Retrovirus-Derived RTL/SIRH: Their Diverse Roles in the Current Eutherian Developmental System and Contribution to Eutherian Evolution
Abstract
:1. Introduction
2. Discovery of PEG10 and PEG11/RTL1 from Genomic Imprinting Research
2.1. Roles of PEG10 and PEG11/RTL1 in Placental Evolution in Mammals
2.2. Roles of PEG11/RTL1 in Muscle Development
2.3. PEG10 and PEG11/RTL1 in Neurological Disorders
3. Retrovirus-Derived RTL/SIRH Genes as Eutherian-Specific Genes
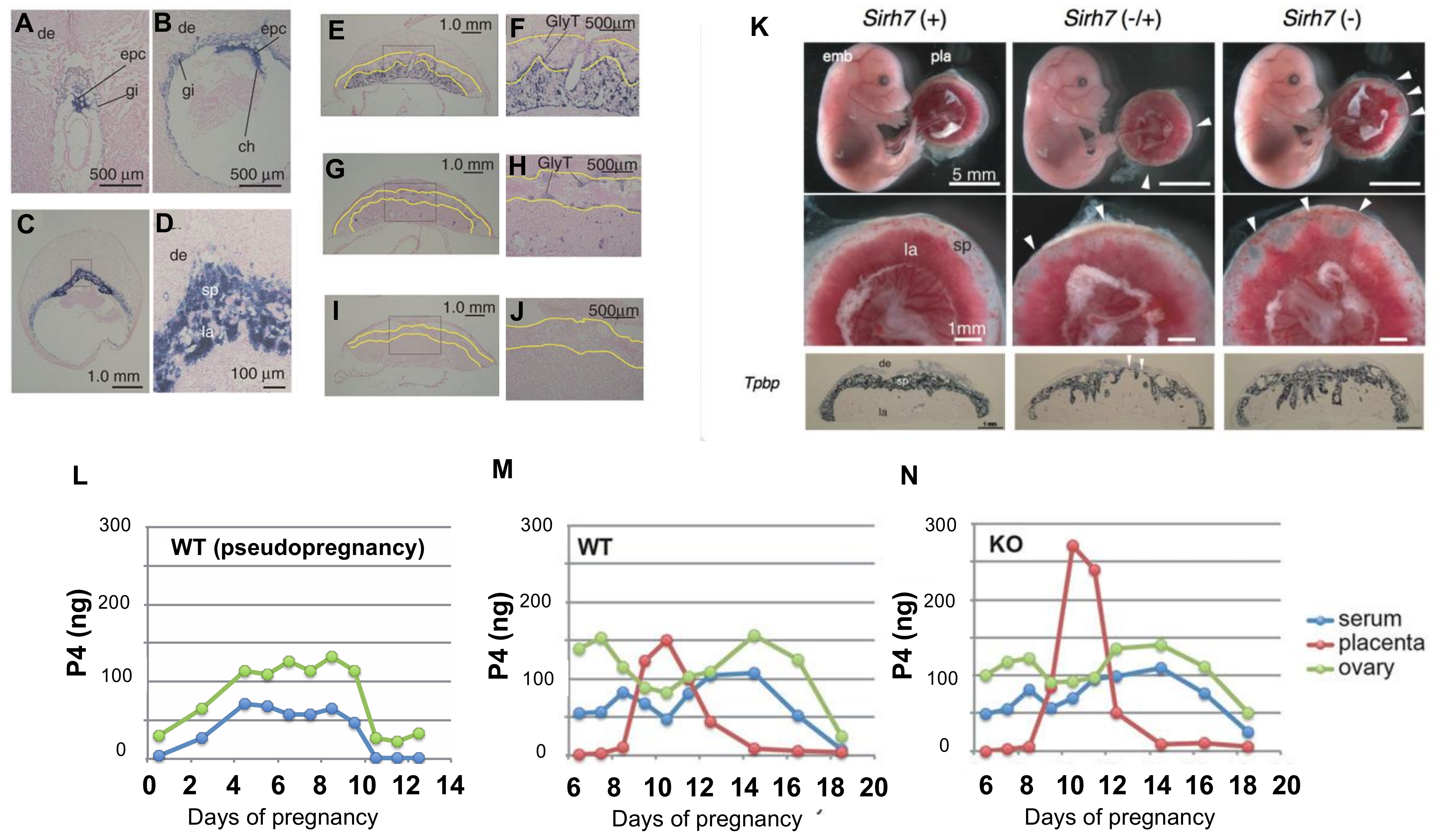
3.1. RTL7/SIRH7 in the Placenta
3.2. RTL4/SIRH11 in the Brain
3.3. RTL8A, 8B, 8C/SIRH5, 6, 4 in the Brain
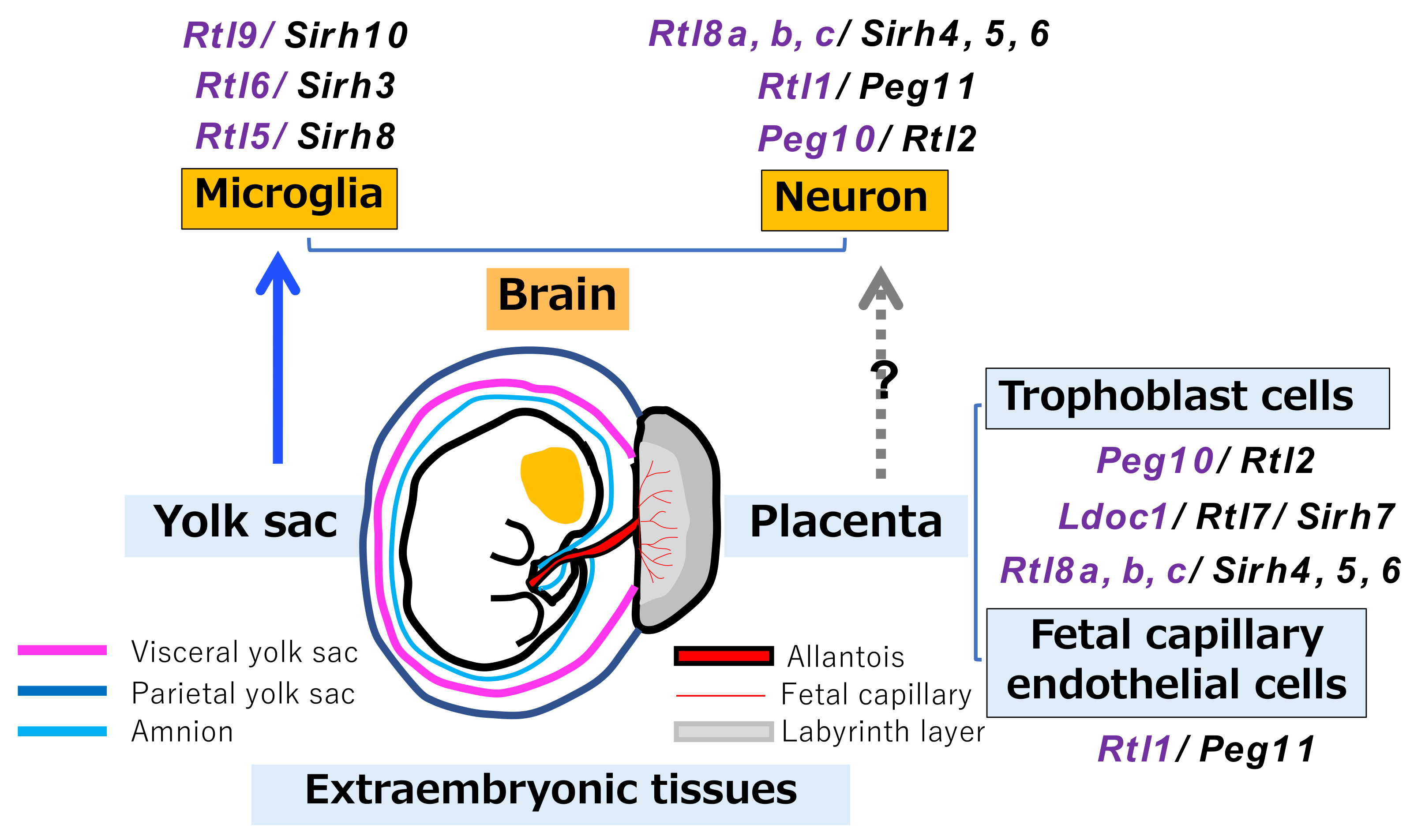
3.4. RTL6/SIRH3, RTL5/SIRH8 and RTL9/SIRH10 Are Microglial Genes in the Brain
3.5. Relationship between RTL/SIRH Genes and Extraembryonic Tissues
4. Two Types of Exapted Genes: Acquired RTL/SIRH Genes and Captured Syncytin Genes
5. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ono, R.; Kobayashi, S.; Wagatsuma, H.; Aisaka, K.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 2001, 73, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Charlier, C.; Segers, K.; Wagenaar, D.; Karim, L.; Berghmans, S.; Jaillon, O.; Shay, T.; Weissenbach, J.; Cockett, N.; Gyapay, G.; et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001, 11, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.-L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.-L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.-P.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.-Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Ono, R.; Nakamura, K.; Inoue, K.; Naruse, M.; Usami, T.; Wakisaka-Saito, N.; Hino, T.; Suzuki-Migishima, R.; Ogonuki, N.; Miki, H.; et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006, 38, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A.; et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 2008, 40, 243–248. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Bawa, O.; Harper, F.; Pierron, G.; Opolon, P.; Heidmann, T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. USA 2009, 106, 12127–12132. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Harper, F.; Guegan, J.; Dessen, P.; Pierron, G.; Heidmann, T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. USA 2011, 108, 1164–1173. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Bénit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef]
- Mallet, F.; Bouton, O.; Prudhomme, S.; Cheynet, V.; Oriol, G.; Bonnaud, B.; Lucotte, G.; Duret, L.; Mandrand, B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. USA 2004, 101, 1731–1736. [Google Scholar] [CrossRef]
- Heidmann, O.; Vernochet, C.; Dupressoir, A.; Heidemann, T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology 2009, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Heidmann, O.; Degrelle, S.A.; Vernochet, C.; Lavialle, C.; Letzelter, C.; Bernard-Stoecklin, S.; Hassanin, A.; Mulot, B.; Guillomot, M.; et al. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. USA 2013, 110, E828–E837. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Vernochet, C.; Malicorne, S.; Souquere, S.; Tzika, A.C.; Goodman, S.M.; Catzeflis, F.; Robinson, T.J.; Milinkovitch, M.C.; Pierron, G.; et al. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc. Natl. Acad. Sci. USA 2014, 111, E4332–E4341. [Google Scholar] [CrossRef]
- Cornelis, G.; Vernochet, C.; Carradec, Q.; Souquere, S.; Mulot, B.; Catzeflis, F.; Nilsson, M.A.; Menzies, B.R.; Renfree, M.B.; Pierron, G.; et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl. Acad. Sci. USA 2015, 112, E487–E496. [Google Scholar] [CrossRef]
- Nakaya, Y.; Koshi, K.; Nakagawa, S.; Hashizume, K.; Miyazawa, T. Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J. Virol. 2013, 87, 10563–10572. [Google Scholar] [CrossRef]
- Sugimoto, J.; Sugimoto, M.; Bernstein, H.; Jinno, Y.; Schust, D. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci. Rep. 2013, 3, 1462. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, J.; Schust, D.J.; Kinjo, T.; Aoki, Y.; Jinno, Y.; Kudo, Y. Suppressyn localization and dynamic expression patterns in primary human tissues support a physiologic role in human placentation. Sci. Rep. 2019, 9, 19502. [Google Scholar] [CrossRef]
- Kitao, K.; Shoji, H.; Miyazawa, T.; Nakagawa, S. Dynamic Evolution of Retroviral Envelope Genes in Egg-Laying Mammalian Genomes. Mol. Biol. Evol. 2023, 40, msad090. [Google Scholar] [CrossRef]
- Kagami, M.; Sekita, Y.; Nishimura, G.; Irie, M.; Kato, F.; Okada, M.; Yamamori, S.; Kishimoto, H.; Nakayama, M.; Tanaka, Y.; et al. Deletions and epimutations affecting the human chromosome 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 2008, 40, 237–242. [Google Scholar] [CrossRef]
- Brandt, J.; Schrauth, S.; Veith, A.-M.; Froschauer, A.; Haneke, T.; Schultheis, C.; Gessler, M.; Leimeister, C.; Volff, J.-N. Transposable elements as a source of genetic innovation: Expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 2005, 345, 101–111. [Google Scholar] [CrossRef]
- Youngson, N.A.; Kocialkowski, S.; Peel, N.; Ferguson-Smith, A.C. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 2005, 61, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Ishino, T.; Ishino, F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 2012, 3, 262. [Google Scholar] [CrossRef] [PubMed]
- Poulter, R.; Butler, M. A retrotransposon family from the pufferfish (fugu) Fugu rubripes. Gene 1998, 215, 241–249. [Google Scholar] [CrossRef]
- Butler, M.; Goodwin, T.; Simpson, M.; Singh, M.; Poulter, R. Vertebrate LTR retrotransposons of the Tf1/Sushi Group. J. Mol. Evol. 2001, 52, 260–274. [Google Scholar] [CrossRef]
- Llorens, C.; Muñoz-Pomer, A.; Bernad, L.; Botella, H.; Moyam, A. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct 2009, 4, 41. [Google Scholar] [CrossRef]
- Naruse, M.; Ono, R.; Irie, M.; Nakamura, K.; Furuse, T.; Hino, T.; Oda, K.; Kashimura, M.; Yamada, I.; Wakana, S.; et al. Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 2014, 141, 4763–4771. [Google Scholar] [CrossRef]
- Irie, M.; Yoshikawa, M.; Ono, R.; Iwafune, H.; Furuse, T.; Yamada, I.; Wakana, S.; Yamashita, Y.; Abe, T.; Ishino, F.; et al. Cognitive function related to the Sirh11/Zcchc16 gene acquired from an LTR retrotransposon in eutherians. PLoS Genet. 2015, 11, e1005521. [Google Scholar] [CrossRef]
- Irie, M.; Koga, A.; Kaneko-Ishino, T.; Ishino, F. An LTR retrotransposon-derived gene displays lineage-specific structural and putative species-specific functional variations in eutherians. Front. Chem. 2016, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Irie, M.; Itoh, J.; Matsuzawa, A.; Ikawa, M.; Kiyonari, H.; Kihara, M.; Suzuki, T.; Hiraoka, Y.; Ishino, F.; Kaneko-Ishino, T. Retrovirus-derived acquired genes, RTL5 and RTL6, are novel constituents of the innate immune system in the eutherian brain. Development 2022, 149, dev200976. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, Y.; Shiura, H.; Ishii, M.; Ono, R.; Endo, T.; Kiyonari, H.; Hirate, Y.; Ito, H.; Kanai-Azuma, M.; Kohda, T.; et al. Targeting retrovirus-derived Rtl8a and Rtl8b causes late onset obesity and neurodevelopmental defects like Prader-Willi syndrome. bioRxiv 2023, 2023-05. [Google Scholar] [CrossRef]
- Ishino, F.; Itoh, J.; Irie, M.; Matsuzawa, A.; Naruse, M.; Suzuki, T.; Hiraoka, Y.; Kaneko-Ishino, T. Retrovirus-derived RTL9 plays an important role in innate antifungal immunity in the eutherian brain. bioRxiv 2023, 2023-06. [Google Scholar] [CrossRef]
- Kitazawa, M.; Hayashi, S.; Imamura, M.; Takeda, S.; Oishi, Y.; Kaneko-Ishino, T.; Ishino, F. Deficiency and overexpression of Rtl1 in the mouse cause distinct muscle abnormalities related to Temple and Kagami-Ogata syndromes. Development 2020, 147, dev185918. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Sutani, A.; Kaneko-Ishino, T.; Ishino, F. The role of eutherian-specific RTL1 in the nervous system and its implications for the Kagami-Ogata and Temple syndromes. Genes Cells 2021, 26, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Shiura, H.; Ono, R.; Tachibana, S.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. PEG10 viral aspartic protease domain is essential for the maintenance of fetal capillary structure in the mouse placenta. Development 2021, 148, dev199564. [Google Scholar] [CrossRef]
- Pandya, N.J.; Wang, C.; Costa, V.; Lopatta, P.; Meier, S.; Zampeta, F.I.; Punt, A.M.; Mientjes, E.; Grossen, P.; Distler, T.; et al. Secreted retrovirus-like GAG-domain-containing protein PEG10 is regulated by UBE3A and is involved in Angelman syndrome pathophysiology. Cell Rep. Med. 2021, 2, 100360. [Google Scholar] [CrossRef]
- Whiteley, A.M.; Prado, M.A.; de Poot, S.A.; Paulo, J.A.; Ashton, M.; Dominguez, S.; Weber, M.; Ngu, H.; Szpyt, J.; Jedrychowski, M.P.; et al. Global proteomics of Ubqln2-based murine models of ALS. J. Biol. Chem. 2021, 296, 100153. [Google Scholar] [CrossRef]
- Black, H.H.; Hanson, J.L.; E Roberts, J.; Leslie, S.N.; Campodonico, W.; Ebmeier, C.C.; Holling, G.A.; Tay, J.W.; Matthews, A.M.; Ung, E.; et al. UBQLN2 restrains the domesticated retrotransposon PEG10 to maintain neuronal health in ALS. Elife 2023, 12, e79452. [Google Scholar] [CrossRef]
- Surani, M.A.; Barton, S.C.; Norris, M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984, 308, 548–550. [Google Scholar] [CrossRef]
- McGrath, J.; Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef]
- Mann, J.R.; Lovell-Badge, R.H. Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature 1984, 310, 66–67. [Google Scholar] [CrossRef]
- Cattanach, B.M.; Kirk, M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985, 315, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Cattanach, B.M.; Beechey, C.V. Autosomal and X-chromosome imprinting. Development 1990, 108, 63–72. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Kohda, T.; Ishino, F. The regulation and biological significance of genomic imprinting in mammals. J. Biochem. 2003, 133, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Bartolomei, M.S.; Ferguson-Smith, A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011, 3, a002592. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Ishino, T.; Ishino, F. Mammalian-specific genomic functions: Newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2015, 91, 511–538. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. Evolution of viviparaity in mammals: What genomic imprinting tells us about mammalian placental evolution. Reprod. Fert. Dev. 2019, 31, 1219–1227. [Google Scholar] [CrossRef]
- Ono, R.; Shiura, H.; Abratani, Y.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 2003, 13, 1696–1705. [Google Scholar] [CrossRef]
- Cockett, N.E.; Jackson, S.P.; Shay, T.L.; Nielsen, D.; Moore, S.S.; Steele, M.R.; Barendse, W.; Green, R.D.; Georges, M. Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc. Natl. Acad. Sci. USA 1994, 91, 3019–3023. [Google Scholar] [CrossRef]
- Kotzot, D. Maternal uniparental disomy 14 dissection of the phenotype with respect to rare autosomal recessively inherited traits, trisomy mosaicism, and genomic imprinting. Ann. Genet. 2004, 47, 251–260. [Google Scholar] [CrossRef]
- Kagami, M.; Nishimura, G.; Okuyama, T.; Hayashidani, M.; Takeuchi, T.; Tanaka, S.; Ishino, F.; Kurosawa, K.; Ogata, T. Segmental and full paternal isodisomy for chromosome 14 in three patients: Narrowing the critical region and implication for the clinical features. Am. J. Med. Genet. 2005, 138A, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Ioannides, Y.; Lokulo-Sodipe, K.; Mackay, D.J.; Davies, J.H.; Temple, I.K. Temple syndrome: Improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: An analysis of 51 published cases. J. Med. Genet. 2014, 51, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Kagami, M.; Kurosawa, K.; Miyazaki, O.; Ishino, F.; Matsuoka, K.; Ogata, T. Comprehensive clinical studies in 34 patients with molecularly defined UPD (14)pat and related conditions (Kagami Ogata syndrome). Eur. J. Hum. Genet. 2015, 23, 1488–1498. [Google Scholar] [CrossRef] [PubMed]
- Kagami, M.; Nagasaki, K.; Kosaki, R.; Horikawa, R.; Naiki, Y.; Saitoh, S.; Tajima, T.; Yorifuji, T.; Numakura, C.; Mizuno, S.; et al. Temple syndrome: Comprehensive molecular and clinical findings in 32 Japanese patients. Genet. Med. 2017, 19, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Tamura, M.; Kaneko-Ishino, T.; Ishino, F. Severe damage to the placental fetal capillary network causes mid- to late fetal lethality and reductionin placental size in Peg11/Rtl1 KO mice. Genes Cells 2017, 22, 174–188. [Google Scholar] [CrossRef]
- Dauber, A.; Cunha-Silva, M.; Macedo, D.B.; Brito, V.N.; Abreu, A.P.; Roberts, S.A.; Montenegro, L.R.; Andrew, M.; Kirby, A.; Weirauch, M.T.; et al. Paternally inherited DLK1 deletion associated with familial central precocious puberty. J. Clin. Endocrinol. Metab. 2017, 102, 1557–1567. [Google Scholar] [CrossRef]
- Macedo, D.B.; Kaiser, U.B. DLK1, Notch signaling and the timing of puberty. Semin. Reprod. Med. 2019, 37, 174–181. [Google Scholar] [CrossRef]
- Byrne, K.; Colgrave, M.L.; Vuocolo, T.; Pearson, R.; Bidwell, C.A.; Cockett, N.E.; Lynn, D.J.; Fleming-Waddell, J.N.; Tellam, R.L. The imprinted retrotransposon-like gene PEG11 (RTL1) is expressed as a full-length protein in skeletal muscle from Callipyge sheep. PLoS ONE 2010, 5, e8638. [Google Scholar] [CrossRef]
- Xu, X.; Ectors, F.; Davis, E.E.; Pirottin, D.; Cheng, H.; Farnir, F.; Hadfield, T.; Cockett, N.; Charlier, C.; Georges, M.; et al. Ectopic expression of retrotransposon-derived PEG11/RTL1 contributes to the callipyge muscular hypertrophy. PLoS ONE 2015, 10, e0140594. [Google Scholar] [CrossRef]
- Volff, J.; Körting, C.; Schartl, M. Ty3/Gypsy retrotransposon fossils in mammalian genomes: Did they evolve into new cellular functions? Mol. Biol. Evol. 2001, 18, 266–270. [Google Scholar] [CrossRef]
- Imakawa, K.; Kusama, K.; Kaneko-Ishino, T.; Nakagawa, S.; Kitao, K.; Miyazawa, T.; Ishino, F. Endogenous retroviruses and placental evolution, development, and diversity. Cells 2022, 11, 2458. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Ono, R.; Narita, T.; Pask, A.J.; Shaw, G.; Wang, C.; Kohda, T.; Alsop, A.E.; Graves, J.A.M.; Kohara, Y.; et al. Retrotransposon Silencing by DNA Methylation Can Drive Mammalian Genomic Imprinting. PLoS Genet. 2007, 3, e55. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.A.; Mungall, A.J.; Matthews, L.; Ryder, E.; Gray, D.J.; Pask, A.J.; Shaw, G.; Graves, J.A.; Rogers, J.; Dunham, I.; et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008, 6, e135. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Terzian, C.; Santamaria, P.; Pelisson, A.; Purd’homme, N.; Bucheton, A. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1994, 91, 1285–1289. [Google Scholar] [CrossRef] [PubMed]
- Song, S.U.; Gerasimova, T.; Kurkulos, M.; Boeke, J.D.; Corces, V.G. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994, 8, 2046–2057. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. The evolutionary advantage in mammals of the complementary monoallelic expression mechanism of genomic imprinting and its emergence from a defense against the insertion into the host genome. Front. Genet. 2022, 13, 832983. [Google Scholar] [CrossRef]
- Shiura, H.; Kitazawa, M.; Ishino, F.; Kaneko-Ishino, T. Roles of retrovirus-derived PEG10 and PEG11/RTL1 in mammalian development and evolution and their involvement in human disease. Front. Cell Dev. Biol. 2023, 11, 1273638. [Google Scholar] [CrossRef]
- Seitz, H.; Youngson, N.; Lin, S.-P.; Dalbert, S.; Paulsen, M.; Bachellerie, J.-P.; Ferguson-Smith, A.C.; Cavaillé, J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 2003, 34, 261–262. [Google Scholar] [CrossRef]
- Davis, E.; Caiment, F.; Tordoir, X.; Cavaillé, J.; Ferguson-Smith, A.; Cockett, N.; Georges, M.; Charlier, C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005, 15, 743–749. [Google Scholar] [CrossRef]
- Ito, M.; Sferruzzi-Perri, A.N.; Edwards, C.A.; Adalsteinsson, B.T.; Allen, S.E.; Loo, T.-H.; Kitazawa, M.; Kaneko-Ishino, T.; Ishino, F.; Stewart, C.L.; et al. A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development 2015, 142, 2425–2430. [Google Scholar] [CrossRef]
- Renfree, M.B. Marsupials: Placental mammals with a difference. Placenta 2010, 31, S21–S26. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, B.M.; Ditzel, N.; Mahmood, A.; Isa, A.; Traustadottir, G.A.; Schilling, A.F.; Ruiz-Hidalgo, M.-J.; Laborda, J.; Amling, M.; Kassem, M. DLK1 is a novel regulator of bone mass that mediates estrogen deficiency–induced bone loss in mice. J. Bone Miner. Res. 2011, 26, 1457–1471. [Google Scholar] [CrossRef] [PubMed]
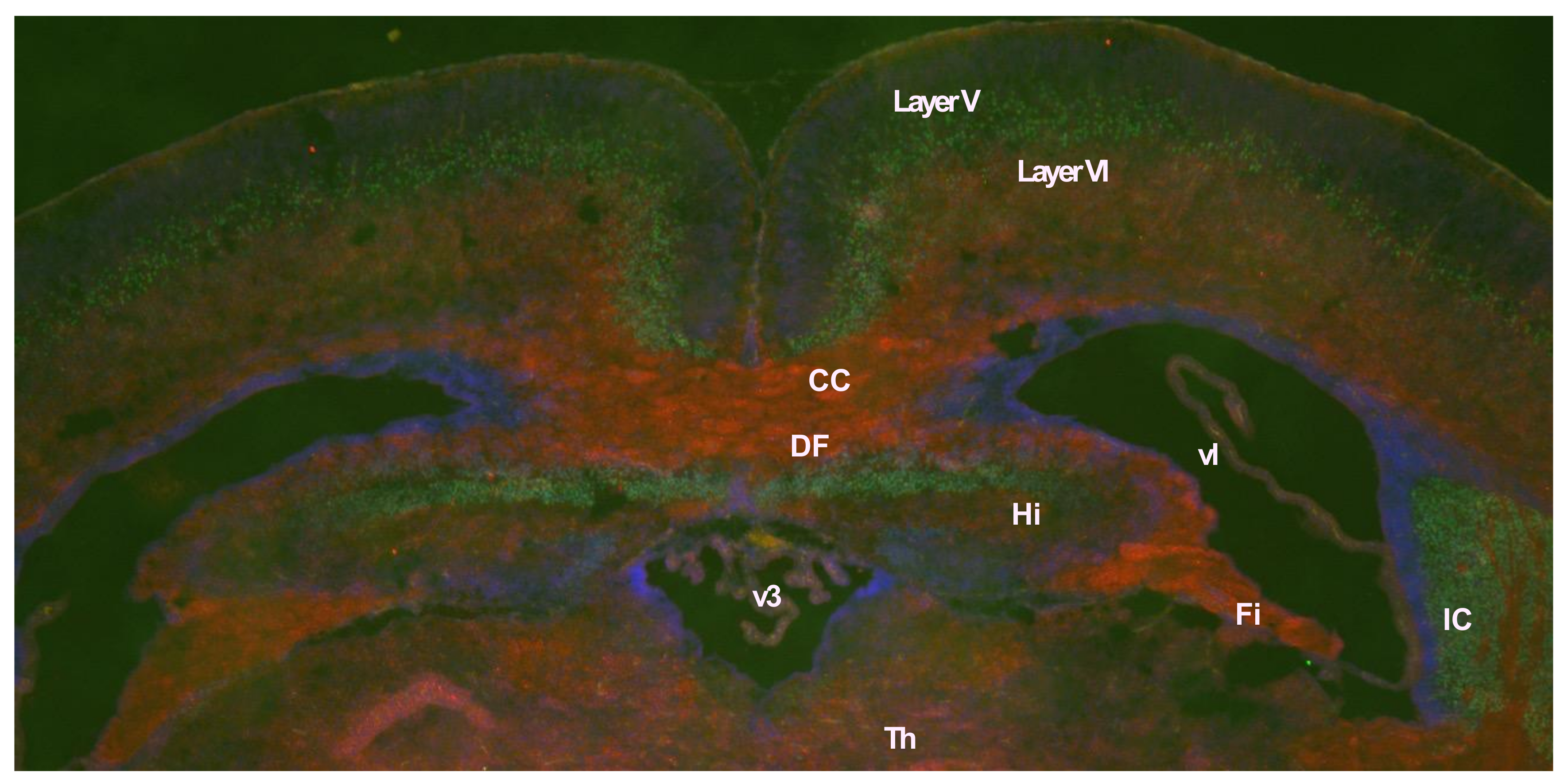
- Mihrshahi, R. The corpus callosum as an evolutionary innovation. J. Exp. Zool. 2006, 306B, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Suárez, R.; Gobius, I.; Richards, L.J. Evolution and development of interhemispheric connections in the vertebrate forebrain. Front. Hum. Neurosci. 2014, 14, 497. [Google Scholar] [CrossRef]
- Fame, R.M.; MacDonald, J.L.; Macklis, J.D. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011, 34, 41–50. [Google Scholar] [CrossRef]
- Aboitiz, F.; Montiel, J. One hundred million years of interhemispheric communication: The history of the corpus callosum. Braz. J. Med. Biol. Res. 2003, 36, 409–420. [Google Scholar] [CrossRef]
- Chou, M.-Y.; Hu, M.-C.; Chen, P.-Y.; Hsu, C.-L.; Lin, T.-Y.; Tan, M.-J.; Lee, C.-Y.; Kuo, M.-F.; Huang, P.-H.; Wu, V.-C.; et al. RTL1/PEG11 imprinted in human and mouse brain mediates anxiety-like and social behaviors and regulates neuronal excitability in the locus coeruleus. Hum. Mol. Genet. 2022, 31, 3161–3180. [Google Scholar] [CrossRef]
- Lin, S.-P.; Youngson, N.; Takada, S.; Seitz, H.; Reik, W.; Paulsen, M.; Cavaille, J.; Ferguson-Smith, A.C. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003, 35, 97–102. [Google Scholar] [CrossRef]
- Nicholls, R.D.; Knepper, J.L. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genom. Hum. Genet. 2001, 2, 153–175. [Google Scholar] [CrossRef]
- Buiting, K.; Williams, C.; Horsthemke, B. Angelman syndrome—Insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016, 12, 584–593. [Google Scholar] [CrossRef]
- Brown, R.H.; Al-Chalabi, A. Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2017, 377, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Chen, W.; Hong, S.T.; Boycott, K.M.; Gorrie, G.H.; Siddique, N.; Yang, Y.; Fecto, F.; Shi, Y.; Zhai, H.; et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011, 477, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Ono, R.; Kuroki, Y.; Naruse, M.; Ishii, M.; Iwasaki, S.; Toyoda, A.; Fujiyama, A.; Shaw, G.; Renfree, M.B.; Kaneko-Ishino, T.; et al. Identification of SIRH12, a retrotransposon-derived gene specific to marsupial mammals. DNA Res. 2011, 18, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Malassine, A.; Frendo, J.-L.; Evain-Brion, D. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 2003, 9, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Arensburg, J.; Payne, A.H.; Orly, J. Expression of steroidogenic genes in maternal and extraembryonic cells during early pregnancy in mice. Endocrinology 1999, 140, 5220–5232. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.G.; Rawn, S.; Davies, A.; Hughes, M.; Cross, J.C. Spatial and temporal expression of the 23 murine Prolactin/Placental Lactogen-related genes is not associated with their position in the locus. BMC Genom. 2008, 9, 352. [Google Scholar] [CrossRef]
- Lyon, M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961, 190, 372–373. [Google Scholar] [CrossRef]
- Takagi, N.; Sasaki, M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 1975, 256, 640–642. [Google Scholar] [CrossRef]
- Murr, S.M.; Stabenfeldt, G.H.; Bradford, G.E.; Geschwind, I.I. Plasma progesterone during pregnancy in the mouse. Endocrinology 1974, 94, 1209–1211. [Google Scholar] [CrossRef]
- Virgo, B.B.; Bellward, G.D. Serum progesterone levels in the pregnant and postpartum laboratory mouse. Endocrinology 1974, 95, 1486–1490. [Google Scholar] [CrossRef]
- Tyndale-Biscoe, H.; Renfree, M. Ovarian function and control. In Reproductive Physiology of Marsupials; Cambridge University Press: Cambridge, UK, 1987; Chapter 6; pp. 203–257. [Google Scholar] [CrossRef]
- Lim, E.T.; Raychaudhuri, S.; Sanders, S.J.; Stevens, C.; Sabo, A.; MacArthur, D.G.; Neale, B.M.; Kirby, A.; Ruderfer, D.M.; Fromer, M.; et al. Rare complete knockouts in humans: Population distribution and significant role in autism spectrum disorders. Neuron 2013, 77, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Aston-Jones, G.; Rajkowski, J.; Cohen, J. Role of locus coeruleus in attention and behavioral flexibility. Biol. Psychiatry 1999, 46, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Berridge, C.W.; Waterhouse, B.D. The locus coeruleus-noradrenergic system: Modulation of behavioral state and state-dependent cognitive processes. Brain Res. Brain Res. Rev. 2003, 42, 33–84. [Google Scholar] [CrossRef] [PubMed]
- Sara, S.J. The locus coeruleus and noradrenergic modulation of cognition. Nat. Rev. Neurosci. 2009, 10, 211–223. [Google Scholar] [CrossRef]
- Bouret, S.; Sara, S.J. Network reset: A simplified overarching theory of locus coeruleus noradrenaline function. Trends Neurosci. 2005, 28, 574–582. [Google Scholar] [CrossRef]
- Lanfray, D.; Richard, D. Emerging Signaling Pathway in Arcuate Feeding-Related Neurons: Role of the Acbd7. Front. Neurosci. 2017, 11, 328. [Google Scholar] [CrossRef]
- Seabrook, L.T.; Borgland, S. The orbitofrontal cortex, food intake and obesity. J. Psychiatry Neurosci. 2020, 45, 304–312. [Google Scholar] [CrossRef]
- Yousefvand, S.; Hamidi, F. Role of paraventricular nucleus in regulation of feeding behaviour and the design of intranuclear neuronal pathway communications. Int. J. Pept. Res. Ther. 2020, 26, 1231–1242. [Google Scholar] [CrossRef]
- Szczepanski, S.M.; Knight, R.T. Insights into human behavior from lesions to the prefrontal cortex. Neuron 2014, 83, 1002–1018. [Google Scholar] [CrossRef]
- Hare, B.D.; Duman, R.S. Prefrontal cortex circuits in depression and anxiety: Contribution of discrete neuronal populations and target regions. Mol. Psychiatry 2020, 25, 2742–2758. [Google Scholar] [CrossRef]
- Pizzagalli, D.A.; Roberts, A.C. Prefrontal cortex and depression. Neuropsychopharmacology 2022, 47, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Phil. Trans. R. Soc. B 2013, 368, 20120507. [Google Scholar] [CrossRef] [PubMed]
- Imakawa, K.; Nakagawa, S.; Miyazawa, T. Baton pass hypothesis: Successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells 2005, 20, 771–788. [Google Scholar] [CrossRef] [PubMed]
- Abed, M.; Verschueren, E.; Budayeva, H.; Liu, P.; Kirkpatrick, D.S.; Reja, R.; Kummerfeld, S.K.; Webster, J.D.; Gierke, S.; Reichelt, M.; et al. The Gag protein PEG10 binds to RNA and regulates trophoblast stem cell lineage specification. PLoS ONE 2019, 14, e0214110. [Google Scholar] [CrossRef]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef]
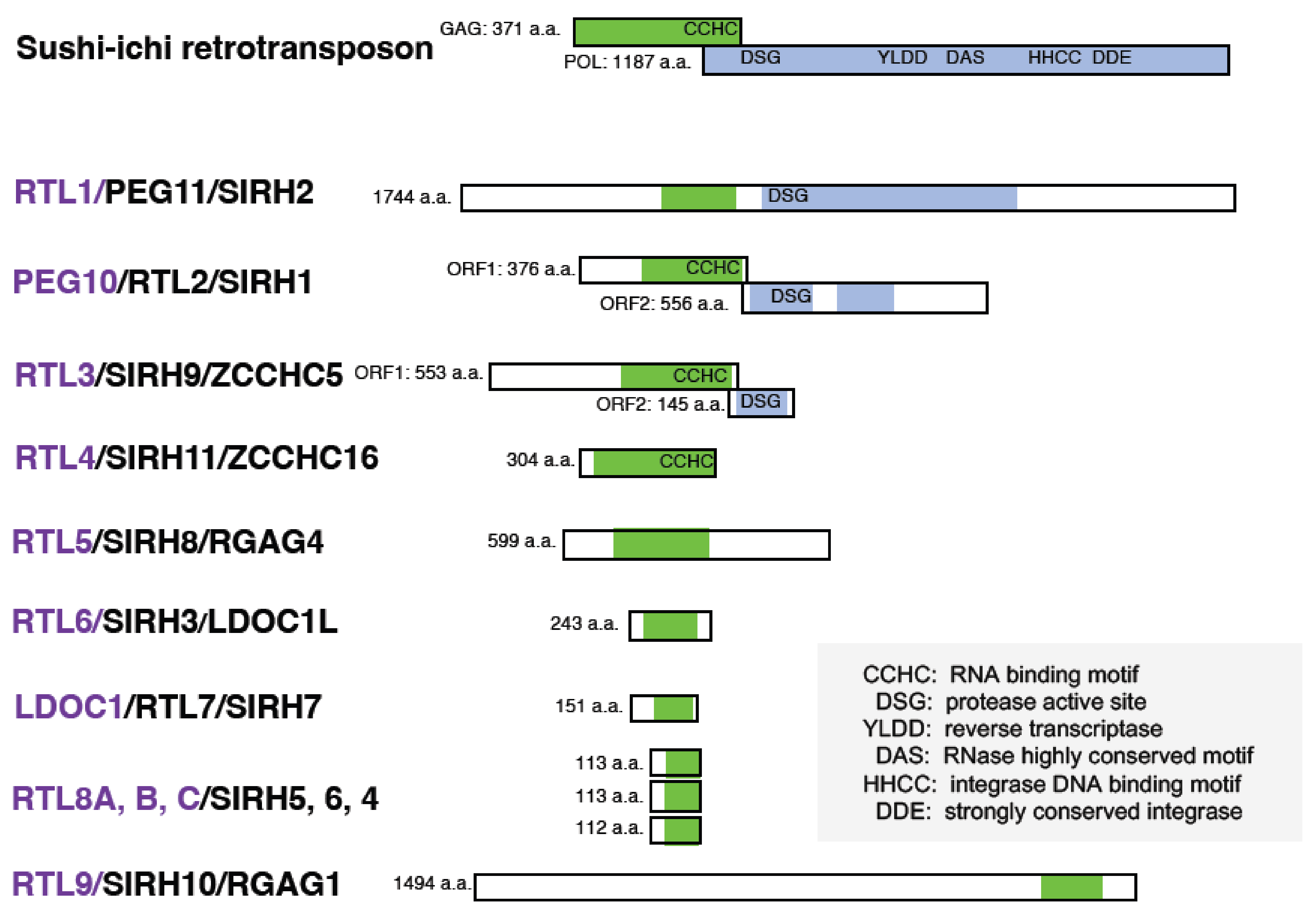


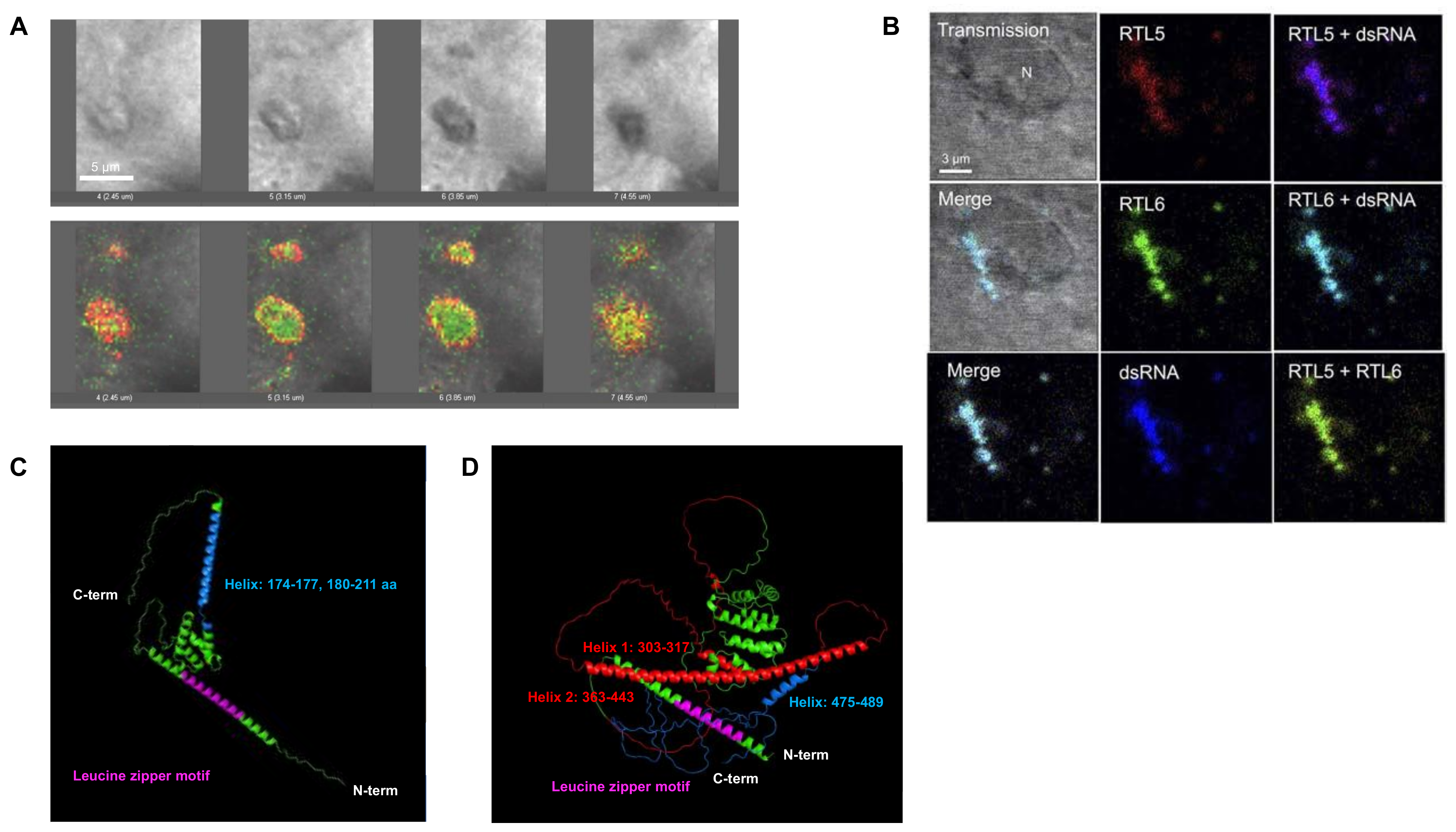
| Formal Gene Name | Aliases | Placenta | Brain | Muscle | Human Diseases | Behavior Abnormalities | Rtl/Sirh KO Mouse | Phenotypes | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Neuron | Microglia | ||||||||||
| 1 | RTL1 | PEG11 | SIRH2 | ◯ | ◯ | ◯ | KOS14, TS14 | Rtl1/Peg11 Pat-KO | Late fetal/neonatal lethality, TS14-like phenotypes in placenta, muscle and brain | ||
| Rtl1/Peg11 Mat-KO | Neonatal lethality, KOS14-like phenotypes in placenta, muscle and brain | ||||||||||
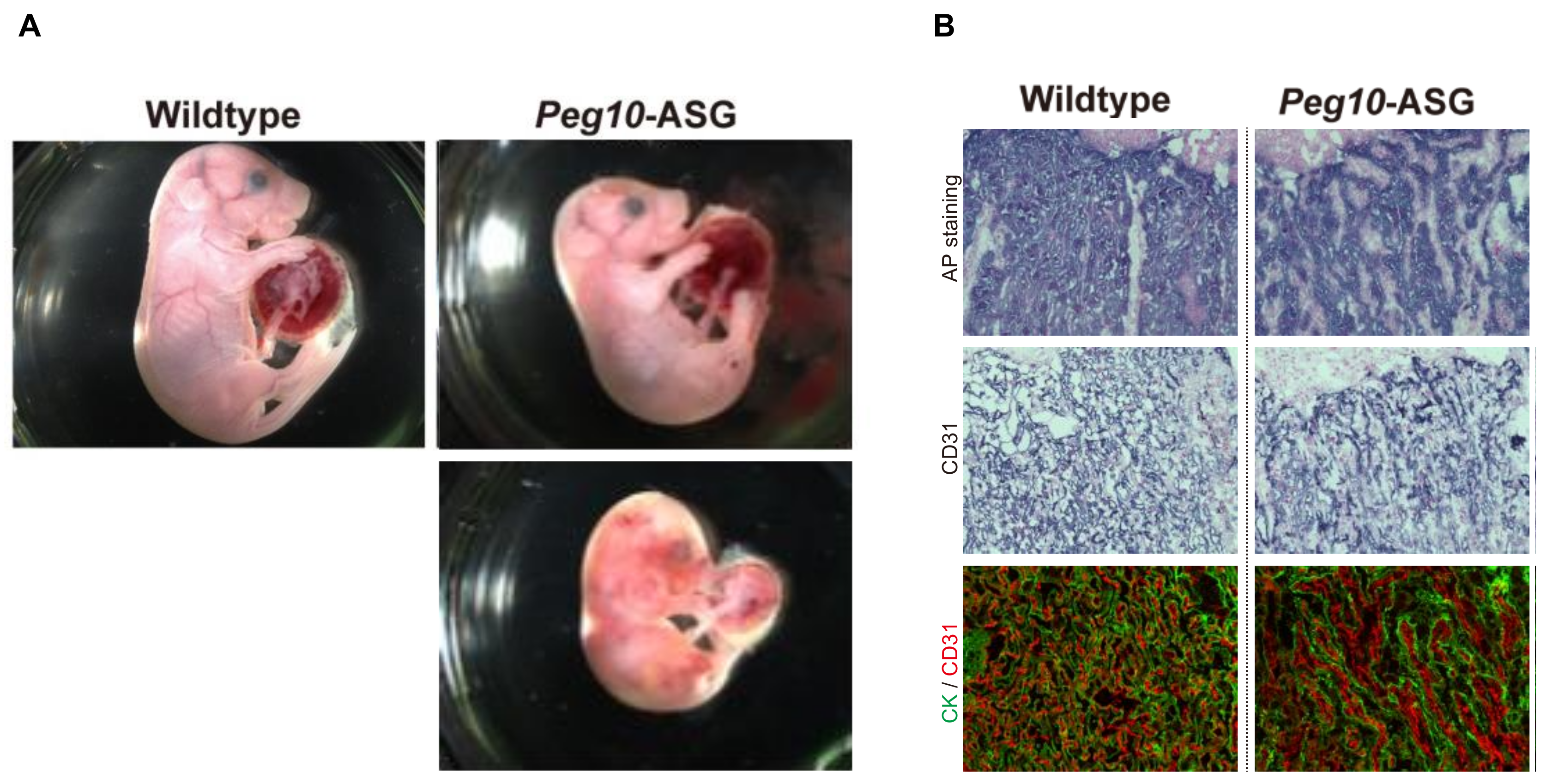
| 2 | PEG10 | RTL2 | SIRH1 | ◯ | ◯ | NK | AS *, ALS * | Peg10 KO | Early embryonic lethality due to severe placental defects | ||
| Peg10 ASG-mutant | Late fetal/neonatal lethality due to placental defects | ||||||||||
| 3 | RTL3 | ZCCHC5 | SIRH9 | – | – | – | – | Rtl3/Sirh9 KO | No data | ||
| 4 | RTL4 | ZCCHC16 | SIRH11 | ND | ◯ | ND | ASD | ◯ | Rtl4/Sirh11 KO | Increased impulsivity, reduced attention and spacial memory possibly due to low recovery rate of NA in FC | |
| 5 | RTL5 | RGAG4 | SIRH8 | NK | ◯ | NK | ◯ * | Rtl5/Sirh8 KO | Defects in innate immunity against viruses (dsRNA) | ||
| 6 | RTL6 | LDOC1L | SIRH3 | NK | ◯ | NK | ◯ * | Rtl6/Sirh3 KO | Defects in innate immunity against bacteria (LPS) | ||
| 7 | LDOC1 | RTL7 | SIRH7 | ◯ | ◯ * | NK | ◯ | Rtl7/Sirh7 KO | Delayed parturition due to abnormal placental endocrine function, poor maternal care | ||
| 8 | RTL8A, B, C | CXXA, B, C | SIRH5, 6, 4 | ◯ | ◯ * | NK | AS *, PWS * | ◯ | Rtl8a, b/Sirh5, 6 DKO | Increased depression-like behavior and anxiety, late onset obesity, poor maternal care | |
| 9 | RTL9 | RGAG1 | SIRH10 | NK | ◯ | NK | ◯ * | Rtl9/Sirh10 KO | Defects in innate immunity against fungi (Zymosan) | ||
| Behavioral Tests | WT vs Pat-KO | WT vs Mat-KO | Relationship between Behavioral Analysis and PEG11/RTL1 Expression |
|---|---|---|---|
| Open Field Test | Decreased spontaneous movement | Impaired functions of the CC and cortical layer V possibly due to reduced information exchange | |
| Elevated Plus Maze Test | Increased anxiety-like behavior | NS | Increased anxiety due to impaired cortex, amygdala and hippocampus |
| Light/Dark Transition Test | Mild increament of anxiety-like behavior | Increased anxiety due to impaired cortex, amygdala and hippocampus | |
| Fear Conditioning Test: hippocampus-dependent memory | NS | Impaired hippocampus-dependent memory | Increased anxiety due to impaired hippocampus-dependent fear responses |
| Fear Conditioning Test: amygdala-dependent memory | Impaired amygdala-dependent memory | Increased anxiety due to impaired amygdala-dependent fear responses | |
| Morris Water Maze Test | NS | Severely impaired spatial memory | Impaired hippocampus-dependent learning |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaneko-Ishino, T.; Ishino, F. Retrovirus-Derived RTL/SIRH: Their Diverse Roles in the Current Eutherian Developmental System and Contribution to Eutherian Evolution. Biomolecules 2023, 13, 1436. https://doi.org/10.3390/biom13101436
Kaneko-Ishino T, Ishino F. Retrovirus-Derived RTL/SIRH: Their Diverse Roles in the Current Eutherian Developmental System and Contribution to Eutherian Evolution. Biomolecules. 2023; 13(10):1436. https://doi.org/10.3390/biom13101436
Chicago/Turabian StyleKaneko-Ishino, Tomoko, and Fumitoshi Ishino. 2023. "Retrovirus-Derived RTL/SIRH: Their Diverse Roles in the Current Eutherian Developmental System and Contribution to Eutherian Evolution" Biomolecules 13, no. 10: 1436. https://doi.org/10.3390/biom13101436
APA StyleKaneko-Ishino, T., & Ishino, F. (2023). Retrovirus-Derived RTL/SIRH: Their Diverse Roles in the Current Eutherian Developmental System and Contribution to Eutherian Evolution. Biomolecules, 13(10), 1436. https://doi.org/10.3390/biom13101436





