Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Circular Dichroism Studies
2.3. Scanning Calorimetry Measurements
2.4. Fluorescence Studies
2.5. Dynamic Light Scattering Studies
2.6. Chemical Crosslinking of Proteins
2.7. Modeling the Full Structure of Human VEGF165 and Its Complex with HE
3. Results and Discussion
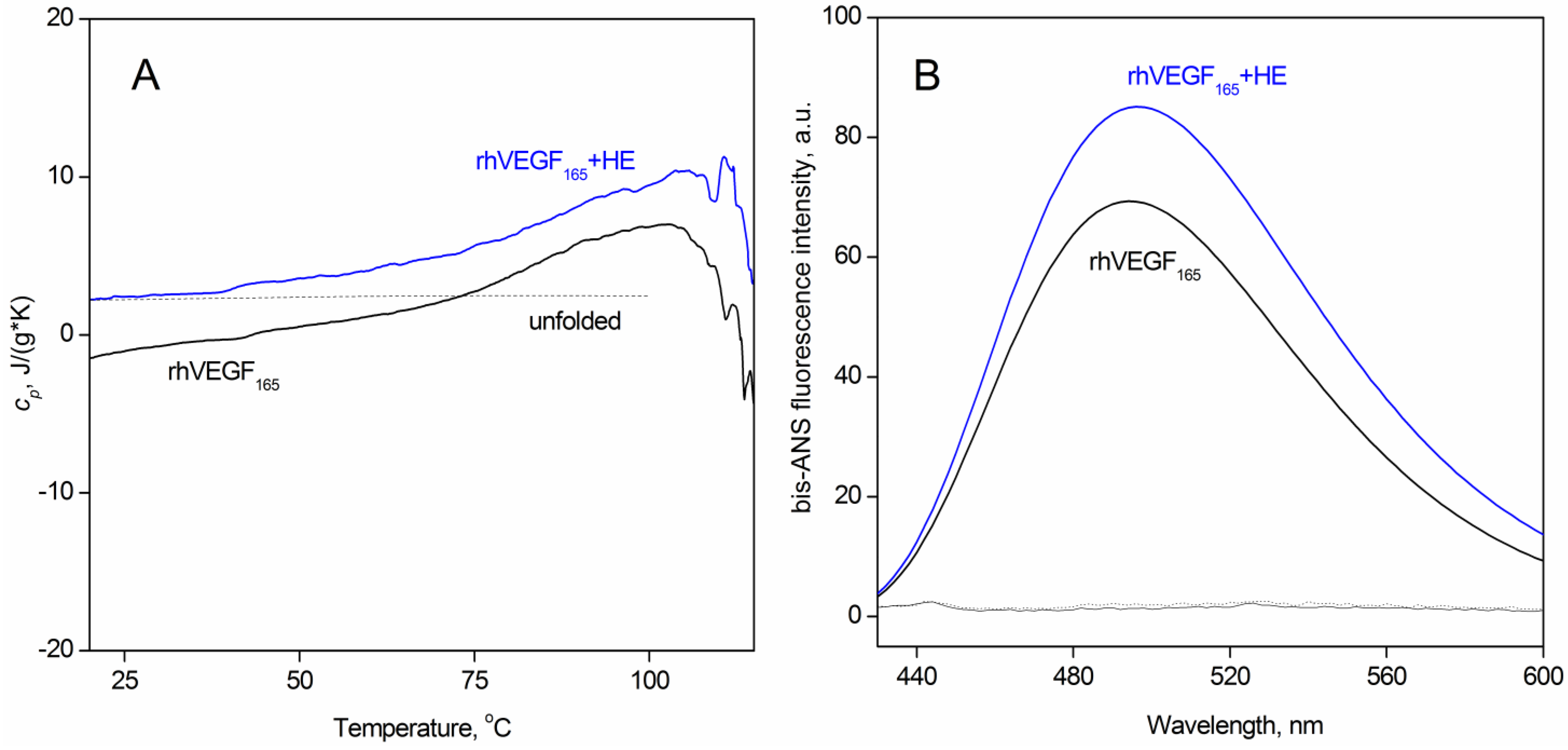
3.1. The Effect of Heparin on the Secondary and Tertiary Structure of VEGF165
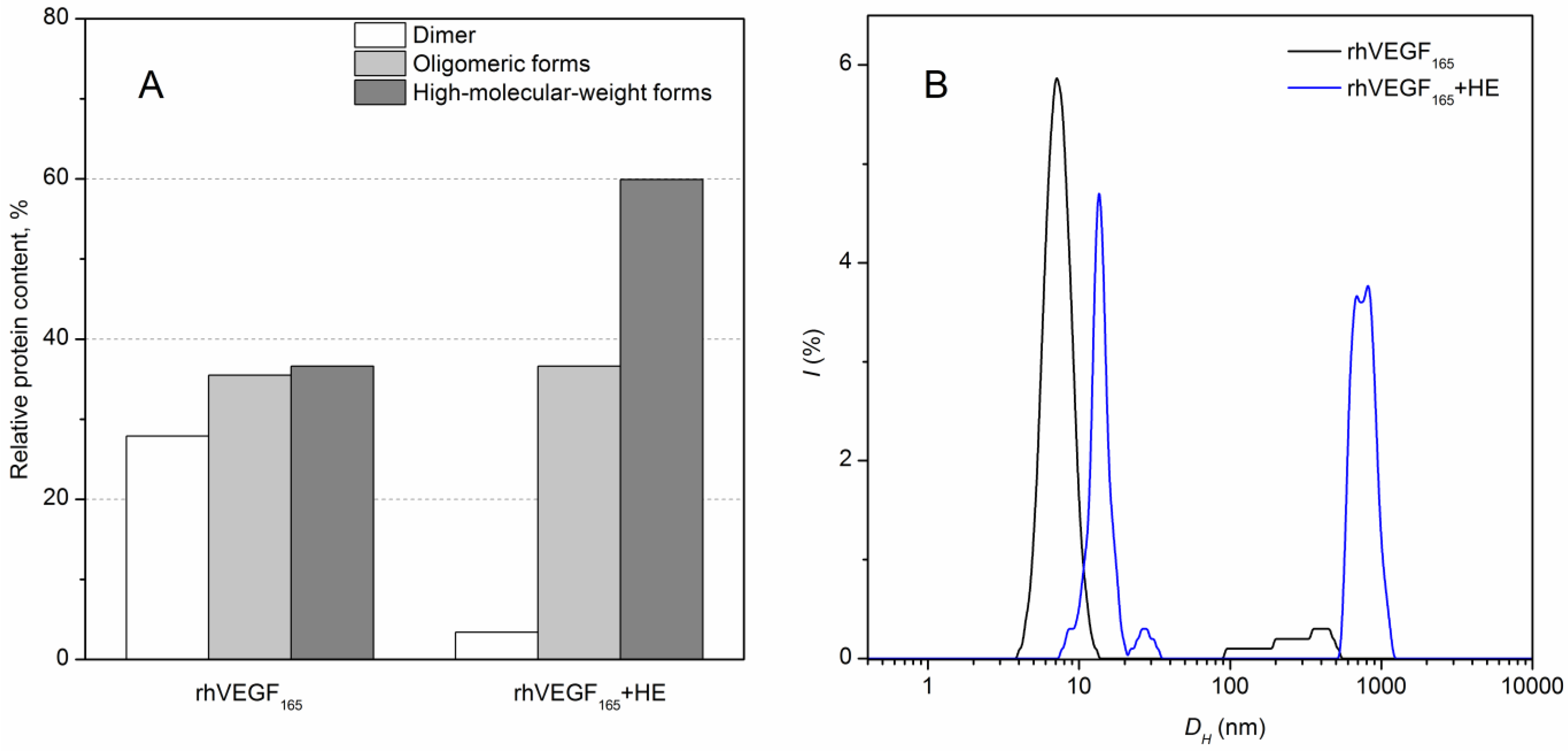
3.2. HE-Dependent Changes in the Quaternary Structure of rhVEGF165
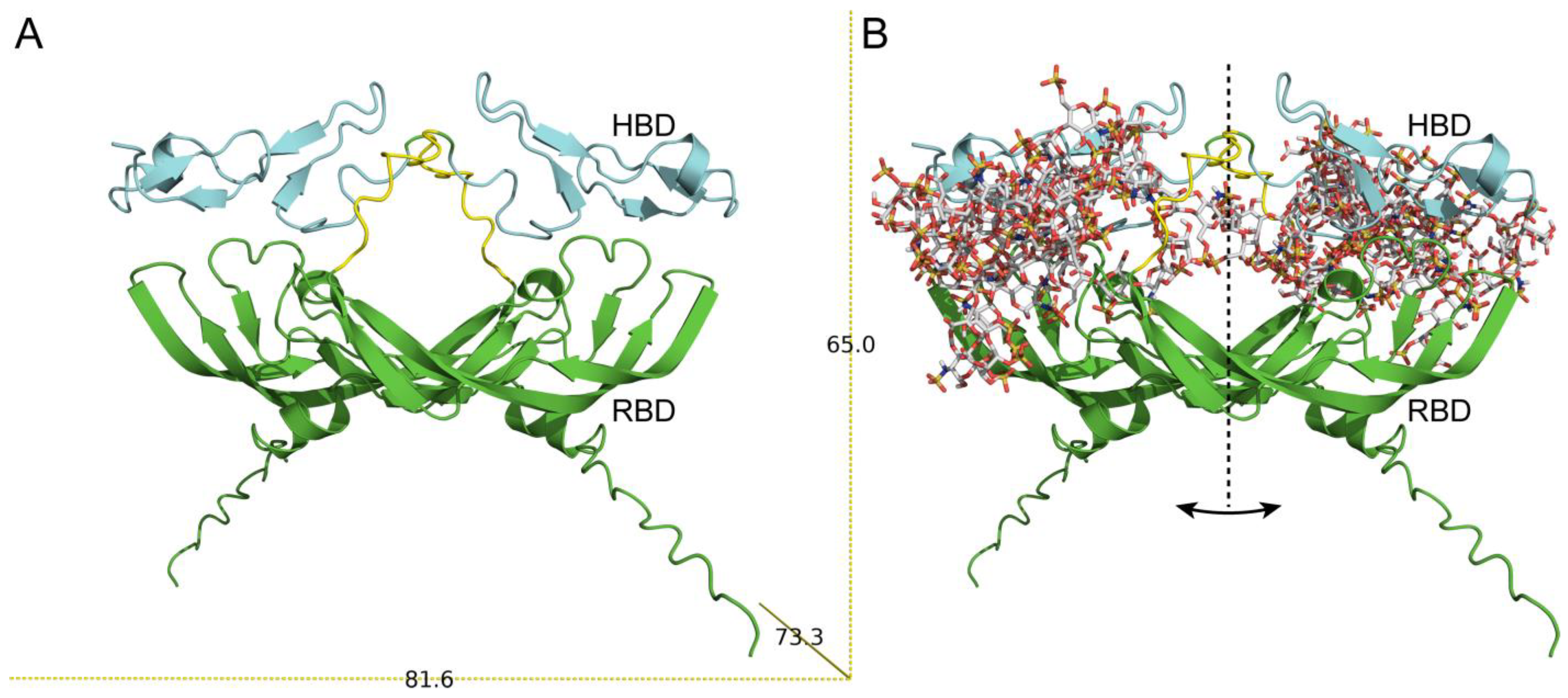
3.3. Model of VEGF165–HE Complex
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamis, A.P.; Shima, D.T. The role of vascular endothelial growth factor in ocular health and disease. Retina 2005, 25, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Carmeliet, P.; Jain, R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature 2011, 473, 298–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kut, C.; Mac Gabhann, F.; Popel, A.S. Where is VEGF in the body? A meta-analysis of VEGF distribution in cancer. Br. J. Cancer 2007, 97, 978–985. [Google Scholar] [CrossRef] [Green Version]
- Holmes, D.I.R.; Zachary, I. The vascular endothelial growth factor (VEGF) family: Angiogenic factors in health and disease. Genome Biol. 2005, 6, 209. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, N.; Gerber, H.-P.; LeCouter, J. The biology of VEGF and its receptors. Nat. Med. 2003, 9, 669–676. [Google Scholar] [CrossRef]
- Ferrara, N. VEGF-A: A critical regulator of blood vessel growth. Eur. Cytokine Netw. 2009, 20, 158–163. [Google Scholar] [CrossRef] [Green Version]
- Nagy, J.A.; Dvorak, A.M.; Dvorak, H.F. VEGF-A and the Induction of Pathological Angiogenesis. Annu. Rev. Pathol. Mech. Dis. 2007, 2, 251–275. [Google Scholar] [CrossRef]
- Olsson, A.-K.; Dimberg, A.; Kreuger, J.; Claesson-Welsh, L. VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 2006, 7, 359–371. [Google Scholar] [CrossRef]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef]
- Krilleke, D.; Ng, Y.-S.E.; Shima, D.T. The heparin-binding domain confers diverse functions of VEGF-A in development and disease: A structure-function study. Biochem. Soc. Trans. 2009, 37, 1201–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gitay-Goren, H.; Soker, S.; Vlodavsky, I.; Neufeld, G. The binding of vascular endothelial growth factor to its receptors is dependent on cell surface-associated heparin-like molecules. J. Biol. Chem. 1992, 267, 6093–6098. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, N.S.; Mancera, R.L. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. [Google Scholar] [CrossRef] [PubMed]
- Morla, S. Glycosaminoglycans and Glycosaminoglycan Mimetics in Cancer and Inflammation. Int. J. Mol. Sci. 2019, 20, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelberg, H. Plasma heparin levels in normal man. Circulation 1961, 23, 578–581. [Google Scholar] [CrossRef] [Green Version]
- Delcombel, R.; Janssen, L.; Vassy, R.; Gammons, M.; Haddad, O.; Richard, B.; Letourneur, D.; Bates, D.; Hendricks, C.; Waltenberger, J.; et al. New prospects in the roles of the C-terminal domains of VEGF-A and their cooperation for ligand binding, cellular signaling and vessels formation. Angiogenesis 2013, 16, 353–371. [Google Scholar] [CrossRef]
- Muller, Y.A.; Christinger, H.W.; Keyt, B.A.; de Vos, A.M. The crystal structure of vascular endothelial growth factor (VEGF) refined to 1.93 A resolution: Multiple copy flexibility and receptor binding. Structure 1997, 5, 1325–1338. [Google Scholar] [CrossRef] [Green Version]
- Muller, Y.A.; Li, B.; Christinger, H.W.; Wells, J.A.; Cunningham, B.C.; de Vos, A.M. Vascular endothelial growth factor: Crystal structure and functional mapping of the kinase domain receptor binding site. Proc. Natl. Acad. Sci. USA 1997, 94, 7192–7197. [Google Scholar] [CrossRef] [Green Version]
- Houck, K.A.; Leung, D.W.; Rowland, A.M.; Winer, J.; Ferrara, N. Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 1992, 267, 26031–26037. [Google Scholar] [CrossRef]
- Fairbrother, W.J.; Champe, M.A.; Christinger, H.W.; Keyt, B.A.; Starovasnik, M.A. Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure 1998, 6, 637–648. [Google Scholar] [CrossRef]
- Keyt, B.A.; Berleau, L.T.; Nguyen, H.V.; Chen, H.; Heinsohn, H.; Vandlen, R.; Ferrara, N. The carboxyl-terminal domain (111-165) of vascular endothelial growth factor is critical for its mitogenic potency. J. Biol. Chem. 1996, 271, 7788–7795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stauffer, M.E.; Skelton, N.J.; Fairbrothe, W.J. Refinement of the solution structure of the heparin-binding domain of vascular endothelial growth factor using residual dipolar couplings. J. Biomol. NMR 2002, 23, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Uciechowska-Kaczmarzyk, U.; Babik, S.; Zsila, F.; Bojarski, K.K.; Beke-Somfai, T.; Samsonov, S.A. Molecular dynamics-based model of VEGF-A and its heparin interactions. J. Mol. Graph. Model. 2018, 82, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Guryanov, I.; Tennikova, T.; Urtti, A. Peptide Inhibitors of Vascular Endothelial Growth Factor A: Current Situation and Perspectives. Pharmaceutics 2021, 13, 1337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; McCallum, S.A.; Xiao, Z.; Zhang, F.; Linhardt, R.J. Binding affinities of vascular endothelial growth factor (VEGF) for heparin-derived oligosaccharides. Biosci. Rep. 2012, 32, 71–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijelath, E.; Namekata, M.; Murray, J.; Furuyashiki, M.; Zhang, S.; Coan, D.; Wakao, M.; Harris, R.B.; Suda, Y.; Wang, L.; et al. Multiple mechanisms for exogenous heparin modulation of vascular endothelial growth factor activity. J. Cell. Biochem. 2010, 111, 461–468. [Google Scholar] [CrossRef] [Green Version]
- Ito, N.; Claesson-Welsh, L. Dual effects of heparin on VEGF binding to VEGF receptor-1 and transduction of biological responses. Angiogenesis 1999, 3, 159–166. [Google Scholar] [CrossRef]
- Dougher, A.M.; Wasserstrom, H.; Torley, L.; Shridaran, L.; Westdock, P.; Hileman, R.E.; Fromm, J.R.; Anderberg, R.; Lyman, S.; Linhardt, R.J.; et al. Identification of a heparin binding peptide on the extracellular domain of the KDR VEGF receptor. Growth Factors 1997, 14, 257–268. [Google Scholar] [CrossRef]
- Ferrara, N.; Hillan, K.J.; Novotny, W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem. Biophys. Res. Commun. 2005, 333, 328–335. [Google Scholar] [CrossRef]
- Tsiros, D.; Sheehy, C.E.; Pecchia, S.; Nugent, M.A. Heparin potentiates Avastin-mediated inhibition of VEGF binding to fibronectin and rescues Avastin activity at acidic pH. J. Biol. Chem. 2019, 294, 17603–17611. [Google Scholar] [CrossRef]
- Ko, S.Y.; Lee, W.; Kenny, H.A.; Dang, L.H.; Ellis, L.M.; Jonasch, E.; Lengyel, E.; Naora, H. Cancer-derived small extracellular vesicles promote angiogenesis by heparin-bound, bevacizumab-insensitive VEGF, independent of vesicle uptake. Commun. Biol. 2019, 2, 386. [Google Scholar] [CrossRef] [Green Version]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [Green Version]
- Farris, F.J.; Weber, G.; Chiang, C.C.; Paul, I.C. Preparation, crystalline structure, and spectral properties of the fluorescent probe 4,4’-bis-1-phenylamino-8-naphthalenesulfonate. J. Am. Chem. Soc. 1978, 100, 4469–4474. [Google Scholar] [CrossRef]
- Fu, L.; Li, G.; Yang, B.; Onishi, A.; Li, L.; Sun, P.; Zhang, F.; Linhardt, R.J. Structural characterization of pharmaceutical heparins prepared from different animal tissues. J. Pharm. Sci. 2013, 102, 1447–1457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sreerama, N.; Venyaminov, S.Y.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 2000, 287, 243–251. [Google Scholar] [CrossRef] [Green Version]
- Privalov, P.L.; Potekhin, S.A. Scanning microcalorimetry in studying temperature-induced changes in proteins. Methods Enzymol. 1986, 131, 4–51. [Google Scholar] [CrossRef] [PubMed]
- Häckel, M.; Hinz, H.J.; Hedwig, G.R. Partial molar volumes of proteins: Amino acid side-chain contributions derived from the partial molar volumes of some tripeptides over the temperature range 10-90 degrees C. Biophys. Chem. 1999, 82, 35–50. [Google Scholar] [CrossRef]
- Häckel, M.; Hinz, H.J.; Hedwig, G.R. A new set of peptide-based group heat capacities for use in protein stability calculations. J. Mol. Biol. 1999, 291, 197–213. [Google Scholar] [CrossRef]
- Privalov, P.L.; Makhatadze, G.I. Heat capacity of proteins. II. Partial molar heat capacity of the unfolded polypeptide chain of proteins: Protein unfolding effects. J. Mol. Biol. 1990, 213, 385–391. [Google Scholar] [CrossRef]
- Makhatadze, G.I.; Privalov, P.L. Heat capacity of proteins. I. Partial molar heat capacity of individual amino acid residues in aqueous solution: Hydration effect. J. Mol. Biol. 1990, 213, 375–384. [Google Scholar] [CrossRef]
- Burstein, E.A.; Permyakov, E.A.; Emelyanenko, V.I.; Bushueva, T.L.; Pechere, J.F. Investigation of some physico-chemical properties of muscular parvalbumins by means of the luminescence of their phenylalanyl residues. Biochim. Biophys. Acta 1975, 400, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002, 11, 739–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemashkalova, E.L.; Permyakov, E.A.; Permyakov, S.E.; Litus, E.A. Modulation of linoleic acid-binding properties of human serum albumin by divalent metal cations. Biometals 2017, 30, 341–353. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Mottarella, S.E.; Beglov, D.; Beglova, N.; Nugent, M.A.; Kozakov, D.; Vajda, S. Docking server for the identification of heparin binding sites on proteins. J. Chem. Inf. Model. 2014, 54, 2068–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Claffey, K.P.; Senger, D.R.; Spiegelman, B.M. Structural requirements for dimerization, glycosylation, secretion, and biological function of VPF/VEGF. Biochim. Biophys. Acta 1995, 1246, 1–9. [Google Scholar] [CrossRef]
- Walter, D.H.; Hink, U.; Asahara, T.; Van Belle, E.; Horowitz, J.; Tsurumi, Y.; Vandlen, R.; Heinsohn, H.; Keyt, B.; Ferrara, N.; et al. The in vivo bioactivity of vascular endothelial growth factor/vascular permeability factor is independent of N-linked glycosylation. Lab. Investig. 1996, 74, 546–556. Available online: http://www.ncbi.nlm.nih.gov/pubmed/8780172 (accessed on 1 November 2022). [PubMed]
- Hawe, A.; Sutter, M.; Jiskoot, W. Extrinsic fluorescent dyes as tools for protein characterization. Pharm. Res. 2008, 25, 1487–1499. [Google Scholar] [CrossRef] [Green Version]
- Ahanger, I.A.; Parray, Z.A.; Nasreen, K.; Ahmad, F.; Hassan, M.I.; Islam, A.; Sharma, A. Heparin Accelerates the Protein Aggregation via the Downhill Polymerization Mechanism: Multi-Spectroscopic Studies to Delineate the Implications on Proteinopathies. ACS Omega 2021, 6, 2328–2339. [Google Scholar] [CrossRef]
- Maïza, A.; Chantepie, S.; Vera, C.; Fifre, A.; Huynh, M.B.; Stettler, O.; Ouidja, M.O.; Papy-Garcia, D. The role of heparan sulfates in protein aggregation and their potential impact on neurodegeneration. FEBS Lett. 2018, 592, 3806–3818. [Google Scholar] [CrossRef]
- Mitternacht, S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Research 2016, 5, 189. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A.; Teichmann, S.A. Relative Solvent Accessible Surface Area Predicts Protein Conformational Changes upon Binding. Structure 2011, 19, 859–867. [Google Scholar] [CrossRef] [Green Version]
- Rambo, R.P.; Tainer, J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 2011, 95, 559–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, J.A. Buried and Accessible Surface Area Control Intrinsic Protein Flexibility. J. Mol. Biol. 2013, 425, 3250–3263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freire, E. Thermal denaturation methods in the study of protein folding. Methods Enzymol. 1995, 259, 144–168. [Google Scholar] [CrossRef] [PubMed]
| Protein | α-Helices, % | β-Sheets, % | Turns, % | Unordered Structure, % |
|---|---|---|---|---|
| rhVEGF165-CHO | 8.8 ± 3.5 | 30.2 ± 4.5 | 22.7 ± 2.8 | 38.4 ± 4.8 |
| rhVEGF165-CHO + HE | 8.1 ± 2.4 | 31.9 ± 2.9 | 22.3 ± 2.1 | 37.7 ± 3.2 |
| rhVEGF165-E. coli | 8.7 ± 2.7 | 29.4 ± 2.8 | 22.3 ± 2.7 | 39.6 ± 5.3 |
| rhVEGF165-E. coli + HE | 7.0 ± 1.7 | 30.0 ± 2.2 | 22.7 ± 2.2 | 40.2 ± 4.6 |
| Total Area, Å2 | BSA, Å2 | SAS, Å2 | SASap, Å2 | SASpol, Å2 | |
|---|---|---|---|---|---|
| VEGF165 | 39,586 | 17,422 | 22,166 | 5713 | 16,453 |
| VEGF165 + HE | 41,094 | 20,154 | 20,940 | 4856 | 11,254 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemashkalova, E.L.; Shevelyova, M.P.; Machulin, A.V.; Lykoshin, D.D.; Esipov, R.S.; Deryusheva, E.I. Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure. Biomolecules 2023, 13, 98. https://doi.org/10.3390/biom13010098
Nemashkalova EL, Shevelyova MP, Machulin AV, Lykoshin DD, Esipov RS, Deryusheva EI. Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure. Biomolecules. 2023; 13(1):98. https://doi.org/10.3390/biom13010098
Chicago/Turabian StyleNemashkalova, Ekaterina L., Marina P. Shevelyova, Andrey V. Machulin, Dmitry D. Lykoshin, Roman S. Esipov, and Evgenia I. Deryusheva. 2023. "Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure" Biomolecules 13, no. 1: 98. https://doi.org/10.3390/biom13010098
APA StyleNemashkalova, E. L., Shevelyova, M. P., Machulin, A. V., Lykoshin, D. D., Esipov, R. S., & Deryusheva, E. I. (2023). Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure. Biomolecules, 13(1), 98. https://doi.org/10.3390/biom13010098






