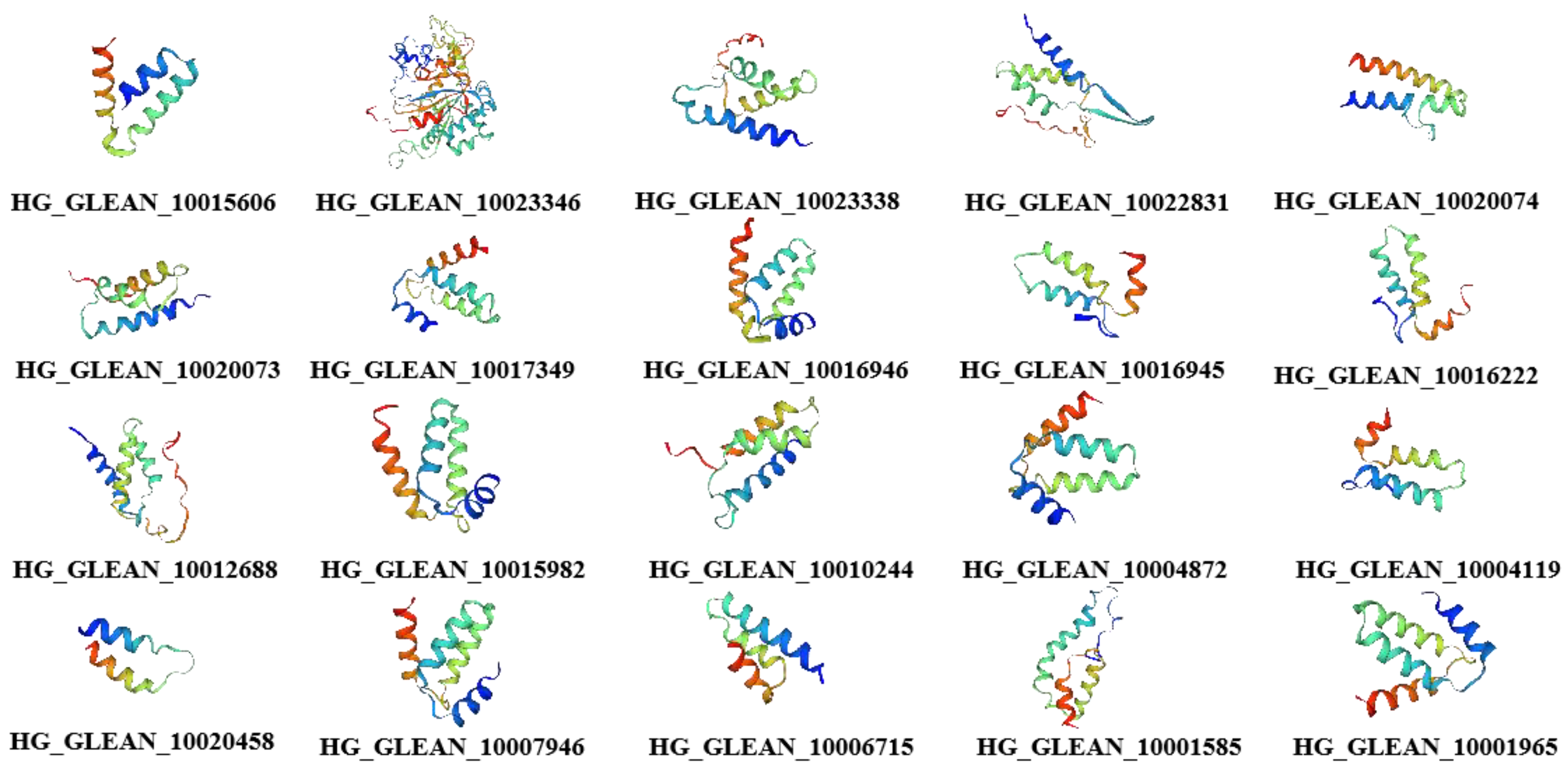
3.3. Structural Characterization of Bottle Gourd OVATE Family Genes
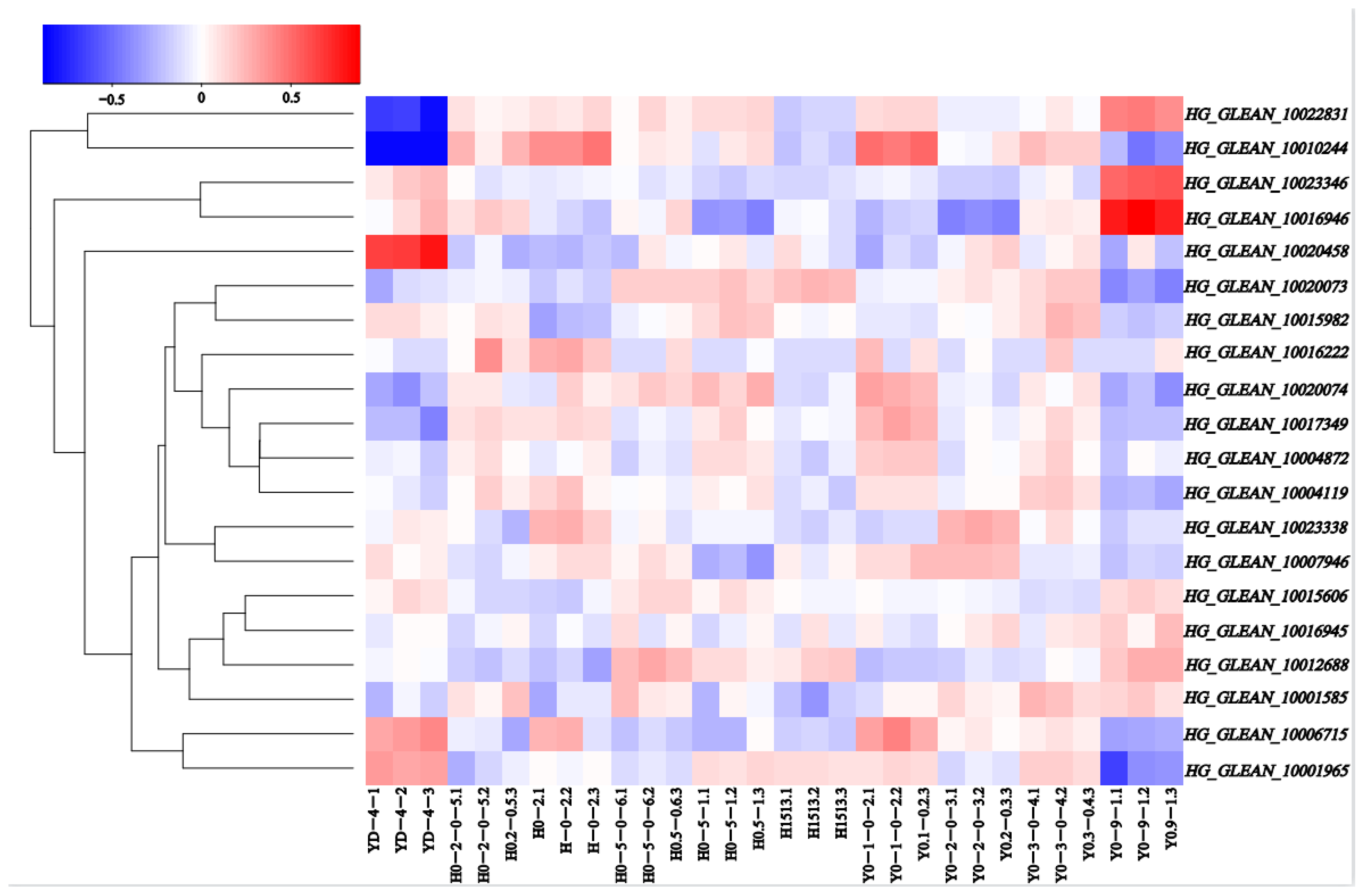
3.4. Expression Profiles of the OVATE Gene Family in the Bottle Gourd
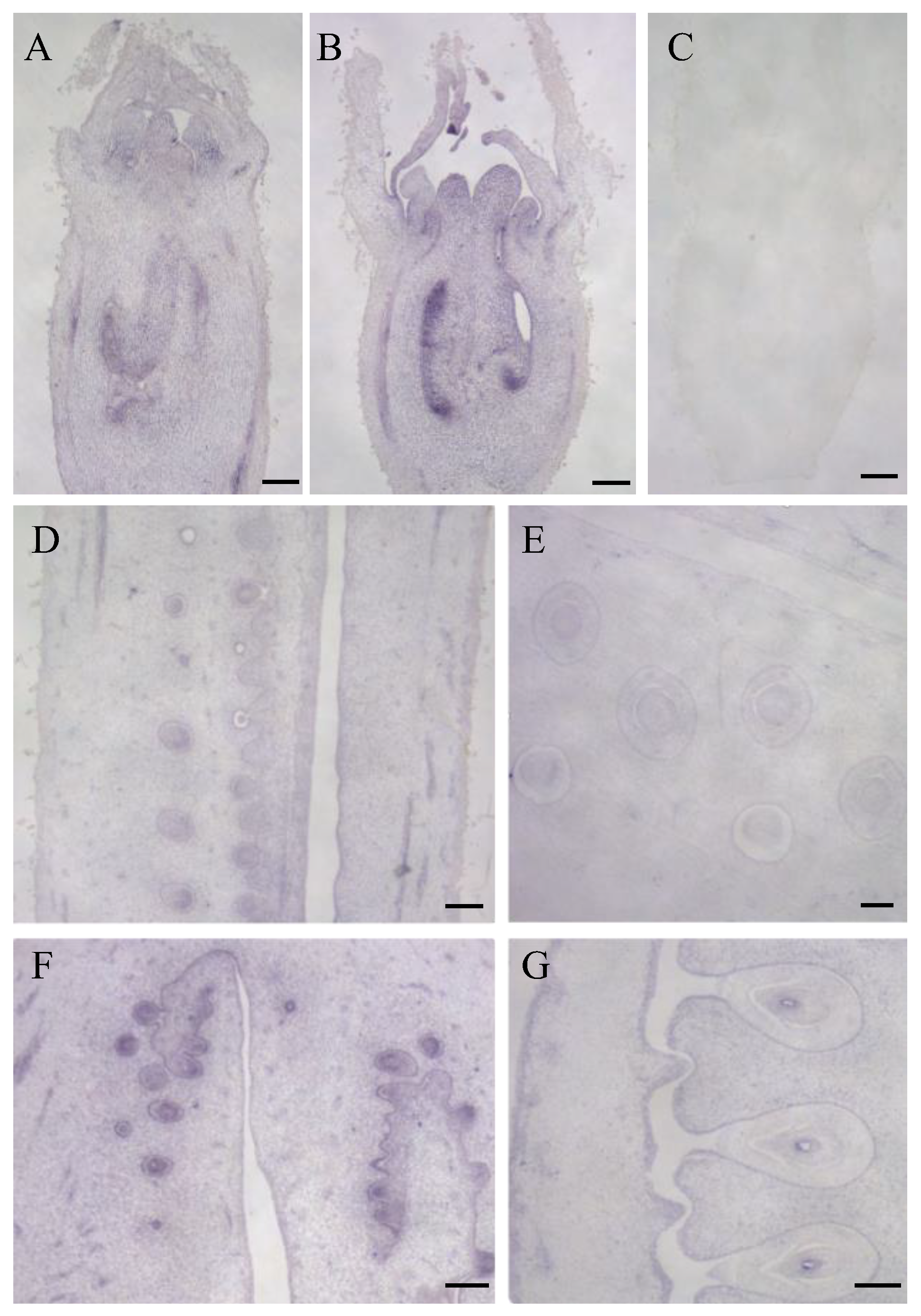
3.5. Expression Characteristics of LsOVATE1 Gene
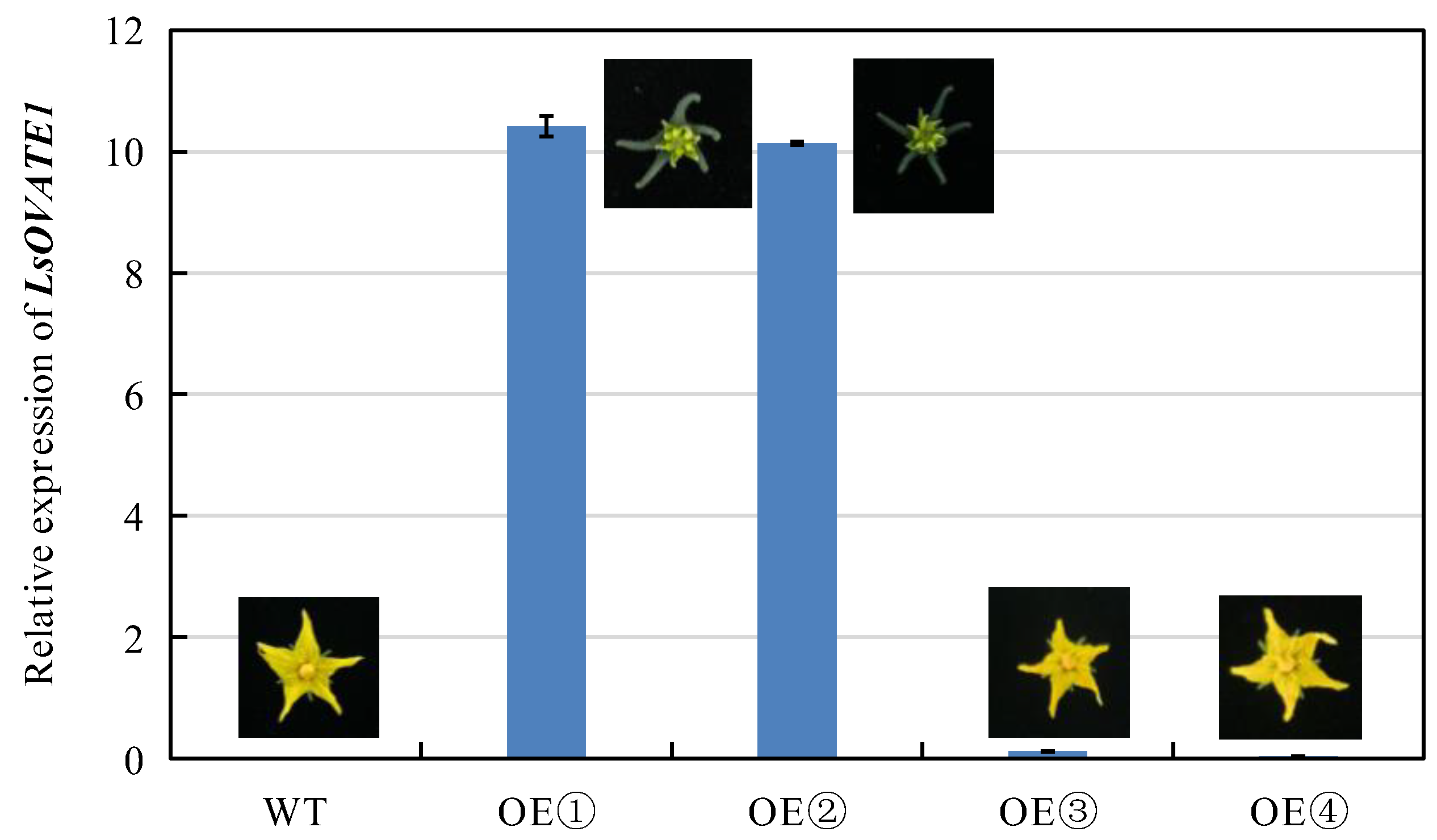
3.6. Functional Validation of LsOVATE1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beevy, S.S.; Kuriachan, P. Chromosome numbers of south Indian Cucurbitaceae and a note on the cytological evolution in the family. J. Cytol. Genet. 1996, 31, 65–71. [Google Scholar]
- Erickson, D.L.; Smith, B.D.; Clarke, A.C.; Sandweiss, D.H.; Tuross, N. An Asian origin for a 10,000-year-old domesticated plant in the Americas. Proc. Natl. Acad. Sci. USA 2005, 102, 18315–18320. [Google Scholar] [CrossRef] [PubMed]
- Heiser, C.B. The Gourd Book: A Thorough and Fascinating Account of Gourds from throughout the World; University of Oklahoma Press: Norman, OK, USA, 1979. [Google Scholar]
- Xu, P.; Xu, S.; Wu, X.; Tao, Y.; Wang, B.; Wang, S.; Qin, D.; Lu, Z.; Li, G. Population genomic analyses from low-coverage RAD-Seq data: A case study on the non-model cucurbit bottle gourd. Plant J. 2014, 77, 430–442. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Tan, G.; Zhou, W.; Wang, G. Gibberellin and the plant growth retardant paclobutrazol altered fruit shape and ripening in tomato. Protoplasma 2020, 257, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, C.; Zong, M.; Qiu, Y.; Liu, Y.; Huang, Y.; Xie, Y.; Zhang, H.; Wang, J. CmFSI8/CmOFP13 gene encoding an OFP family protein controls fruit shape in melon (Cucumis melo L.). J. Exp. Bot. 2021, 5, 1370–1384. [Google Scholar] [CrossRef]
- Pan, Y.; Liang, X.; Gao, M.; Liu, H.; Meng, H.; Weng, Y.; Cheng, Z. Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 2017, 130, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.G.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2020, 133, 1–21. [Google Scholar] [CrossRef]
- Ercisli, S.; Esitken, A.; Turkkal, C.; Orhan, E. The allelopathic effects of juglone and walnut leaf extracts on yield, growth, chemical and PNE composition of strawberry cv. Fern. Plant Soil Environ. 2005, 51, 283–387. [Google Scholar] [CrossRef]
- Bolaric, S.; Müller, I.D.; Vokurka, A.; Cepo, D.V.; Ruscic, M.; Srecec, S.; Kremer, D. Morphological and molecular characterization of Croatian carob tree (Ceratonia siliqua L.) germplasm. Turk. J. Agric. For. 2021, 45, 807–818. [Google Scholar] [CrossRef]
- Milosevic, T.; Milosevic, N.; Glisic, I. Early tree performances, precocity and fruit quality attributes of newly introduced apricot cultivars grown under western Serbian conditions. Turk. J. Agric. For. 2021, 45, 819–833. [Google Scholar] [CrossRef]
- Han, J.L. Functional Identification of Arabidopsis Transcription Factor AtOFP19. Master’s Thesis, Northeast Agricultural University, Harbin, China, 2021. [Google Scholar] [CrossRef]
- Liu, J.; Van Eck, J.; Cong, B.; Tanksley, S.D. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 2002, 99, 13302. [Google Scholar] [CrossRef]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; Knaap, E.V.D. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2018, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Cong, B.; Barrero, L.S.; Tanksley, S.D. Regulatory change in yabby-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 2008, 40, 800–804. [Google Scholar] [CrossRef] [PubMed]
- Munos, S.; Ranc, N.; Botton, E.; Berard, A.; Rolland, S.; Duffe, P.; Carretero, Y.; Le Paslier, M.; Delalande, C.; Bouzayen, M.; et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near wuschel. Plant Physiol. 2011, 156, 2244–2254. [Google Scholar] [CrossRef]
- Wang, L.; Cao, C.; Zheng, S.; Zhang, H.; Liu, P.; Ge, Q.; Li, J.; Ren, Z. Transcriptomic analysis of short-fruit 1 (SF1) reveals new insights into the variation of fruit-related traits in cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef]
- Li, S.; Pan, Y.; Wen, C.; Li, Y.; Liu, X.; Zhang, X.; Behera, T.; Xing, G.; Weng, Y. Integrated analysis in bi-parental and natural populations reveals Csclavata3 (CsCLV3) underlying carpel number variations in cucumber. Theor. Appl. Genet. 2016, 129, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Che, G.; Gu, R.; Zhao, J.; Liu, X.; Song, X.; Zi, H.; Cheng, Z.; Shen, J.; Wang, Z.; Liu, R.; et al. Gene regulatory network of carpel number variation in cucumber. Development 2020, 147, dev.184788. [Google Scholar] [CrossRef] [PubMed]
- Hackbusch, J.; Richter, K.; Müller, J.; Salamini, F.; Uhrig, J.F. A central role of Arabidopsis thaliana OVATE family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 4908–4912. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chang, Y.; Guo, J.; Chen, J. Arabidopsis Ovate Family Protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 2007, 50, 858–872. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Walia, H.; Begcy, K.; Sarath, G. Rice OVATE family protein 2 (OFP2) alters hormonal homeostasis and vasculature development. Plant Sci. 2015, 241, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Hui, Y.; Jiang, W.; Liu, Q.; Zhang, H.; Mingxin, P.; Chen, Z.; Bian, M. Expression pattern and subcellular localization of the OVATE protein family in rice. PLoS ONE 2017, 10, e0118966. [Google Scholar] [CrossRef]
- Tsaballa, A.; Pasentsis, K.; Darzentas, N.; Tsaftaris, A.S. Multiple evidence for the role of an OVATE-like gene in determining fruit shape in pepper. BMC Plant Biol. 2011, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, B.; Keyhaninejad, N.; Rodríguez, G.; Kim, H.J.; Chakrabarti, M.; Illa-Berenguer, E.; Taitano, N.K.; Gonzalo, M.J.; Díaz, A.; et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018, 9, 4734. [Google Scholar] [CrossRef]
- Guan, J.; Xu, Y.; Yu, Y.; Fu, J.; Ren, F.; Guo, J.; Zhao, J.; Jiang, Q.; Wei, J.; Xie, H. Genome structure variation analyses of peach reveal population dynamics and a 1.67 Mb causal inversion for fruit shape. Genome Biology. 2021, 22, 13. [Google Scholar] [CrossRef]
- Spinner, L.; Gadeyne, A.; Belcram, K.; Goussot, M.; Moison, M.; Duroc, Y.; Eeckhout, D.; De Winne, N.; Schaefer, E.; Van De Slijke, E.; et al. A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 2013, 4, 1863. [Google Scholar] [CrossRef]
- Schaefer, E.; Belcram, K.; Uyttewaal, M.; Duroc, Y.; Goussot, M.; Legland, D.; Laruelle, E.; de Tauzia-Moreau, M.-L.; Pastuglia, M.; Bouchez, D. The preprophase band of microtubules controls the robustness of division orientation in plants. Science 2017, 356, 186–189. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, G.; Kim, I.J.; Park, J.; Kwak, S.; Choi, G.; Chung, W. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef]
- Drevensek, S.; Goussot, M.; Duroc, Y.; Christodoulidou, A.; Steyaert, S.; Schaefer, E.; Duvernois, E.; Grandjean, O.; Vantard, M.; Bouchez, D.; et al. The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell 2012, 24, 178–191. [Google Scholar] [CrossRef]
- Lazzaro, M.D.; Wu, S.; Snouffer, A.; Wang, Y.; van der Knaap, E. Plant organ shapes are regulated by protein interactions and associations with microtubules. Front. Plant Sci. 2018, 9, 1766. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Ahmed, M.A. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Wang, G.; Lovato, A.; Polverari, A.; Wang, M.; Liang, Y.H.; Ma, Y.C.; Cheng, Z.M. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 2014, 14, 219. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆Ct) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- De Hoon, M.J.; Imoto, S.; Nolan, J.; Miyano, S. Open source clustering software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef]
- Saldanha, A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ding, L.; Zhang, X. RNA in situ hybridization technology for vegetable crops. J. Shanxi Agric. Univ. (Nat. Sci. Ed.) 2013, 33, 42–45. [Google Scholar] [CrossRef]
- Haseloff, J.; Siemering, K.R.; Prasher, D.C.; Hodge, S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 1997, 94, 2122–2127. [Google Scholar] [CrossRef]
- Sun, S.; Kang, X.; Xing, X.; Xu, X.; Cheng, J.; Zheng, S.; Xing, G. Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol. Biotechnol. Equip. 2015, 29, 861–868. [Google Scholar] [CrossRef]
- Wang, S.; Chang, Y.; Guo, J.; Zeng, Q.; Ellis, B.E.; Chen, J. Arabidopsis OVATE Family Proteins, a Novel Transcriptional Repressor Family, Control Multiple Aspects of Plant Growth and Development. PLoS ONE 2011, 6, e23896. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Fisher, E.M.C. Abstracts of papers presented at the sixteenth Genetics Society’s Mammalian Genetics and Development Workshop held at the Institute of Child Health, University College London on 21 and 22 November 2005. Genet. Res. 2006, 88, 67–76. [Google Scholar] [CrossRef][Green Version]
- Guo, Y.; Li, N.; Li, S.; Han, Y.; Duan, M.; Ma, F. Identification and Expression Analysis of OFP Gene Family in Setavia italica. J. Shan Xi Agric. Sci. 2020, 48, 467–476. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef]
- Song, X. Cloning and functional analysis of fruit shape-related gene in grape. Nanjing Agric. Univ. 2014, 7, 82. [Google Scholar]
- Han, J.; Wang, H.; Zhang, D.; Chang, Y. Preliminary study on the function of Arabidopsis transcription factor AtOFP19. North China J. Agric. 2021, 36, 56–63. [Google Scholar] [CrossRef]
- Zhang, S.; Yao, J.; Wang, L.; Wu, N.; van Nocker, S.; Li, Z.; Gao, M.; Wang, X. Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. Cell Mol. Biol. 2022, 111, 1565–1579. [Google Scholar] [CrossRef]
- Kim, K.; Hwang, J.; Han, D.; Park, M.; Kim, S.; Choi, D.; Kim, Y.; Lee, G.; Kim, S.; Park, Y. Major Quantitative Trait Loci and Putative Candidate Genes for Powdery Mildew Resistance and Fruit-Related Traits Revealed by an Intraspecific Genetic Map for Watermelon (Citrullus lanatus var. lanatus). PLoS ONE 2015, 10, e0145665. [Google Scholar] [CrossRef]
- Wei, Q.; Fu, W.X.; Wang, Y.Z.; Qin, X.D.; Wang, J.; Li, J.; Lou, Q.F.; Chen, J.F. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci. Rep. 2016, 6, 27496. [Google Scholar] [CrossRef]
- Tan, J.Y.; Tao, Q.Y.; Niu, H.H.; Zhang, Z.; Li, D.D.; Gong, Z.H.; Weng, Y.Q.; Li, Z. A novel allele of monoecious (m) locus is responsible for elongated fruit shape and perfect flowers in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2015, 128, 2483–2493. [Google Scholar] [CrossRef]
- Zhao, J.; Jiang, L.; Che, G.; Pan, Y.; Li, Y.; Hou, Y.; Zhao, W.; Zhong, Y.; Ding, L.; Yan, S.; et al. A Functional Allele of CsFUL1 Regulates Fruit Length through Repressing CsSUP and Inhibiting Auxin Transport in Cucumber. Plant Cell 2019, 31, 1289–1307. [Google Scholar] [CrossRef] [PubMed]
- Karlova, R.; Rosin, F.M.; Busscher-Lange, J.; Parapunova, V.; Do, P.T.; Fernie, A.R.; Fraser, P.D.; Baxter, C.; Angenent, G.C.; de Maagd, R.A. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011, 23, 923–941. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Zhou, Z.; Wang, Q.; Guo, J.; Zhao, P.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, X.; et al. Genome-wide association study of 12 agronomic traits in peach. Nat. Commun. 2016, 7, 13246. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, Y.; Sun, F.; Wu, R.; Du, H.; Wang, Y.; Jiang, L.; Wu, X.; Wu, X.; Yang, L.; et al. Long-read genome assembly and genetic architecture of fruit shape in the bottle gourd. Plant J. Cell Mol. Biol. 2021, 107, 956–968. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Chromosome | Start | End | pI | Molecular Weight (kDa) |
|---|---|---|---|---|---|
| HG_GLEAN_10012688 | 1 | 23,417,053 | 23,417,667 | 6.34 | 23.1 |
| HG_GLEAN_10015606 | 2 | 28,062,254 | 28,063,057 | 4.88 | 30.0 |
| HG_GLEAN_10015982 | 3 | 1,914,641 | 1,924,905 | 9.51 | 51.7 |
| HG_GLEAN_10016222 | 3 | 3,622,948 | 3,623,433 | 6.82 | 18.9 |
| HG_GLEAN_10016945 | 3 | 9,539,752 | 9,540,195 | 7.90 | 16.8 |
| HG_GLEAN_10016946 | 3 | 9,555,872 | 9,556,693 | 4.55 | 30.6 |
| HG_GLEAN_10017349 | 3 | 13,489,371 | 13,490,354 | 9.80 | 36.7 |
| HG_GLEAN_10020458 | 4 | 32,000,706 | 32,001,644 | 9.33 | 22.4 |
| HG_GLEAN_10020073 | 4 | 28,549,054 | 28,549,566 | 9.67 | 18.9 |
| HG_GLEAN_10020074 | 4 | 28,561,476 | 28,562,183 | 5.15 | 25.6 |
| HG_GLEAN_10022831 | 5 | 28,760,144 | 28,760,947 | 5.48 | 29.2 |
| HG_GLEAN_10023338 | 5 | 33,196,658 | 33,197,443 | 8.20 | 29.6 |
| HG_GLEAN_10023346 | 5 | 33,258,189 | 33,258,818 | 8.70 | 79.0 |
| HG_GLEAN_10010244 | 6 | 20,205,559 | 20,206,137 | 8.88 | 21.6 |
| HG_GLEAN_10006715 | 7 | 21,324,837 | 21,325,556 | 9.96 | 27.3 |
| HG_GLEAN_10004119 | 8 | 13,885,568 | 13,886,230 | 4.66 | 25.4 |
| HG_GLEAN_10004872 | 8 | 21,090,898 | 21,091,833 | 9.72 | 30.1 |
| HG_GLEAN_10001585 | 9 | 18,363,582 | 18,364,110 | 9.45 | 22.9 |
| HG_GLEAN_10007946 | 10 | 17,697,582 | 17,698,373 | 9.78 | 30.0 |
| HG_GLEAN_10001965 | 11 | 2,189,256 | 2,190,179 | 8.11 | 35.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, Z.; Wu, X.; Wang, J.; Wu, X.; Wang, B.; Lu, Z.; Ye, Z.; Li, G.; Wang, Y. Identification of Bottle Gourd (Lagenaria siceraria) OVATE Family Genes and Functional Characterization of LsOVATE1. Biomolecules 2023, 13, 85. https://doi.org/10.3390/biom13010085
Feng Z, Wu X, Wang J, Wu X, Wang B, Lu Z, Ye Z, Li G, Wang Y. Identification of Bottle Gourd (Lagenaria siceraria) OVATE Family Genes and Functional Characterization of LsOVATE1. Biomolecules. 2023; 13(1):85. https://doi.org/10.3390/biom13010085
Chicago/Turabian StyleFeng, Zishan, Xiaohua Wu, Jian Wang, Xinyi Wu, Baogen Wang, Zhongfu Lu, Zihong Ye, Guojing Li, and Ying Wang. 2023. "Identification of Bottle Gourd (Lagenaria siceraria) OVATE Family Genes and Functional Characterization of LsOVATE1" Biomolecules 13, no. 1: 85. https://doi.org/10.3390/biom13010085
APA StyleFeng, Z., Wu, X., Wang, J., Wu, X., Wang, B., Lu, Z., Ye, Z., Li, G., & Wang, Y. (2023). Identification of Bottle Gourd (Lagenaria siceraria) OVATE Family Genes and Functional Characterization of LsOVATE1. Biomolecules, 13(1), 85. https://doi.org/10.3390/biom13010085





