Insights into COVID-19: Perspectives on Drug Remedies and Host Cell Responses
Abstract
:1. Introduction
1.1. Classification of Coronaviruses
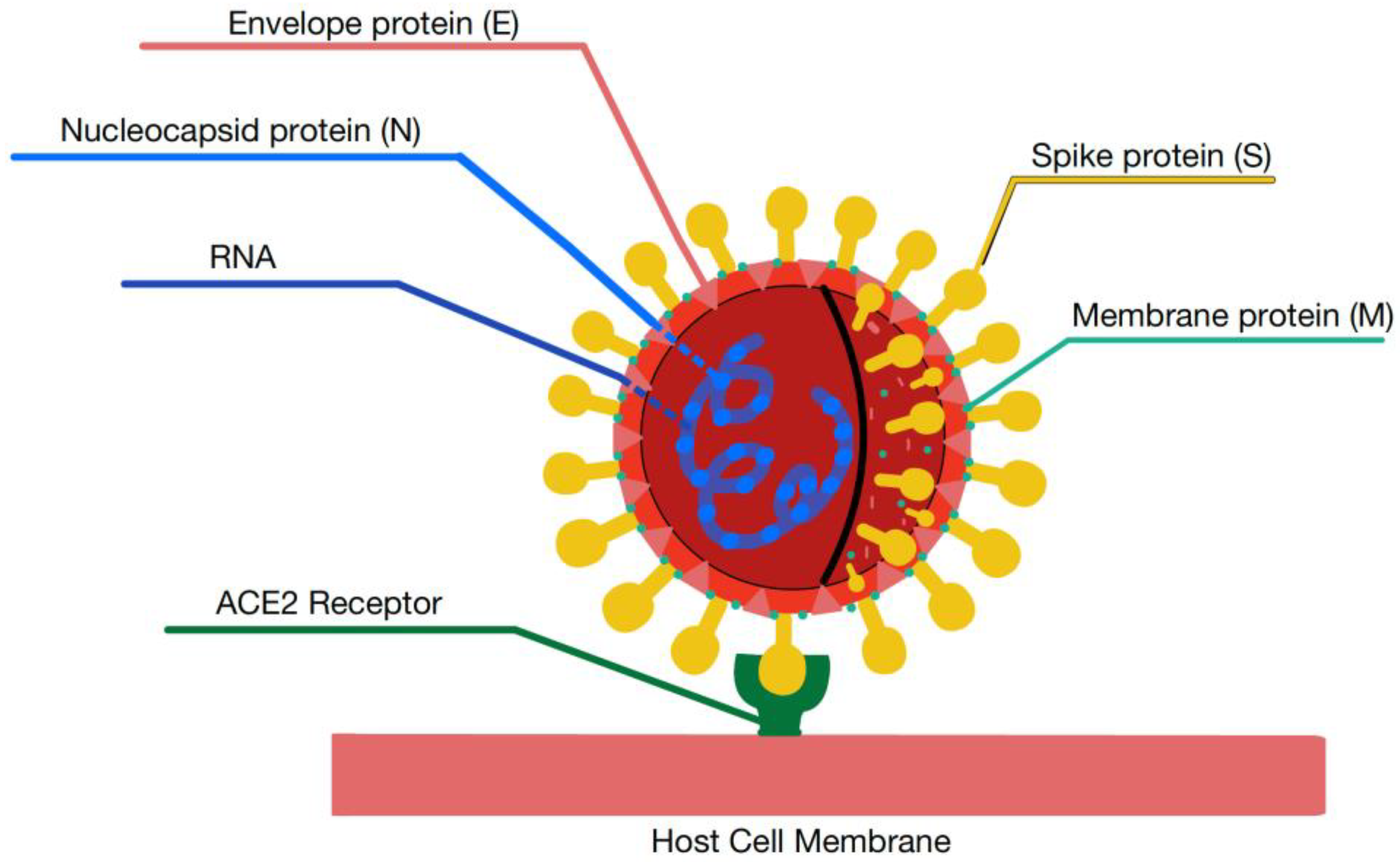
1.2. Structure of Coronaviruses
1.3. Nonstructural Proteins in CoVs
2. Viral Entry and Host Cell Responses
2.1. Infection and Transmission
2.2. ACE2 Receptor and Mediation of Virus Host Cell Entry
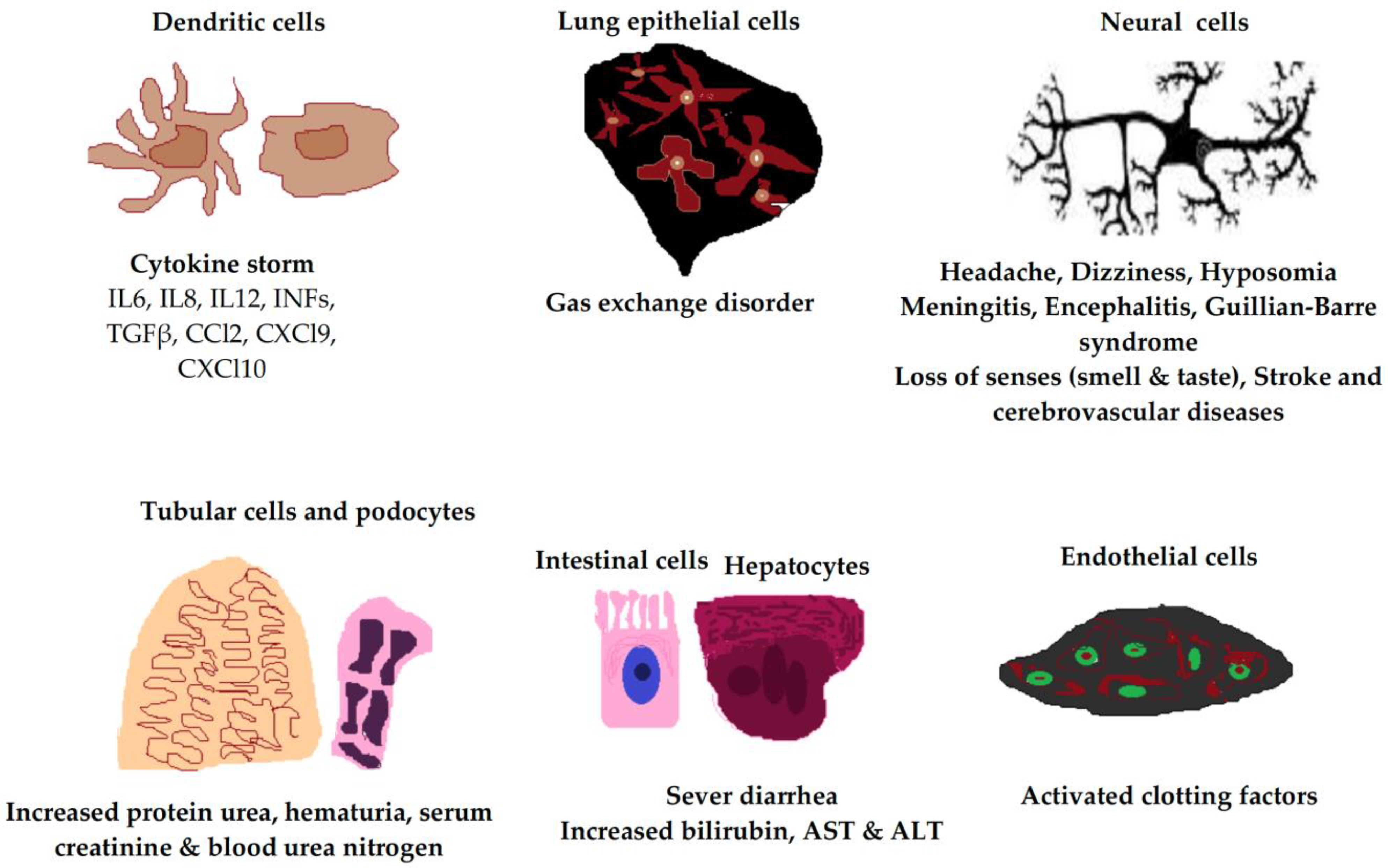
2.3. Host Cells Responses to SARS-CoV-2
2.3.1. SARS-CoV-2 in Immune Cells
2.3.2. Endothelial Cells
2.3.3. Respiratory Tract Epithelial Cells
2.3.4. Tubular Cells and Podocytes
2.3.5. Hepatocytes and Intestinal Cells
2.3.6. Neural Cells
3. Potential Drug Candidates and Clinical Trials for COVID-19 Treatment
3.1. Potential Candidates
3.1.1. Antimalarial Agents
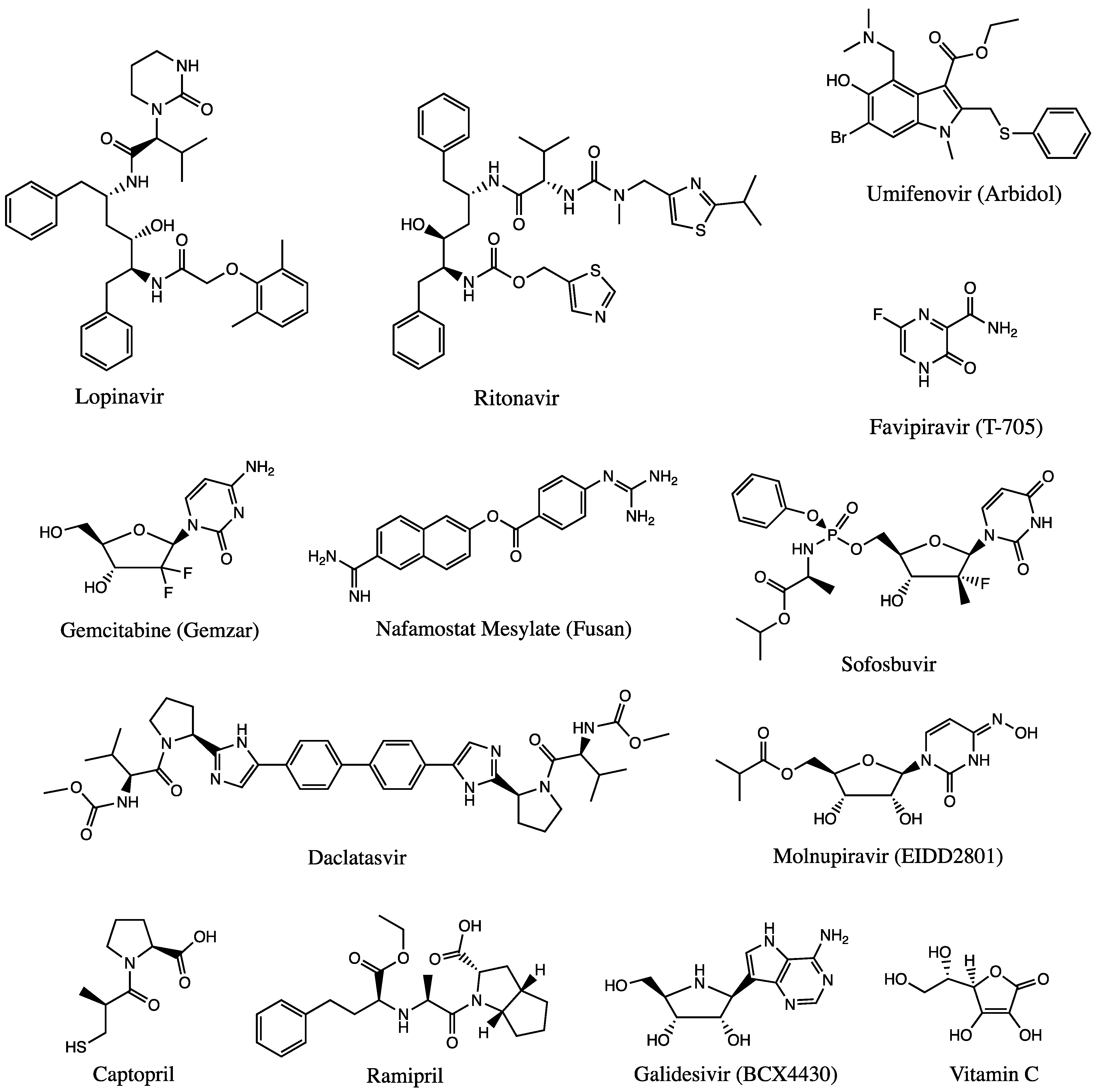
3.1.2. Antiviral Agents
3.1.3. Other Agents and Therapies
3.1.4. Antibiotic/Antibacterial
3.1.5. ACE2 Inhibitors
3.1.6. Anti-Inflammatories
4. Pathways as Potential Targets
Clathrin-Mediated Endocytosis
5. Computer-Aided Drug Design
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malik, Y.S.; Sircar, S.; Bhat, S.; Sharun, K.; Dhama, K.; Dadar, M.; Tiwari, R.; Chaicumpa, W. Emerging novel coronavirus (2019-nCoV)—Current scenario, evolutionary perspective based on genome analysis and recent developments. Vet. Q. 2020, 40, 68–76. [Google Scholar] [CrossRef] [PubMed]
- WHO. 2023. Available online: https://covid19.who.int/ (accessed on 18 September 2023).
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2020, 54, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and Coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- Ye, Z.W.; Yuan, S.; Yuen, K.S.; Fung, S.Y.; Chan, C.P.; Jin, D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020, 16, 1686–1697. [Google Scholar] [CrossRef]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and Chloroquine Effectively Inhibit the Recently Emerged Novel Coronavirus (2019-nCoV) In Vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef]
- Hillen, H.S. Structure and function of SARS-CoV-2 polymerase. Curr. Opin. Virol. 2021, 48, 82–90. [Google Scholar] [CrossRef]
- Huston, N.C.; Wan, H.; Strine, M.S.; Tavares, R.d.C.A.; Wilen, C.B.; Pyle, A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell 2021, 81, 584–598.e5. [Google Scholar] [CrossRef]
- Parks, J.M.; Smith, J.C. How to Discover Antiviral Drugs Quickly. N. Engl. J. Med. 2020, 382, 2261–2264. [Google Scholar] [CrossRef]
- Pandey, A.; Nikam, A.N.; Shreya, A.B.; Mutalik, S.P.; Gopalan, D.; Kulkarni, S.; Padya, B.S.; Fernandes, G.; Mutalik, S.; Prassl, R. Potential therapeutic targets for combating SARS-CoV-2: Drug repurposing, clinical trials and recent advancements. Life Sci. 2020, 256, 117883. [Google Scholar] [CrossRef] [PubMed]
- Publishing, Harvard Health. COVID-19 Basics. Harvard Health Last Updated 2021. Available online: https://www.health.harvard.edu/diseases-and-conditions/covid-19-basics (accessed on 18 September 2023).
- Cardone, M.; Yano, M.; Rosenberg, A.S.; Puig, M. Lessons Learned to Date on COVID-19 Hyperinflammatory Syndrome: Considerations for Interventions to Mitigate SARS-CoV-2 Viral Infection and Detrimental Hyperinflammation. Front. Immunol. 2020, 11, 1131. [Google Scholar] [CrossRef] [PubMed]
- Ilmjärv, S.; Abdul, F.; Acosta-Gutiérrez, S.; Estarellas, C.; Galdadas, I.; Casimir, M.; Alessandrini, M.; Gervasio, F.L.; Krause, K.-H. Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant. Sci. Rep. 2021, 11, 13705. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Thao, T.T.N.; Hoffmann, D.; Taddeo, A.; Ebert, N.; Labroussaa, F.; Pohlmann, A.; King, J.; Steiner, S.; Kelly, J.N.; et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 2021, 592, 122–127. [Google Scholar] [CrossRef]
- Jiang, M.; Kolehmainen, P.; Kakkola, L.; Maljanen, S.; Melén, K.; Smura, T.; Julkunen, I.; Österlund, P. SARS-CoV-2 Isolates Show Impaired Replication in Human Immune Cells but Differential Ability to Replicate and Induce Innate Immunity in Lung Epithelial Cells. Microbiol. Spectr. 2021, 9, e00774-21. [Google Scholar] [CrossRef]
- Laine, L.; Skön, M.; Väisänen, E.; Julkunen, I.; Österlund, P. SARS-CoV-2 variants Alpha, Beta, Delta and Omicron show a slower host cell interferon response compared to an early pandemic variant. Front. Immunol. 2022, 13, 1016108. [Google Scholar] [CrossRef]
- Belik, M.; Jalkanen, P.; Lundberg, R.; Reinholm, A.; Laine, L.; Väisänen, E.; Skön, M.; Tähtinen, P.A.; Ivaska, L.; Pakkanen, S.H.; et al. Comparative analysis of COVID-19 vaccine responses and third booster dose-induced neutralizing antibodies against Delta and Omicron variants. Nat. Commun. 2022, 13, 2476. [Google Scholar] [CrossRef]
- Jalkanen, P.; Kolehmainen, P.; Haveri, A.; Huttunen, M.; Laine, L.; Österlund, P.; Tähtinen, P.A.; Ivaska, L.; Maljanen, S.; Reinholm, A.; et al. Vaccine-Induced Antibody Responses against SARS-CoV-2 Variants-Of-Concern Six Months after the BNT162b2 COVID-19 mRNA Vaccination. Microbiol. Spectr. 2022, 10, e0225221. [Google Scholar] [CrossRef]
- Ekström, N.; Haveri, A.; Solastie, A.; Virta, C.; Österlund, P.; Nohynek, H.; Nieminen, T.; Ivaska, L.; Tähtinen, P.A.; Lempainen, J.; et al. Strong Neutralizing Antibody Responses to SARS-CoV-2 Variants Following a Single Vaccine Dose in Subjects with Previous SARS-CoV-2 Infection. Open Forum Infect. Dis. 2022, 9, ofac625. [Google Scholar] [CrossRef]
- Awad, K. COVID-19: End or fade of the pandemic? Lupine Online J. Med. Sci. 2023, 6, 638–639. [Google Scholar]
- Gorkhali, R.; Koirala, P.; Rijal, S.; Mainali, A.; Baral, A.; Bhattarai, H.K. Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins. Bioinform. Biol. Insights 2021, 15, 11779322211025876. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Shen, H.-M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Sriram, K.; Insel, P. What Is the ACE2 Receptor? American Society for Biochemistry and Molecular Biology. 16 May 2020. Available online: https://www.asbmb.org/asbmb-today/science/051620/what-is-the-ace2-receptor (accessed on 18 September 2023).
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Mahmudpour, M.; Roozbeh, J.; Keshavarz, M.; Farrokhi, S.; Nabipour, I. COVID-19 cytokine storm: The anger of inflammation. Cytokine 2020, 133, 155151. [Google Scholar] [CrossRef] [PubMed]
- Goodman Brenda. Cytokine Storms May Be Fueling Some COVID Deaths. WebMD 2020. Available online: www.webmd.com/lung/news/20200417/cytokine-storms-may-be-fueling-some-covid-deaths (accessed on 18 September 2023).
- Katopodis, P.; Randeva, H.S.; Spandidos, D.A.; Saravi, S.; Kyrou, I.; Karteris, E. Host cell entry mediators implicated in the cellular tropism of SARS-CoV-2, the pathophysiology of COVID-19 and the identification of microRNAs that can modulate the expression of these mediators (Review). Int. J. Mol. Med. 2021, 49, 20. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2021, 6, 80. [Google Scholar] [CrossRef]
- Pruimboom, L. SARS-CoV 2; Possible alternative virus receptors and pathophysiological determinants. Med. Hypotheses 2020, 146, 110368. [Google Scholar] [CrossRef]
- Awad, K.; Maghraby, A.S.; Abd-Elshafy, D.N.; Bahgat, M.M. Carbohydrates Metabolic Signatures in Immune Cells: Response to Infection. Front. Immunol. 2022, 13, 912899. [Google Scholar] [CrossRef]
- Felsenstein, S.; Herbert, J.A.; McNamara, P.S.; Hedrich, C.M. COVID-19: Immunology and treatment options. Clin. Immunol. 2020, 215, 108448. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Network, H.L. SARS-CoV-2 Entry Genes Are Most Highly Expressed in Nasal Goblet and Ciliated Cells within Human Airways. arXiv 2020, arXiv:2003.06122v1. [Google Scholar]
- Xu, H.; Zhong, L.; Deng, J.; Peng, J.; Dan, H.; Zeng, X.; Li, T.; Chen, Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Resnik, R.; Mingorance, F.L.; Rivera, F.; Mitchell, F.; Gonzalez, C.D.; Vaccaro, M.I. Autophagy in Inflammatory Response against SARS-CoV-2. Int. J. Mol. Sci. 2023, 24, 4928. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Wang, R.; Feng, Z.; Zhang, J.; Yang, H.; Tan, Y.; Wang, H.; Wang, C.; Liu, L.; et al. Human kidney is a target for novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Nat. Commun. 2021, 12, 2506. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef]
- Schulert, G.S.; Grom, A.A. Pathogenesis of macrophage activation syndrome and potential for cytokine- directed therapies. Annu. Rev. Med. 2015, 66, 145–159. [Google Scholar] [CrossRef]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J.; on behalf of the HLH Across Speciality Collaboration, UK. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Zhou, Z.; Ren, L.; Zhang, L.I.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Overly exuberant innate immune response SARS- CoV-2 infection. Cell Host Microbe 2020. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and exacerbated inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fu, B.; Zheng, X.; Wang, D.; Zhao, C.; Qi, Y.; Sun, R.; Tian, Z.; Xu, X.; Wei, H. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020, 7, 998–1002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, R.; Lei, L.; Liu, H.; Wang, Y.; Wang, Y.; Qian, H.; Dai, T.; Zhang, T.; Lai, Y.; et al. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. medRxiv 2020. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Enz, N.; Vliegen, G.; De Meester, I.; Jungraithmayr, W. CD26/DPP4—A potential biomarker and target for cancer therapy. Pharmacol. Ther. 2019, 198, 135–159. [Google Scholar] [CrossRef]
- Samsami, M.; Fatemi, A.; Khoshnoud, R.J.; Kohansal, K.; Hussen, B.M.; Soghala, S.; Taheri, M.; Ghafouri-Fard, S. Abnormal Transcript Levels of Cytokines among Iranian COVID-19 Patients. J. Mol. Neurosci. 2022, 72, 27–36. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shahin, N.N.; Awad, K.; Maghraby, A.S.; Abd-Elshafy, D.N.; Bahgat, M.M. Glycolytic and immunological alterations in human U937 monocytes in response to H1N1 infection. IUBMB Life 2020, 72, 2481–2498. [Google Scholar] [CrossRef]
- de Wit, E.; van Doremalen, N.; Falzarano, D.; Munster, V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016, 14, 523–534. [Google Scholar] [CrossRef]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shahin, N.N.; Maghraby, A.S.; Kirschfink, M.; Abd-Elshafy, D.N.; Awad, K.; Bahgat, M.M. H1N1 Infection Reduces Glucose Level in Human U937 Monocytes Culture. Viral Immunol. 2020, 33, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Atkin-Smith, G.K.; Duan, M.; Chen, W.; Poon, H. The induction and consequences of Influenza A virus-induced cell death. Cell Death Dis. 2018, 9, 1002. [Google Scholar] [CrossRef] [PubMed]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, M.; Schneider, K.; Weber, F.; Weidmann, M.; Hufert, F.T. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 2006, 87, 1953–1960. [Google Scholar] [CrossRef] [PubMed]
- Saeedifar, A.M.; Ghorban, K.; Ganji, A.; Mosayebi, G.; Gholami, M.; Dadmanesh, M.; Rouzbahani, N.H. Evaluation of Tcell exhaustion based on the expression of EOMES, Tbet and co-inhibitory receptors in severe and non-severe COVID-19 patients. Gene Rep. 2023, 31, 101747–101753. [Google Scholar] [CrossRef]
- Gu, J.; Gong, E.; Zhang, B.; Zheng, J.; Gao, Z.; Zhong, Y.; Zou, W.; Zhan, J.; Wang, S.; Xie, Z.; et al. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005, 202, 415–424. [Google Scholar] [CrossRef]
- Flammer, A.J.; Anderson, T.; Celermajer, D.S.; Creager, M.A.; Deanfield, J.; Ganz, P.; Hamburg, N.M.; Lüscher, T.F.; Shechter, M.; Taddei, S.; et al. The Assessment of Endothelial Function: From research into clinical practice. Circulation 2012, 126, 753–767. [Google Scholar] [CrossRef]
- Bonetti, P.O.; Lerman, L.O.; Lerman, A. Endothelial dysfunction—A marker of atherosclerotic risk. Arterioscl. Throm Vas. 2003, 23, 168–175. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Horton, R. Offline: COVID-19—Bewilderment and candour. Lancet 2020, 395, 1178. [Google Scholar] [CrossRef]
- Cyranoski, D. Why children avoid the worst coronavirus complications might lie in their arteries. Nature 2020, 582, 324–325. [Google Scholar] [CrossRef]
- Xu, S.W.; Ilyas, I.; Weng, J.P. Endothelial dysfunction in COVID-19: An overview of evidence, biomarkers, mechanisms and potential therapies. Acta Pharmacol. Sin. 2023, 44, 695–709. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Chen, K.; Zou, J.; Han, P.; Hao, J.; Han, Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Becavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Blume, C.; Jackson, C.L.; Spalluto, C.M.; Legebeke, J.; Nazlamova, L.; Conforti, F.; Perotin, J.-M.; Frank, M.; Butler, J.; Crispin, M.; et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021, 53, 205–214. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Cheung, M.-C.; Perera, R.A.P.M.; Ng, K.-C.; Bui, C.H.T.; Ho, J.C.W.; Ng, M.M.T.; Kuok, D.I.T.; Shih, K.C.; Tsao, S.-W.; et al. Tropism, Replication Competence, and Innate Immune Responses of The Coronavirus SARS-CoV-2 in Human Respiratory Tract and Conjunctiva: An Analysis in Ex-Vivo and In-Vitro Cultures. Lancet Respir. Med. 2020, 8, 687–695. [Google Scholar] [CrossRef]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Munnink, B.B.O.; De Meulder, D.; Van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef]
- Tsuji, K.; Yamada, S.; Hirai, K.; Asakura, H.; Kanda, Y. Development of alveolar and airway cells from human iPS cells: Toward SARS-CoV-2 research and drug toxicity testing. J. Toxicol. Sci. 2021, 46, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.J. Thoughts on the alveolar phase of COVID-19. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L115–L120. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, R.; Wang, K.; Zhang, M.; Wang, Z.; Dong, L.; Li, J.; Yao, Y.; Ge, S.; Xu, G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020, 97, 829–838. [Google Scholar] [CrossRef]
- Martinez-Rojas, M.A.; Vega-Vega, O.; Bobadilla, N.A. Is the kidney a target of SARS-CoV-2? Am. J. Physiol. Ren. Physiol. 2020, 318, F1454–F1462. [Google Scholar] [CrossRef] [PubMed]
- Armaly, Z.; Kinaneh, S.; Skorecki, K. Renal Manifestations of COVID-19: Physiology and Pathophysiology. J. Clin. Med. 2021, 10, 1216. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Yang, M.; Wan, C.; Yi, L.-X.; Tang, F.; Zhu, H.-Y.; Yi, F.; Yang, H.-C.; Fogo, A.B.; Nie, X.; et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020, 98, 219–227. [Google Scholar] [CrossRef]
- Agarwal, A.; Chen, A.; Ravindran, N.; To, C.; Thuluvath, P.J. Gastrointestinal and Liver Manifestations of COVID-19. J. Clin. Exp. Hepatol. 2020, 10, 263–265. [Google Scholar] [CrossRef]
- Yeo, C.; Kaushal, S.; Yeo, D. Enteric involvement of coronaviruses: Is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020, 5, 335–337. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, Y.; Pan, Y.; Zhao, Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020, 525, 135–140. [Google Scholar] [CrossRef]
- Liang, W.; Feng, Z.; Rao, S.; Xiao, C.; Xue, X.; Lin, Z.; Zhang, Q.; Qi, W. Diarrhoea may be underestimated: A missing link in 2019 novel coronavirus. Gut 2020, 69, 1141–1143. [Google Scholar] [CrossRef]
- Ong, J.; Young, B.E.; Ong, S. COVID-19 in gastroenterology: A clinical perspective. Gut 2020, 69, 1144–1145. [Google Scholar] [CrossRef] [PubMed]
- Young, B.E.; Ong, S.W.X.; Kalimuddin, S.; Low, J.G.; Tan, S.Y.; Loh, J.; Ng, O.T.; Marimuthu, K.; Ang, L.W.; Mak, T.M.; et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA 2020, 323, 1488–1494. [Google Scholar] [CrossRef]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.C.; et al. Clinical Characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Dufour, J.-F.; Marjot, T.; Becchetti, C.; Tilg, H. COVID-19 and liver disease. Gut 2022, 71, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.R.; Wangzhou, A.; Ghneim, N.; Yousuf, M.S.; Paige, C.; Tavares-Ferreira, D.; Mwirigi, J.M.; Shiers, S.; Sankaranarayanan, I.; McFarland, A.J.; et al. A pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain Behav. Immun. 2020, 89, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Kang, H.; Li, S.; Zhao, X. Understanding the neurotropic characteristics of SARS-CoV-2: From neurological manifestations of COVID-19 to potential neurotropic mechanisms. J. Neurol. 2020, 267, 2179–2184. [Google Scholar] [CrossRef]
- Aguilar-Valles, A.; Haji, N.; De Gregorio, D.; Matta-Camacho, E.; Eslamizade, M.J.; Popic, J.; Sharma, V.; Cao, R.; Rummel, C.; Tanti, A.; et al. Translational control of depression-like behavior via phosphorylation of eukaryotic translation initiation factor 4E. Nat. Commun. 2018, 9, 2459. [Google Scholar] [CrossRef]
- Bostanciklioglu, M.; Temiz, E. Severe Acute Respiratory Syndrome Coronavirus 2 Is Penetrating to Dementia Research. Curr. Neurovasc. Res. 2020, 17, 342–343. [Google Scholar] [CrossRef]
- Netland, J.; Meyerholz, D.K.; Moore, S.; Cassell, M.; Perlman, S. Severe Acute Respiratory Syndrome Coronavirus Infection Causes Neuronal Death in the Absence of Encephalitis in Mice Transgenic for Human ACE2. J. Virol. 2008, 82, 7264–7275. [Google Scholar] [CrossRef]
- Reinhold, D.; Farztdinov, V.; Yan, Y.; Meisel, C.; Sadlowski, H.; Kühn, J.; Perschel, F.H.; Endres, M.; Düzel, E.; Vielhaber, S.; et al. The brain reacting to COVID-19: Analysis of the cerebrospinal fluid proteome, RNA and inflammation. J. Neuroinflamm. 2023, 20, 30. [Google Scholar] [CrossRef]
- Chen, R.; Wang, K.; Yu, J.; Howard, D.; French, L.; Chen, Z.; Wen, C.; Xu, Z. The spatial and cell-type distribution of SARS-CoV-2 receptor ACE2 in the human and mouse brains. Front. Neurol. 2021, 11, 573095. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Lazartigues, E. Angiotensin-converting enzyme 2 in the brain: Properties and future directions. J. Neurochem. 2008, 107, 1482–1494. [Google Scholar] [CrossRef] [PubMed]
- Doobay, M.F.; Talman, L.S.; Obr, T.D.; Tian, X.; Davisson, R.L.; Lazartigues, E.; Santos, R.A.S.; Sampaio, W.O.; Alzamora, A.C.; Motta-Santos, D.; et al. Differential expression of neuronal ACE2 in transgenic mice with overexpression of the brain renin-angiotensin system. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007, 292, R373–R381. [Google Scholar] [CrossRef]
- Xiao, L.; Haack, K.K.V.; Zucker, I.H. Angiotensin II regulates ACE and ACE2 in neurons through p38 mitogen-activated protein kinase and extracellular signal-regulated kinase 1/2 signaling. Am. J. Physiol. Cell Physiol. 2013, 304, C1073–C1079. [Google Scholar] [CrossRef]
- Zubair, A.S.; McAlpine, L.S.; Gardin, T.; Farhadian, S.; Kuruvilla, D.E.; Spudich, S. Neuropathogenesis and Neurologic Manifestations of the Coronaviruses in the Age of Coronavirus Disease 2019. JAMA Neurol. 2020, 77, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dubé, M.; Talbot, P.J. Human Coronaviruses and Other Respiratory Viruses: Underestimated Opportunistic Pathogens of the Central Nervous System? Viruses 2019, 12, 14. [Google Scholar] [CrossRef]
- Swanson, P.A.; McGavern, D.B. Viral diseases of the central nervous system. Curr. Opin. Virol. 2015, 11, 44–54. [Google Scholar] [CrossRef]
- Desforges, M.; Le Coupanec, A.; Stodola, J.K.; Meessen-Pinard, M.; Talbot, P.J. Human coronaviruses: Viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res. 2014, 194, 145–158. [Google Scholar] [CrossRef]
- Guo, Y.; Korteweg, C.; McNutt, M.A.; Gu, J. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008, 133, 4–12. [Google Scholar] [CrossRef]
- Baig, A.M.; Khaleeq, A.; Ali, U.; Syeda, H. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host–Virus Interaction, and Proposed Neurotropic Mechanisms. ACS Chem. Neurosci. 2020, 11, 995–998. [Google Scholar] [CrossRef]
- Uversky, V.N.; Elrashdy, F.; Aljadawi, A.; Ali, S.M.; Khan, R.H.; Redwan, E.M. Severe acute respiratory syndrome coronavirus 2 infection reaches the human nervous system: How? J. Neurosci. Res. 2021, 99, 750–777. [Google Scholar] [CrossRef] [PubMed]
- Ponde, N.O.; Shoger, K.E.; Khatun, M.S.; Sarkar, M.K.; Dey, I.; Taylor, T.C.; Cisney, R.N.; Arunkumar, S.P.; Gudjonsson, J.E.; Kolls, J.K.; et al. SARS-CoV-2 ORF8 Mediates Signals in Macrophages and Monocytes through MyD88 Independently of the IL-17 Receptor. J. Immunol. 2023, 211, 252–260. [Google Scholar] [CrossRef] [PubMed]
- van Riel, D.; Verdijk, R.; Kuiken, T. The olfactory nerve: A shortcut for influenza and other viral diseases into the central nervous system. J. Pathol. 2015, 235, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, D.; Zhou, H.; Liu, J.; Chen, S. Guillain-Barre syndrome associated with SARS-CoV-2 infection: Causality or coincidence? Lancet Neurol. 2020, 19, 383–384. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, X.; Chen, Z.; Duan, J.; Hashimoto, K.; Yang, L.; Liu, C.; Yang, C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav. Immun. 2020, 87, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Qiang, T.; Liang, C.; Ren, X.; Jia, M.; Zhang, J.; Li, J.; Wan, M.; Wen, X.Y.; Li, H.; et al. RNA-dependent RNA polymerase (RdRp) inhibitors: The current landscape and repurposing for the COVID-19 pandemic. Eur. J. Med. Chem. 2021, 213, 113201. [Google Scholar] [CrossRef]
- Ianevski, A.; Zusinaite, E.; Kuivanen, S.; Strand, M.; Lysvand, H.; Teppor, M.; Kakkola, L.; Paavilainen, H.; Laajala, M.; Kallio-Kokko, H.; et al. Novel activities of safe-in-human broad-spectrum antiviral agents. Antivir. Res. 2018, 154, 174–182. [Google Scholar] [CrossRef]
- Kumar, D.; Trivedi, N. Disease-drug and drug-drug interaction in COVID-19: Risk and assessment. Biomed. Pharmacother. 2021, 139, 111642. [Google Scholar] [CrossRef]
- Identification of an Existing Japanese Pancreatitis Drug, Nafamostat, Which Is Expected to Prevent the Transmission of New Coronavirus Infection (COVID-19). The University of Tokyo. 2020. Available online: https://www.u-tokyo.ac.jp/focus/en/articles/z0508_00083.html (accessed on 18 September 2023).
- Li, K.; Meyerholz, D.K.; Bartlett, J.A.; McCray, P.B. The TMPRSS2 Inhibitor Nafamostat Reduces SARS-CoV-2 Pulmonary Infection in Mouse Models of COVID-19. Mbio 2021, 12, e0097021. [Google Scholar] [CrossRef]
- Christian, A.; Rolain, R.; Colson, P.; Raoult, D. New Insights on the Antiviral Effects of Chloroquine against Coronavirus: What to Expect for COVID-19. Int. J. Antimicrob. Agents 2020, 55, 105938. [Google Scholar] [CrossRef]
- Ou, T.; Mou, H.; Zhang, L.; Ojha, A.; Choe, H.; Farzan, M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021, 17, e1009212. [Google Scholar] [CrossRef] [PubMed]
- Goldman, F.D.; Gilman, A.L.; Hollenback, C.; Kato, R.M.; Premack, B.A.; Rawlings, D.J. Hydroxychloroquine Inhibits Calcium Signals in T Cells: A New Mechanism to Explain Its Immunomodulatory Properties. Blood, Content Repository Only! 14 December 2020. Available online: https://www.sciencedirect.com/science/article/pii/S0006497120640113?via%3Dihub (accessed on 18 September 2023).
- Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Chloroquine and Hydroxychloroquine. US FDA 2020. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-chloroquine-and (accessed on 18 September 2023).
- Pardo, J.; Shukla, A.M.; Chamarthi, G.; Gupte, A. The journey of remdesivir: From Ebola to COVID-19. Drugs Context 2020, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Remdesivir-List Results. Home-Clinical Trials.gov 2020. Available online: Clinicaltrials.gov/ct2/results?term=remedesivir (accessed on 18 September 2023).
- Dangerfield, T.L.; Huang, N.Z.; Johnson, K.A. Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral RNA-Dependent RNA Polymerase. iScience 2020, 23, 101849. [Google Scholar] [CrossRef]
- Goldman, J.D.; Lye, D.C.; Hui, D.S.; Marks, K.M.; Bruno, R.; Montejano, R.; Spinner, C.D.; Galli, M.; Ahn, M.-Y.; Nahass, R.G.; et al. Remdesivir for 5 or 10 Days in Patients with Severe COVID-19. N. Engl. J. Med. 2020, 383, 1827–1837. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef]
- Yousefi, B.; Valizadeh, S.; Ghaffari, H.; Vahedi, A.; Karbalaei, M.; Eslami, M. A global treatments for coronaviruses including COVID-19. J. Cell. Physiol. 2020, 235, 9133–9142. [Google Scholar] [CrossRef]
- Martinez, M.A. Compounds with Therapeutic Potential against Novel Respiratory 2019 Coronavirus. Antimicrob. Agents Chemother. 2020, 64, e00399-20. [Google Scholar] [CrossRef]
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Chan, K.S.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Momattin, H.; Dib, J.; Memish, Z.A. Ribavirin and interferon therapy in patients infected with the Middle East respiratory syndrome coronavirus: An observational study. Int. J. Infect. Dis. 2014, 20, 42–46. [Google Scholar] [CrossRef]
- Hung, I.F.-N.; Lung, K.-C.; Tso, E.Y.-K.; Liu, R.; Chung, T.W.-H.; Chu, M.-Y.; Ng, Y.-Y.; Lo, J.; Chan, J.; Tam, A.R.; et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: An open-label, randomised, phase 2 trial. Lancet 2020, 395, 1695–1704. [Google Scholar] [CrossRef]
- Information on COVID-19 Treatment, Prevention and Research. NIH US 2021. Available online: www.covid19treatmentguidelines.nih.gov/ (accessed on 18 September 2023).
- Sallard, E.; Lescure, F.-X.; Yazdanpanah, Y.; Mentre, F.; Peiffer-Smadja, N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020, 178, 104791. [Google Scholar] [CrossRef] [PubMed]
- Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. Treatment of SARS with human interferons. Lancet 2003, 362, 293–294. [Google Scholar] [CrossRef] [PubMed]
- Falzarano, D.; de Wit, E.; Rasmussen, A.L.; Feldmann, F.; Okumura, A.; Scott, D.P.; Brining, D.; Bushmaker, T.; Martellaro, C.; Baseler, L.; et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat. Med. 2013, 19, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Chan, K.-H.; Kao, R.Y.; To, K.K.; Zheng, B.-J.; Li, C.P.; Li, P.T.; Dai, J.; Mok, F.K.; Chen, H.; et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Infect. 2013, 67, 606–616. [Google Scholar] [CrossRef]
- Kalil, A.C.; Mehta, A.K.; Patterson, T.F.; Erdmann, N.; Gomez, C.A.; Jain, M.K.; Wolfe, C.R.; Ruiz-Palacios, G.M.; Kline, S.; Pineda, J.R.; et al. Efficacy of interferon beta-1a plus remdesivir compared with remdesivir alone in hospitalised adults with COVID-19: A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Respir. Med. 2021, 9, 1365–1376. [Google Scholar] [CrossRef]
- WHO Solidarity Trial Consortium; Pan, H.; Peto, R.; Henao-Restrepo, A.M.; Preziosi, M.P.; Sathiyamoorthy, V. Repurposed Antiviral Drugs for COVID-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. [Google Scholar] [CrossRef]
- Wang, B.; Li, D.; Liu, T.; Wang, H.; Luo, F.; Liu, Y. Subcutaneous injection of IFN alpha-2b for COVID-19: An observational study. BMC Infect. Dis. 2020, 20, 723. [Google Scholar] [CrossRef]
- Pandit, A.; Bhalani, N.; Bhushan, B.S.; Koradia, P.; Gargiya, S.; Bhomia, V.; Kansagra, K. Efficacy and safety of pegylated interferon alfa-2b in moderate COVID-19: A phase II, randomized, controlled, open-label study. Int. J. Infect. Dis. 2021, 105, 516–521. [Google Scholar] [CrossRef]
- Yao, T.; Qian, J.; Zhu, W.; Wang, Y.; Wang, G. A systematic review of lopinavir therapy for SARS coronavirus and MERS coronavirus—A possible reference for coronavirus disease-19 treatment option. J. Med. Virol. 2020, 92, 556–563. [Google Scholar] [CrossRef]
- Srivastava, K.; Singh, M.K. Drug repurposing in COVID-19: A review with past, present and future. Metab. Open 2021, 12, 100121. [Google Scholar] [CrossRef]
- Neil, O. Triple Antiviral Combo May Speed COVID-19 Recovery. Medscape. 2020. Available online: www.medscape.com/viewarticle/930336 (accessed on 18 September 2023).
- Deng, J.; Zhou, F.; Hou, W.; Heybati, K.; Ali, S.; Chang, O.; Silver, Z.; Dhivagaran, T.; Ramaraju, H.B.; Wong, C.Y.; et al. Efficacy of lopinavir–ritonavir combination therapy for the treatment of hospitalized COVID-19 patients: A meta-analysis. Future Virol. 2022, 17, 169–189. [Google Scholar] [CrossRef] [PubMed]
- Leneva, I.; Kartashova, N.; Poromov, A.; Gracheva, A.; Korchevaya, E.; Glubokova, E.; Borisova, O.; Shtro, A.; Loginova, S.; Shchukina, V.; et al. Antiviral Activity of Umifenovir In Vitro against a Broad Spectrum of Coronaviruses, Including the Novel SARS-CoV-2 Virus. Viruses 2021, 13, 1665. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus (COVID-19) ACEi/ARB Investigation. NIH 2020. ClinicalTrials.gov. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT04330300 (accessed on 18 September 2023).
- Deng, L.; Li, C.; Zeng, Q.; Liu, X.; Li, X.; Zhang, H.; Hong, Z.; Xia, J. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J. Infect. 2020, 81, e1–e5. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xie, Z.; Lin, W.; Cai, W.; Wen, C.; Guan, Y.; Mo, X.; Wang, J.; Wang, Y.; Peng, P.; et al. Efficacy and Safety of Lopinavir/Ritonavir or Arbidol in Adult Patients with Mild/Moderate COVID-19: An Exploratory Randomized Controlled Trial. Med 2020, 1, 105–113.e4. [Google Scholar] [CrossRef]
- Trivedi, N.; Verma, A.; Kumar, D. Possible treatment and strategies for COVID-19: Review and assessment. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12593–12608. [Google Scholar] [CrossRef]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013, 100, 446–454. [Google Scholar] [CrossRef]
- Shiraki, K.; Daikoku, T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020, 209, 107512. [Google Scholar] [CrossRef]
- Madelain, V.; Nguyen, T.H.T.; Olivo, A.; de Lamballerie, X.; Guedj, J.; Taburet, A.-M.; Mentré, F. Ebola Virus Infection: Review of the Pharmacokinetic and Pharmacodynamic Properties of Drugs Considered for Testing in Human Efficacy Trials. Clin. Pharmacokinet. 2016, 55, 907–923. [Google Scholar] [CrossRef]
- Sangawa, H.; Komeno, T.; Nishikawa, H.; Yoshida, A.; Takahashi, K.; Nomura, N.; Furuta, Y. Mechanism of Action of T-705 Ribosyl Triphosphate against Influenza Virus RNA Polymerase. Antimicrob. Agents Chemother. 2013, 57, 5202–5208. [Google Scholar] [CrossRef]
- Srinivasan, K.; Rao, M. Understanding the clinical utility of favipiravir (T-705) in coronavirus disease of 2019: A review. Ther. Adv. Infect. Dis. 2021, 8, 2049936121106301. [Google Scholar] [CrossRef]
- Shah, P.L.; Orton, C.M.; Grinsztejn, B.; Donaldson, G.C.; Ramírez, B.C.; Tonkin, J.; Santos, B.R.; Cardoso, S.W.; Ritchie, A.I.; Conway, F.; et al. Favipiravir in patients hospitalised with COVID-19 (PIONEER trial): A multicentre, open-label, phase 3, randomised controlled trial of early intervention versus standard care. Lancet Respir. Med. 2023, 11, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Zhang, Q.-Y.; Li, X.-D.; Xiong, J.; Xiao, S.-Q.; Wang, Z.; Zhang, Z.-R.; Deng, C.-L.; Yang, X.-L.; Wei, H.-P.; et al. Gemcitabine, Lycorine and Oxysophoridine Inhibit Novel Coronavirus (SARS-CoV-2) in Cell Culture. Emerg. Microbes Infect. 2020, 9, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Gemcitabine Hydrochloride for the Treatment of SARS-CoV-2—Creative Biolabs. Creative BioLabs 2020. Available online: sars-cov-2.creative-biolabs.com/gemcitabine-hydrochloride-for-the-treatment-of-sars-cov2.htm (accessed on 18 September 2023).
- Jang, Y.; Shin, J.S.; Lee, M.K.; Jung, E.; An, T.; Kim, U.-I.; Kim, K.; Kim, M. Comparison of Antiviral Activity of Gemcitabine with 2′-Fluoro-2′-Deoxycytidine and Combination Therapy with Remdesivir against SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 1581. [Google Scholar] [CrossRef]
- Chen, X.; Xu, Z.; Zeng, S.; Wang, X.; Liu, W.; Qian, L.; Wei, J.; Yang, X.; Shen, Q.; Gong, Z.; et al. The Molecular Aspect of Antitumor Effects of Protease Inhibitor Nafamostat Mesylate and Its Role in Potential Clinical Applications. Front. Oncol. 2019, 9, 852. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Schroeder, S.; Kleine-Weber, H.; Müller, M.A.; Drosten, C.; Pöhlmann, S. Nafamostat Mesylate Blocks Activation of SARS-CoV-2: New Treatment Option for COVID-19. Antimicrob. Agents Chemother. 2020, 64, e00754-20. [Google Scholar] [CrossRef]
- Doi, K.; the COVID-UTH Study Group; Ikeda, M.; Hayase, N.; Moriya, K.; Morimura, N. Nafamostat Mesylate Treatment in Combination with Favipiravir for Patients Critically Ill with COVID-19: A Case Series. Crit. Care 2020, 24, 392. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Mitre, M.P.; Tong, S.Y.C.; Denholm, J.T.; Dore, G.J.; Bowen, A.C.; Lewin, S.R.; Venkatesh, B.; Hills, T.E.; McQuilten, Z.; Paterson, D.L.; et al. Nafamostat Mesylate for Treatment of COVID-19 in Hospitalised Patients: A Structured, Narrative Review. Clin. Pharmacokinet. 2022, 61, 1331–1343. [Google Scholar] [CrossRef]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
- Shabani, M.; Ehdaei, B.S.; Fathi, F.; Dowran, R. A mini-review on sofosbuvir and daclatasvir treatment in coronavirus disease 2019. New Microbes New Infect. 2021, 42, 100895. [Google Scholar] [CrossRef]
- Maia, I.S.; Marcadenti, A.; Veiga, V.C.; Miranda, T.A.; Gomes, S.P.; Carollo, M.B.; Negrelli, K.L.; Gomes, J.O.; Tramujas, L.; Abreu-Silva, E.O.; et al. Antivirals for adult patients hospitalised with SARS-CoV-2 infection: A randomised, phase II/III, multicentre, placebo-controlled, adaptive study, with multiple arms and stages. COALITION COVID-19 BRAZIL IX—REVOLUTIOn trial. Lancet Reg. Health Am. 2023, 20, 100466. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Zhou, S.; Graham, R.L.; Pruijssers, A.J.; Agostini, M.L.; Leist, S.R.; Schäfer, A.; Dinnon, K.H., 3rd; Stevens, L.J.; et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020, 12, eabb5883. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Pang, Z.; Li, M.; Lou, F.; An, X.; Zhu, S.; Song, L.; Tong, Y.; Fan, H.; Fan, J. Molnupiravir and Its Antiviral Activity Against COVID-19. Front. Immunol. 2022, 13, 855496. [Google Scholar] [CrossRef] [PubMed]
- Agostini, M.L.; Pruijssers, A.J.; Chappell, J.D.; Gribble, J.; Lu, X.; Andres, E.L.; Bluemling, G.R.; Lockwood, M.A.; Sheahan, T.P.; Sims, A.C.; et al. Small-Molecule Antiviral β-d-N4-Hydroxycytidine Inhibits a Proofreading-Intact Coronavirus with a High Genetic Barrier to Resistance. J. Virol. 2019, 93, e01348-19. [Google Scholar] [CrossRef] [PubMed]
- Victor, A. Bringing the Fight to COVID-19 with Accelerated Clinical Trials of Merck’s Molnupiravir. Insights From Our Labs to Yours, 9 December 2021. Available online: https://ddblog.labcorp.com/2021/12/bringing-the-fight-to-covid-19-with-accelerated-clinical-trials-of-mercks-molnupiravir/ (accessed on 18 September 2023).
- Fischer, W.; Eron, J.J.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.; Mollan, K.R.; Wolfe, C.R.; et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2022, 14, eabl7430. [Google Scholar] [CrossRef]
- Johnson, D.M.; Brasel, T.; Massey, S.; Garron, T.; Grimes, M.; Smith, J.; Torres, M.; Wallace, S.; Villasante-Tezanos, A.; Beasley, D.W.; et al. Evaluation of molnupiravir (EIDD-2801) efficacy against SARS-CoV-2 in the rhesus macaque model. Antivir. Res. 2023, 209, 105492. [Google Scholar] [CrossRef]
- Taylor, R.; Kotian, P.; Warren, T.; Panchal, R.; Bavari, S.; Julander, J.; Dobo, S.; Rose, A.; El-Kattan, Y.; Taubenheim, B.; et al. BCX4430—A broad-spectrum antiviral adenosine nucleoside analog under development for the treatment of Ebola virus disease. J. Infect. Public Health 2016, 9, 220–226. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef]
- Julander, J.G.; Demarest, J.F.; Taylor, R.; Gowen, B.B.; Walling, D.M.; Mathis, A.; Babu, Y. An update on the progress of galidesivir (BCX4430), a broad-spectrum antiviral. Antivir. Res. 2021, 195, 105180. [Google Scholar] [CrossRef]
- Mei, M.; Tan, X. Current Strategies of Antiviral Drug Discovery for COVID-19. Front. Mol. Biosci. 2021, 8, 671263. [Google Scholar] [CrossRef]
- BioCryst Stops COVID-19 Work to Target Other Viral R&D. NC Biotech. 28 December 2020. Available online: https://www.ncbiotech.org/news/biocryst-stops-covid-19-work-target-other-viral-rd (accessed on 18 September 2023).
- Is IV Vitamin C Effective against COVID-19? Linus Pauling Institute 2020. Available online: lpi.oregonstate.edu/COVID19/IV-VitaminC-virus (accessed on 18 September 2023).
- Kow, C.S.; Hasan, S.S.; Ramachandram, D.S. The effect of vitamin C on the risk of mortality in patients with COVID-19: A systematic review and meta-analysis of randomized controlled trials. Inflammopharmacology 2023, 31, 1–6, Epub ahead of print. [Google Scholar] [CrossRef]
- Moore, A.; Khanna, D. The Role of Vitamin C in Human Immunity and Its Treatment Potential Against COVID-19: A Review Article. Cureus 2023, 15, e33740. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Ahmet, A.; Ward, L.; Krishnamoorthy, P.; Mandelcorn, E.D.; Leigh, R.; Brown, J.P.; Cohen, A.; Kim, H. A practical guide to the monitoring and management of the complications of systemic corticosteroid therapy. Allergy Asthma Clin. Immunol. 2013, 9, 30. [Google Scholar] [CrossRef]
- Dexamethasone: MedlinePlus Drug Information. Medline Plus US National Library of Medicine 2021. Available online: medlineplus.gov/druginfo/meds/a682792.html (accessed on 18 September 2023).
- RECOVERY Collaborative Group; Horby, P.; Lim, W.S.; Emberson, J.R.; Mafham, M.; Bell, J.L.; Linsell, L.; Staplin, N.; Brightling, C.; Ustianowski, A.; et al. Dexamethasone in Hospitalized Patients with COVID-19. N. Engl. J. Med. 2021, 384, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Coronavirus Disease (COVID-19): Corticosteroids, Including Dexamethasone. 2023. Available online: https://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-covid-19-dexamethasone (accessed on 18 September 2023).
- Ssentongo, P.; Yu, N.; Voleti, N.; Reddy, S.; Ingram, D.; Chinchilli, V.M.; Paules, C.I. Optimal Duration of Systemic Corticosteroids in Coronavirus Disease 2019 Treatment: A Systematic Review and Meta-analysis. Open Forum Infect. Dis. 2023, 10, ofad105. [Google Scholar] [CrossRef] [PubMed]
- Lilly Earns First Authorization for Novel Antibody Therapy for COVID-19. c&en 2021. Available online: https://cen.acs.org/pharmaceuticals/biologics/Lilly-earns-first-authorization-novel/98/i44 (accessed on 18 September 2023).
- Dougan, M.; Nirula, A.; Azizad, M.; Mocherla, B.; Gottlieb, R.L.; Chen, P.; Hebert, C.; Perry, R.; Boscia, J.; Heller, B.; et al. Bamlanivimab plus Etesevimab in Mild or Moderate COVID-19. N. Engl. J. Med. 2021, 385, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Commissioner, Office of the. “Coronavirus (COVID-19) Update: FDA Revokes Emergency Use Authorization for Monoclonal Antibody Bamlanivimab”. U.S. Food and Drug Administration, FDA. 2021. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-revokes-emergency-use-authorization-monoclonal-antibody-bamlanivimab (accessed on 18 September 2023).
- Center for Drug Evaluation and Research. FDA Authorizes Bamlanivimab and Etesevimab for COVID-19. U.S. Food and Drug Administration, FDA. 16 September 2021. Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-bamlanivimab-and-etesevimab-monoclonal-antibody-therapy-post-exposure-prophylaxis (accessed on 18 September 2023).
- Emergency Use Authorization (EUA) for the Treatment or Post-Exposure Prophylaxis of COVID-19. Bamlanivimab and Etesevimab EUA|Lilly COVID-19 Products. Available online: https://www.covid19.lilly.com/bam-ete (accessed on 18 September 2023).
- Azithromycin: Medline Plus Drug Information. Medline Plus US National Library of Medicine. 2021. Available online: medlineplus.gov/druginfo/meds/a697037.html (accessed on 18 September 2023).
- Cavalcanti, A.B.; Zampieri, F.G.; Rosa, R.G.; Azevedo, L.C.; Veiga, V.C.; Avezum, A.; Damiani, L.P.; Marcadenti, A.; Kawano-Dourado, L.; Lisboa, T.; et al. Hydroxychloroquine with or without Azithromycin in Mild-to-Moderate COVID-19. N. Engl. J. Med. 2020, 383, 2041–2052. [Google Scholar] [CrossRef]
- Kamel, A.M.; Monem, M.S.A.; Sharaf, N.A.; Magdy, N.; Farid, S.F. Efficacy and safety of azithromycin in COVID-19 patients: A systematic review and meta-analysis of randomized clinical trials. Rev. Med. Virol. 2022, 32, e2258. [Google Scholar] [CrossRef]
- What Antibiotics Kill COVID-19 (Coronavirus)? Drugs.com. 1 December 2021. Available online: https://www.drugs.com/medical-answers/antibiotics-kill-coronavirus-3534867/ (accessed on 18 September 2023).
- Durojaiye, A.B.; Clarke, J.-R.D.; Stamatiades, G.A.; Wang, C. Repurposing cefuroxime for treatment of COVID-19: A scoping review of in silico studies. J. Biomol. Struct. Dyn. 2021, 39, 4547–4554. [Google Scholar] [CrossRef]
- Al-Khafaji, K.; AL-DuhaidahawiL, D.; Tok, T.T. Using integrated computational approaches to identify safe and rapid treatment for SARS-CoV-2. J. Biomol. Struct. Dyn. 2020, 39, 3387–3395. [Google Scholar] [CrossRef]
- Banerjee, R.; Perera, L.; Tillekeratne, L.V. Potential SARS-CoV-2 main protease inhibitors. Drug Discov. Today 2021, 26, 804–816. [Google Scholar] [CrossRef]
- Koulgi, S.; Jani, V.; Uppuladinne, M.; Sonavane, U.; Nath, A.K.; Darbari, H.; Joshi, R. Drug repurposing studies targeting SARS-CoV-2: An ensemble docking approach on drug target 3C-like protease (3CLpro). J. Biomol. Struct. Dyn. 2020, 39, 5735–5755. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.S.; Mundra, S. Increasing Consumption of Antibiotics during the COVID-19 Pandemic: Implications for Patient Health and Emerging Anti-Microbial Resistance. Antibiotics 2022, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Gao, Y.; Cao, W.; Liu, J.; Guan, H.; Zhang, H.; Shi, Y.; Lv, W.; Cheng, L. Retrospective analysis of relationships among the dose regimen, trough concentration, efficacy, and safety of teicoplanin in Chinese patients with moder-ate-severe Gram-positive infections. Infect. Drug Resist. 2018, 11, 29–36. [Google Scholar] [CrossRef]
- Zhou, N.; Pan, T.; Zhang, J.; Li, Q.; Zhang, X.; Bai, C.; Huang, F.; Peng, T.; Zhang, J.; Liu, C.; et al. Glycopeptide Antibiotics Potently Inhibit Cathepsin L in the Late Endosome/Lysosome and Block the Entry of Ebola Virus, Middle East Respiratory Syndrome Coronavirus (MERS-CoV), and Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV). J. Biol. Chem. 2016, 291, 9218–9232. [Google Scholar] [CrossRef] [PubMed]
- Yasar, Z.; Yemisen, M.; Yasar, H.; Ertaş, A.; Meric, K.; Sahin, S. Can treatment with teicoplanin improve the prognosis of COVID-19 patients? Int. J. Clin. Pract. 2021, 75, e14752. [Google Scholar] [CrossRef]
- Ma, L.; Li, Y.; Shi, T.; Zhu, Z.; Zhao, J.; Xie, Y.; Wen, J.; Guo, S.; Wang, J.; Ding, J.; et al. Teicoplanin derivatives block spike protein mediated viral entry as pan-SARS-CoV-2 inhibitors. Biomed. Pharmacother. 2023, 158, 114213. [Google Scholar] [CrossRef]
- Sharif-Askari, N.S.; Sharif-Askari, F.S.; Al Heialy, S.; Hamoudi, R.; Kashour, T.; Hamid, Q.; Halwani, R. Cardiovascular medi-cations and regulation of COVID-19 receptors expression. Int. J. Cardiol. Hypertens. 2020, 6, 100034. [Google Scholar] [CrossRef]
- Biyani, C.S.; Palit, V.; Daga, S. The Use of Captopril—Angiotensin Converting Enzyme (ACE) Inhibitor for Cystinuria During COVID-19 Pandemic. Urology 2020, 141, 182–183. [Google Scholar] [CrossRef]
- Pedrosa, M.A.; Valenzuela, R.; Garrido-Gil, P.; Labandeira, C.M.; Navarro, G.; Franco, R.; Labandeira-Garcia, J.L.; Rodriguez-Perez, A.I. Experimental data using candesartan and captopril indicate no double-edged sword effect in COVID-19. Clin. Sci. 2021, 135, 465–481. [Google Scholar] [CrossRef]
- Oudit, G.Y.; Wang, K.; Viveiros, A.; Kellner, M.J.; Penninger, J.M. Angiotensin-converting enzyme 2—At the heart of the COVID-19 pandemic. Cell 2023, 186, 906–922. [Google Scholar] [CrossRef]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Del Pozo, C.H.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef] [PubMed]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angioten-sin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Ajmera, V.; Thompson, W.K.; Smith, D.M.; Malhotra, A.; Mehta, R.L.; Tolia, V.; Yin, J.; Sriram, K.; Insel, P.A.; Collier, S.; et al. RAMIC: Design of a randomized, double-blind, placebo-controlled trial to evaluate the efficacy of ramipril in patients with COVID-19. Contemp. Clin. Trials 2021, 103, 106330. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, M.; Patel, J.; Ahmad, F. Ramipril [Updated 26 July 2021]. Treasure Island 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK537119/ (accessed on 18 September 2023).
- Singh, R.; Rathore, S.S.; Khan, H.; Bhurwal, A.; Sheraton, M.; Ghosh, P.; Anand, S.; Makadia, J.; Ayesha, F.; Mahapure, K.S.; et al. Mortality and Severity in COVID-19 Patients on ACEIs and ARBs—A Systematic Review, Meta-Analysis, and Meta-Regression Analysis. Front. Med. 2022, 8, 703661. [Google Scholar] [CrossRef]
- Nakhaie, S.; Yazdani, R.; Shakibi, M.; Torabian, S.; Pezeshki, S.; Bazrafshani, M.S.; Azimi, M.; Salajegheh, F. The effects of antihypertensive medications on severity and outcomes of hypertensive patients with COVID-19. J. Hum. Hypertens. 2022, 37, 1–8. [Google Scholar] [CrossRef]
- Song, S.-N.J.; Yoshizaki, K. Tocilizumab for treating rheumatoid arthritis: An evaluation of pharmacokinetics/pharmacodynamics and clinical efficacy. Expert Opin. Drug Metab. Toxicol. 2015, 11, 307–316. [Google Scholar] [CrossRef]
- Xu, X.; Han, M.; Li, T.; Sun, W.; Wang, D.; Fu, B.; Zhou, Y.; Zheng, X.; Yang, Y.; Li, X.; et al. Effective Treatment of Severe COVID-19 Patients with Tocilizumab. Proc. Natl. Acad. Sci. USA 2020, 117, 10970–10975. [Google Scholar] [CrossRef]
- FDA Approves Genentech’s Actemra for the Treatment of COVID-19 in Hospitalized Adults. Business Wire. News Release. 21 December 2022. Available online: https://www.businesswire.com/news/home/20221221005002/en (accessed on 22 December 2022).
- Montazersaheb, S.; Khatibi, S.M.H.; Hejazi, M.S.; Tarhriz, V.; Farjami, A.; Sorbeni, F.G.; Farahzadi, R.; Ghasemnejad, T. COVID-19 infection: An overview on cytokine storm and related interventions. Virol. J. 2022, 19, 92. [Google Scholar] [CrossRef]
- Kyriakopoulos, C.; Ntritsos, G.; Gogali, A.; Milionis, H.; Evangelou, E.; Kostikas, K. Tocilizumab administration for the treatment of hospitalized patients with COVID-19: A systematic review and meta-analysis. Respirology 2021, 26, 1027–1040. [Google Scholar] [CrossRef]
- Baskar, S.; Klein, A.L.; Zeft, A. The Use of IL-1 Receptor Antagonist (Anakinra) in Idiopathic Recurrent Pericarditis: A Narrative Review. Cardiol. Res. Pract. 2016, 2016, 7840724. [Google Scholar] [CrossRef]
- Khani, E.; Shahrabi, M.; Rezaei, H.; Pourkarim, F.; Afsharirad, H.; Solduzian, M. Current evidence on the use of anakinra in COVID-19. Int. Immunopharmacol. 2022, 111, 109075. [Google Scholar] [CrossRef] [PubMed]
- Dahms, K.; Mikolajewska, A.; Ansems, K.; Metzendorf, M.-I.; Benstoem, C.; Stegemann, M. Anakinra for the treatment of COVID-19 patients: A systematic review and meta-analysis. Eur. J. Med. Res. 2023, 28, 100. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorganic Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Trougakos, I.P.; Stamatelopoulos, K.; Terpos, E.; Tsitsilonis, O.E.; Aivalioti, E.; Paraskevis, D.; Kastritis, E.; Pavlakis, G.N.; Dimopoulos, M.A. Insights to SARS-CoV-2 life cycle, pathophysiology, and rationalized treatments that target COVID-19 clinical complications. J. Biomed. Sci. 2021, 28, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Plaze, M.; Attali, D.; Prot, M.; Petit, A.-C.; Blatzer, M.; Vinckier, F.; Levillayer, L.; Chiaravalli, J.; Perin-Dureau, F.; Cachia, A.; et al. Inhibition of the replication of SARS-CoV-2 in human cells by the FDA-approved drug chlorpromazine. Int. J. Antimicrob. Agents 2021, 57, 106274. [Google Scholar] [CrossRef]
- Hufsky, F.; Lamkiewicz, K.; Almeida, A.; Aouacheria, A.; Arighi, C.; Bateman, A.; Baumbach, J.; Beerenwinkel, N.; Brandt, C.; Cacciabue, M.; et al. Computational strategies to combat COVID-19: Useful tools to accelerate SARS-CoV-2 and coronavirus research. Briefings Bioinform. 2021, 22, 642–663. [Google Scholar] [CrossRef]
- Wang, J. Fast Identification of Possible Drug Treatment of Coronavirus Disease-19 (COVID-19) through Computational Drug Repurposing Study. J. Chem. Inf. Model. 2020, 60, 3277–3286. [Google Scholar] [CrossRef]
- Eskandari, V. Repurposing the natural compounds as potential therapeutic agents for COVID-19 based on the molecular docking study of the main protease and the receptor-binding domain of spike protein. J. Mol. Model. 2022, 28, 153. [Google Scholar] [CrossRef]
- Ahsan, T.; Sajib, A.A. Repurposing of approved drugs with potential to interact with SARS-CoV-2 receptor. Biochem. Biophys. Rep. 2021, 26, 100982. [Google Scholar] [CrossRef]
- Borbone, N.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Nucleoside Analogs and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses. Molecules 2021, 26, 986. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Attar, F.; Bloukh, S.H.; Sharifi, M.; Nabi, F.; Bai, Q.; Khan, R.H.; Falahati, M. A review on the interaction of nucleoside analogues with SARS-CoV-2 RNA dependent RNA polymerase. Int. J. Biol. Macromol. 2021, 181, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Clinical Questions about COVID-19: Questions and Answers. CDC. 2020. Available online: www.cdc.gov/coronavirus/2019-ncov/hcp/faq.html (accessed on 18 September 2023).

| Year | Virus Name | Genera | Host/Origin/Intermediate Origin | Cell Receptor | Symptoms |
|---|---|---|---|---|---|
| 1952 | TGEV | α-CoVs | Pig | Porcine APN | Mild Respiratory tract infection and Enteric infection |
| 1966 | HCoV-229E | Human/Bat/Alpaca | Human APN | Mild Respiratory tract infection | |
| 1988 | HCoV-NL63 | Human/Bat/Unkown | ACE2 | Mild Respiratory tract infection | |
| 1961 | MHV-A59 | β-CoVs | Mouse | Murine CEACAMI | Acute pneumonia and severe lung injuries |
| 1967 | HcoV-OC43 | Human/Bat/Cow | Neu5, 9Ac-2-containing moiety | Mild Respiratory tract infection | |
| 2003 | SARS-CoV | Human/Bat/Palm Civets | ACE2 | Severe acute respiratory syndrome, 10% mortality | |
| 2004 | HKU-1 | Human/Mouse/Unkown | HLA | Pneumonia | |
| 2008 | CCoV | Dog/Dog | Canine APN | Diarrhea, Enteric infection | |
| 2012 | MERS-CoV | Human/Bat/Camel | CD26 (DPP4) | Severe acute respiratory syndrome, 37% mortality | |
| 2015 | Bat-SI, CoVZC21 | Bat/Bat | n/a | n/a | |
| 2017 | Bat-SI, CoVZC45 | Bat/Bat | n/a | n/a | |
| 2019 | SARS-CoV-2 | Human/Bat/Unkown | ACE2 | Severe lower respiratory tract infection | |
| 1935 | IBV | γ-CoVs | Chicken/Avian | S glycoprotein | Severe respiratory disease |
| 2008 | SW1 | Whale | n/a | Pulmonary disease, terminal acute liver failure | |
| 2007 | HKU11 | δ-CoVs | Bulbul/Pycocotus/Jocosus | n/a | Respiratory disease collected from respiratory tract of dead birds |
| 2007 | HKU17 | Sparrow/Passer/Montanus | n/a | Respiratory disease collected from respiratory tract of dead birds |
| Nonstructural Proteins | Functions |
|---|---|
| Nsp1 | Degradation of cellular mRNA, inhibition of interferon (IFN) signaling |
| Nsp2 | Unknown |
| Nsp3 | Cleavage of PLP polypeptides, inhibition of IFN signaling |
| Nsp4 | Formation of DMV |
| Nsp5 | Cleavage of 3CLpro, Mpro, polypeptides, inhibition of IFN signaling |
| Nsp6 | Restriction of autophagosome expansion, formation of DMV |
| Nsp7 | Co-factor of holo-RdRp |
| Nsp8 | Co-factor of holo-RdRp |
| Nsp9 | Capping and binding of RNA |
| Nsp10 | Scaffold protein for nsp14 and nsp16 |
| Nsp11 | Unknown |
| Nsp12 | Primer-dependent RdRp |
| Nsp13 | RNA helicase, 5′-triphosphatase |
| Nsp14 | Exoribonuclease, N7 Mtase |
| Nsp15 | Endoribonuclease, evasion of dsRNA sensors |
| Nsp16 | RNA-cap-2′-O-methyltransferase, inhibition of MDA5 recognition, negative regulation of innate immunity |
| Drug | Classification | Target | Treatment of/Usage | FDA Approved for COVID-19 | EUA for COVID-19 | Comments |
|---|---|---|---|---|---|---|
| Actemra (Tocilizumab) | Monoclonal Antibody | Interleukin-6 receptor antagonist | Arthritis, SSc-Interstitial lung disease, Cytokine Release Syndrome | Yes | for adults receiving systemic corticosteroids and require supplemental oxygen, non-invasive/invasive mechanical ventilation, or extracorporeal membrane oxygenation | |
| Veklury (Remdesivir) | Antiviral | RdRp inhibitor | Ebola, SARS, and MERS | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 | |
| Olumiant (Baricitinib) | Immune Modulator | Janus kinase inhibitor | Rheumatoid arthritis | Yes | for hospitalized adults requiring supplemental oxygen, non-invasive/invasive mechanical ventilation, or extracorporeal membrane oxygenation | |
| Paxlovid (Nirmatrelvir & Ritonavir) | Antiviral | Protease inhibitor, CYP3A inhibitor | HIV | No | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 |
| Lagevrio (Molnupiravir) | Antiviral | Viral mutagenesis | Influenza | No | Yes | for adults who have mild-to- moderate and high risk for progression to severe COVID-19 |
| Kineret (Anakinra) | Immune Modulator | Interleukin-1 receptor antagonist | Rheumatoid Arthritis, Cryopyrin-Associated Periodic Syndromes, Deficiency of Interleukin-1 receptor antagonist | No | Yes | for adults with pneumonia requiring supplemental oxygen and at risk for of progressing to severe respiratory failure |
| Gohibic (Vilobelimab) | Immune Modulator | C5a receptor blocker | n/a | No | Yes | for adults when initiated within 48 h of receiving invasive mechanical ventilation or extracorporeal membrane oxygenation |
| REGEN-COV (Casirivimab & Imdevimab) | Monoclonal Antibody | SARS-CoV-2 spike protein binding domain receptor | n/a | No | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 |
| Sotrovimab | Monoclonal Antibody | SARS-CoV-2 spike protein binding domain receptor | n/a | No | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 |
| * Bamlanivimab & Etesevimab | Monoclonal Antibody | SARS-CoV-2 spike protein binding domain receptor | n/a | No | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 |
| Bebtelovimab | Monoclonal Antibody | SARS-CoV-2 spike protein binding domain receptor | n/a | No | Yes | for adults and pediatric patients (with age and weight limitation), have mild-to-moderate and high risk for progression to severe COVID-19 |
| Evusheld (Tixagevimab with Cilgavimab) | Monoclonal Antibody | SARS-CoV-2 spike protein binding domain receptor | n/a | No | Yes | for certain adults and pediatric patients as pre-exposure prophylaxis of COVID-19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awad, A.M.; Hansen, K.; Del Rio, D.; Flores, D.; Barghash, R.F.; Kakkola, L.; Julkunen, I.; Awad, K. Insights into COVID-19: Perspectives on Drug Remedies and Host Cell Responses. Biomolecules 2023, 13, 1452. https://doi.org/10.3390/biom13101452
Awad AM, Hansen K, Del Rio D, Flores D, Barghash RF, Kakkola L, Julkunen I, Awad K. Insights into COVID-19: Perspectives on Drug Remedies and Host Cell Responses. Biomolecules. 2023; 13(10):1452. https://doi.org/10.3390/biom13101452
Chicago/Turabian StyleAwad, Ahmed M., Kamryn Hansen, Diana Del Rio, Derek Flores, Reham F. Barghash, Laura Kakkola, Ilkka Julkunen, and Kareem Awad. 2023. "Insights into COVID-19: Perspectives on Drug Remedies and Host Cell Responses" Biomolecules 13, no. 10: 1452. https://doi.org/10.3390/biom13101452
APA StyleAwad, A. M., Hansen, K., Del Rio, D., Flores, D., Barghash, R. F., Kakkola, L., Julkunen, I., & Awad, K. (2023). Insights into COVID-19: Perspectives on Drug Remedies and Host Cell Responses. Biomolecules, 13(10), 1452. https://doi.org/10.3390/biom13101452







