MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications
Abstract
:1. Introduction
2. Histone-Modifying Enzymes in Chromatin Regulation
- (1)
- Oncogene Activation and Tumor Suppressor Silencing: Histone acetylation and methylation are often associated with gene regulation, influencing the activation or repression of genes, including oncogenes that drive tumorigenesis [112,205]. Mutations or overexpression of HATs and HMTs can lead to the hyperactivation of oncogenes, contributing to uncontrolled cell growth [138,143,206]. Conversely, the silencing of tumor suppressor genes through histone deacetylation and methylation is a hallmark of many cancers. HDACs and histone demethylases can contribute to the epigenetic silencing of genes that regulate cell cycle control and DNA repair [143,201,207].
- (2)
- Epigenetic Plasticity and Drug Resistance: Cancer cells often exhibit epigenetic plasticity, allowing them to adapt to changing environments and develop resistance to therapies [208]. Histone-modifying enzymes contribute to this plasticity by maintaining specific chromatin states that promote drug resistance, making them attractive targets for therapeutic intervention [208,209,210].
- (3)
- Diagnostic and Therapeutic Targets: Aberrant histone modifications and their associated enzymes can serve as potential diagnostic markers for certain cancer types. Furthermore, targeting histone-modifying enzymes with small molecule inhibitors holds promise as a therapeutic strategy to restore normal gene expression patterns in cancer cells [201,211,212].
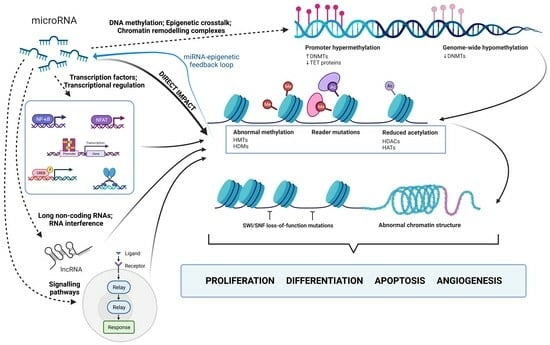
3. MicroRNA-Mediated Regulation of Histone Modifications
- (1)
- Transcription Factors and Transcriptional Regulation: miRNAs can indirectly regulate chromatin remodeling by targeting transcription factors (TFs) or co-regulators involved in chromatin modification [107,313]. When miRNAs target TFs controlling the expression of histone-modifying enzymes, downstream changes in histone modifications occur [107,313,314,315]. MiRNAs, like miRNA-200, indirectly influence histone modifications through interactions with transcription factors. In this case, miRNA-200 interacts with ZEB1 and ZEB2, repressors of E-cadherin, impacting cell adhesion and promoting metastasis [316]. Another example is miR-29b, repressed by the MYC protein in KIT-mutation-associated leukemia. This leads to increased Sp1 expression, which activates KIT gene transcription. Synthetic miR-29b inhibitors disrupt this network, reducing KIT expression and inhibiting leukemia growth [317].
- (2)
- Signaling Pathways and Epigenetic Modulators: Signaling pathways, including Wnt, Notch, and TGF-β, are modulated, which in turn affects epigenetic regulators [12,318,319]. For instance, miR-29 and miR-206 impact the TGF-β pathway and HDAC4 expression, crucial for myogenic genes. Reduced miR-29 and miR-206 levels lead to increased HDAC4 expression by inhibiting its translation. They also regulate the Smad3 levels, a key TGF-β pathway component, impacting muscle cell differentiation. MiR-29 and miR-206 counteract TGF-β’s negative effects on cell commitment, and their overexpression inhibits rhabdomyosarcoma development [320,321]. Interestingly, rhabdomyosarcoma tumors exhibit elevated TGF-β and Smad4, coinciding with our findings that increased TGF-β signaling suppresses these miRNAs, affecting cellular differentiation [320,321,322,323,324].
- (3)
- Long Non-Coding RNAs (lncRNAs) and RNA Interference: LncRNAs act as intermediaries between miRNAs and chromatin remodeling [218,325]. When an miRNA represses an lncRNA, it increases the expression of genes targeted by the lncRNA. LncRNAs interact with chromatin modifiers, indirectly affecting histone modifications [106,218,326]. This intricate regulatory network connects miRNAs and chromatin remodeling, as some miRNAs and lncRNAs share target genes. For example, the lncRNA HOTAIR plays a significant role in cancer progression by affecting prognosis, staging, and multiple cellular processes through miRNA modulation. HOTAIR primarily influences chromatin remodeling and epigenetic changes by acting as a scaffold for histone-modifying protein complexes. It facilitates gene silencing through H3K27 methylation and H3K4 demethylation and is known to promote metastasis by epigenetically silencing the tumor suppressor gene miR-34a. Various miRNAs, including miR-7, miR-206, miR-218, miR-20a-5p, miR-126-5p, and miR-146a-5p, are involved in regulating HOTAIR’s effects [327,328,329,330].
- (4)
- DNA Methylation and Epigenetic Crosstalk: DNA methylation, closely linked to histone modifications, is indirectly influenced by miRNAs targeting DNMTs or DNA demethylation factors. These changes in DNA methylation impact chromatin structure, thereby altering histone modifications and gene expression [331,332,333]. For example, miR-101 inhibits DNMT3A expression, leading to increased DNA methylation in tumor-suppressing gene promoters, affecting chromatin accessibility for histone-modifying enzymes. This results in altered histone modifications, ultimately influencing gene expression. MiR-101 also reverses PRDM16 gene promoter hypomethylation by modifying histones, which are mediated through direct targets such as EZH2, EED, and DNMT3A, suggesting their role in cancer contexts [334].
- (5)
- Chromatin Remodeling Complexes: miRNAs indirectly regulate chromatin remodeling by targeting complex components, impacting chromatin structure and access to DNA. For instance, miR-124 and miR-9 inhibit BAF complex (SWI/SNF) activity by reducing BAF subunit expression [335]. The BAF complex plays a key role in chromatin remodeling and gene expression regulation [334]. Reduced microRNA levels lead to increased BAF expression, affecting complex activity, and ultimately influencing gene expression through chromatin structure [335].
4. Functional Consequences of miRNA-Histone Enzyme Interplay
5. Therapeutic Potential of MicroRNA-Histone Pathways
6. Challenges and Future Directions
7. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ranganathan, K.; Sivasankar, V. MicroRNAs—Biology and clinical applications. J. Oral. Maxillofac. Pathol. 2014, 18, 229–234. [Google Scholar] [PubMed]
- Ratti, M.; Lampis, A.; Ghidini, M.; Salati, M.; Mirchev, M.B.; Valeri, N.; Hahne, J.C. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as new tools for cancer therapy: First steps from bench to bedside. Target. Oncol. 2020, 15, 261–278. [Google Scholar] [PubMed]
- Guzel, E.; Okyay, T.M.; Yalcinkaya, B.; Karacaoglu, S.; Gocmen, M.; Akcakuyu, M.H. Tumor suppressor and oncogenic role of long non-coding RNAs in cancer. North. Clin. Istanb. 2020, 7, 81–86. [Google Scholar] [PubMed]
- Toden, S.; Zumwalt, T.J.; Goel, A. Non-coding RNAs and potential therapeutic targeting in cancer. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188491. [Google Scholar]
- Kovalski, J.R.; Kuzuoglu-Ozturk, D.; Ruggero, D. Protein synthesis control in cancer: Selectivity and therapeutic targeting. EMBO J. 2022, 41, e109823. [Google Scholar]
- Oliveto, S.; Mancino, M.; Manfrini, N.; Biffo, S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017, 8, 45–56. [Google Scholar]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar]
- Gu, S.; Jin, L.; Zhang, F.; Sarnow, P.; Kay, M.A. Biological basis for restriction of microRNA targets to the 3’ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009, 16, 144–150. [Google Scholar]
- Szczepanek, J.; Skorupa, M.; Tretyn, A. MicroRNA as a Potential Therapeutic Molecule in Cancer. Cells 2022, 11, 1008. [Google Scholar]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar]
- Frixa, T.; Donzelli, S.; Blandino, G. Oncogenic microRNAs: Key players in malignant transformation. Cancers 2015, 7, 2466–2485. [Google Scholar] [PubMed]
- Otmani, K.; Lewalle, P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front. Oncol. 2021, 11, 708765. [Google Scholar] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [PubMed]
- Wu, A.; Wu, K.; Li, J.; Mo, Y.; Lin, Y.; Wang, Y.; Shen, X.; Li, S.; Li, L.; Yang, Z. Let-7a inhibits migration, invasion and epithelial-mesenchymal transition by targeting HMGA2 in nasopharyngeal carcinoma. J. Transl. Med. 2015, 13, 105. [Google Scholar]
- Guo, M.; Zhao, X.; Yuan, X.; Jiang, J.; Li, P. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in cervical cancer. Oncotarget 2017, 8, 28226–28236. [Google Scholar]
- Yang, N.; Kaur, S.; Volinia, S.; Greshock, J.; Lassus, H.; Hasegawa, K.; Liang, S.; Leminen, A.; Deng, S.; Smith, L.; et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008, 68, 10307–10314. [Google Scholar]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar]
- Worringer, K.A.; Rand, T.A.; Hayashi, Y.; Sami, S.; Takahashi, K.; Tanabe, K.; Narita, M.; Srivastava, D.; Yamanaka, S. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 2014, 14, 40–52. [Google Scholar]
- Chang, C.J.; Hsu, C.C.; Chang, C.H.; Tsai, L.L.; Chang, Y.C.; Lu, S.W.; Yu, C.H.; Huang, H.S.; Wang, J.J.; Tsai, C.H.; et al. Let-7d functions as novel regulator of epithelial-mesenchymal transition and chemoresistant property in oral cancer. Oncol. Rep. 2011, 26, 1003–1010. [Google Scholar]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Rassenti, L.Z.; Balatti, V.; Ghia, E.M.; Palamarchuk, A.; Tomasello, L.; Fadda, P.; Pekarsky, Y.; Widhopf, G.F., 2nd; Kipps, T.J.; Croce, C.M. MicroRNA dysregulation to identify therapeutic target combinations for chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2017, 114, 10731–10736. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, C.; Cui, M.; Zhang, H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021, 10, 3358–3372. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Song, Y.; Yu, H.; Luo, X. Identification of lncRNA TRPM2-AS/miR-140-3p/PYCR1 axis’s proliferates and anti-apoptotic effect on breast cancer using co-expression network analysis. Cancer Biol. Ther. 2019, 20, 760–773. [Google Scholar] [CrossRef]
- Dong, W.; Yao, C.; Teng, X.; Chai, J.; Yang, X.; Li, B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016, 37, 2973–2985. [Google Scholar] [CrossRef]
- Lan, H.; Chen, W.; He, G.; Yang, S. miR-140-5p inhibits ovarian cancer growth partially by repression of PDGFRA. Biomed. Pharmacother. 2015, 75, 117–122. [Google Scholar] [CrossRef]
- Yuan, Y.; Shen, Y.; Xue, L.; Fan, H. miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS ONE 2013, 8, e73604. [Google Scholar] [CrossRef]
- Elnaggar, G.N.; El-Hifnawi, N.M.; Ismail, A.; Yahia, M.; Elshimy, R.A.A. Micro RNA-148a Targets Bcl-2 in Patients with Non-Small Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Lombard, A.P.; Mooso, B.A.; Libertini, S.J.; Lim, R.M.; Nakagawa, R.M.; Vidallo, K.D.; Costanzo, N.C.; Ghosh, P.M.; Mudryj, M. miR-148a dependent apoptosis of bladder cancer cells is mediated in part by the epigenetic modifier DNMT1. Mol. Carcinog. 2016, 55, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Lv, X.; Su, L.; Li, J.; Yu, Y.; Gu, Q.; Yan, M.; Zhu, Z.; Liu, B. MiR-148a Functions as a Tumor Suppressor by Targeting CCK-BR via Inactivating STAT3 and Akt in Human Gastric Cancer. PLoS ONE 2016, 11, e0158961. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Huang, Q.; Ren, X.; Hu, H.; Sheng, H.; Lai, M. MiR-148a promotes apoptosis by targeting Bcl-2 in colorectal cancer. Cell Death Differ. 2011, 18, 1702–1710. [Google Scholar] [CrossRef]
- Zhang, R.; Li, M.; Zang, W.; Chen, X.; Wang, Y.; Li, P.; Du, Y.; Zhao, G.; Li, L. MiR-148a regulates the growth and apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour Biol. 2014, 35, 837–844. [Google Scholar] [CrossRef]
- Arivazhagan, R.; Lee, J.; Bayarsaikhan, D.; Kwak, P.; Son, M.; Byun, K.; Salekdeh, G.H.; Lee, B. MicroRNA-340 inhibits the proliferation and promotes the apoptosis of colon cancer cells by modulating REV3L. Oncotarget 2018, 9, 5155–5168. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Sun, Y.; Liu, Q. MicroRNA-340 Induces Apoptosis and Inhibits Metastasis of Ovarian Cancer Cells by Inactivation of NF-κB1. Cell. Physiol. Biochem. 2016, 38, 1915–1927. [Google Scholar] [CrossRef]
- Xie, W.; Qin, W.; Kang, Y.; Zhou, Z.; Qin, A. MicroRNA-340 Inhibits Tumor Cell Proliferation and Induces Apoptosis in Endometrial Carcinoma Cell Line RL 95-2. Med. Sci. Monit. 2016, 22, 1540–1546. [Google Scholar] [CrossRef]
- Yu, J.; Wang, R.; Chen, J.; Wu, J.; Dang, Z.; Zhang, Q.; Li, B. miR-340 Inhibits Proliferation and Induces Apoptosis in Gastric Cancer Cell Line SGC-7901, Possibly via the AKT Pathway. Med. Sci. Monit. 2017, 23, 71–77. [Google Scholar] [CrossRef]
- Zhang, L.L.; Xie, F.J.; Tang, C.H.; Xu, W.R.; Ding, X.S.; Liang, J. miR-340 suppresses tumor growth and enhances chemosensitivity of colorectal cancer by targeting RLIP76. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2875–2886. [Google Scholar]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef]
- Feng, H.; Ge, F.; Du, L.; Zhang, Z.; Liu, D. MiR-34b-3p represses cell proliferation, cell cycle progression and cell apoptosis in non-small-cell lung cancer (NSCLC) by targeting CDK4. J. Cell. Mol. Med. 2019, 23, 5282–5291. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Lin, C.P.; Ribeiro, M.C.; Biton, A.; Lai, G.; He, X.; Bu, P.; Vogel, H.; Jablons, D.M.; Keller, A.C.; et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes. Dev. 2014, 28, 438–450. [Google Scholar] [CrossRef] [PubMed]
- Raver-Shapira, N.; Marciano, E.; Meiri, E.; Spector, Y.; Rosenfeld, N.; Moskovits, N.; Bentwich, Z.; Oren, M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 2007, 26, 731–743. [Google Scholar] [CrossRef]
- Rokavec, M.; Oner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef]
- Wang, X.; Li, J.; Dong, K.; Lin, F.; Long, M.; Ouyang, Y.; Wei, J.; Chen, X.; Weng, Y.; He, T.; et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell Signal 2015, 27, 443–452. [Google Scholar] [CrossRef]
- Guo, L.; Chen, C.; Shi, M.; Wang, F.; Chen, X.; Diao, D.; Hu, M.; Yu, M.; Qian, L.; Guo, N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 2013, 32, 5272–5282. [Google Scholar] [CrossRef]
- Pon, J.R.; Marra, M.A. MEF2 transcription factors: Developmental regulators and emerging cancer genes. Oncotarget 2016, 7, 2297–2312. [Google Scholar] [CrossRef]
- He, M.Q.; Wan, J.F.; Zeng, H.F.; Tang, Y.Y.; He, M.Q. miR-133a-5p suppresses gastric cancer through TCF4 down-regulation. J. Gastrointest. Oncol. 2021, 12, 1007–1019. [Google Scholar] [CrossRef]
- Dong, Y.; Zhao, J.; Wu, C.W.; Zhang, L.; Liu, X.; Kang, W.; Leung, W.W.; Zhang, N.; Chan, F.K.; Sung, J.J.; et al. Tumor suppressor functions of miR-133a in colorectal cancer. Mol. Cancer Res. 2013, 11, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Ren, J.; Liu, K.; Tang, L.M. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J. Gastroenterol. 2015, 21, 2949–2958. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Dang, X.; Li, W.; Ma, Q. miR-133a functions as a tumor suppressor and directly targets FSCN1 in pancreatic cancer. Oncol. Res. 2013, 21, 353–363. [Google Scholar] [CrossRef]
- Song, X.; Shi, B.; Huang, K.; Zhang, W. miR-133a inhibits cervical cancer growth by targeting EGFR. Oncol. Rep. 2015, 34, 1573–1580. [Google Scholar] [CrossRef]
- Morita, S.; Horii, T.; Kimura, M.; Ochiya, T.; Tajima, S.; Hatada, I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci. 2013, 14, 14647–14658. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Rossi, M.; Raimondi, L.; Pitari, M.R.; Botta, C.; Tagliaferri, P.; Tassone, P. miR-29s: A family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget 2015, 6, 12837–12861. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics 2019, 12, 173–194. [Google Scholar] [CrossRef]
- Amodio, N.; Stamato, M.A.; Gullà, A.M.; Morelli, E.; Romeo, E.; Raimondi, L.; Pitari, M.R.; Ferrandino, I.; Misso, G.; Caraglia, M.; et al. Therapeutic Targeting of miR-29b/HDAC4 Epigenetic Loop in Multiple Myeloma. Mol. Cancer Ther. 2016, 15, 1364–1375. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Hu, Z.; Wang, Z.; Zhu, T.; Wang, G.; Gao, B.; Wang, J.; Deng, X. miR-125a-5p promotes gastric cancer growth and invasion by regulating the Hippo pathway. J. Clin. Lab. Anal. 2021, 35, e24078. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Li, S.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epi-drug in cancer therapy. Mol. Cancer 2020, 19, 79. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Tong, L.; Ao, Y.; Zhang, G.; Liu, Y.; Zhang, H. Upregulation of TRIAP1 by the lncRNA MFI2-AS1/miR-125a-5p Axis Promotes Thyroid Cancer Tumorigenesis. Onco Targets Ther. 2020, 13, 6967–6974. [Google Scholar] [CrossRef]
- Miska, E.A. How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 2005, 15, 563–568. [Google Scholar] [CrossRef]
- Liu, H.; Ma, Y.; Liu, C.; Li, P.; Yu, T. Reduced miR-125a-5p level in non-small-cell lung cancer is associated with tumour progression. Open Biol. 2018, 8, 180118. [Google Scholar] [CrossRef]
- Chen, H.Y.; Lin, Y.M.; Chung, H.C.; Lang, Y.D.; Lin, C.J.; Huang, J.; Wang, W.C.; Lin, F.M.; Chen, Z.; Huang, H.D.; et al. miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012, 72, 3631–3641. [Google Scholar] [CrossRef]
- Yu, Q.F.; Liu, P.; Li, Z.Y.; Zhang, C.F.; Chen, S.Q.; Li, Z.H.; Zhang, G.Y.; Li, J.C. MiR-103/107 induces tumorigenicity in bladder cancer cell by suppressing PTEN. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8616–8623. [Google Scholar] [PubMed]
- Xiong, B.; Lei, X.; Zhang, L.; Fu, J. miR-103 regulates triple negative breast cancer cells migration and invasion through targeting olfactomedin 4. Biomed. Pharmacother. 2017, 89, 1401–1408. [Google Scholar] [CrossRef]
- Zeng, B.; Li, Z.; Chen, R.; Guo, N.; Zhou, J.; Zhou, Q.; Lin, Q.; Cheng, D.; Liao, Q.; Zheng, L.; et al. Epigenetic regulation of miR-124 by Hepatitis C Virus core protein promotes migration and invasion of intrahepatic cholangiocarcinoma cells by targeting SMYD3. FEBS Lett. 2012, 586, 3271–3278. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, N.; Li, M.Y.; Du, M.F. Long non-coding RNA ZEB2-AS1 regulates osteosarcoma progression by acting as a molecular sponge of miR-107 to modulate SALL4 expression. Am. J. Transl. Res. 2021, 13, 1140–1154. [Google Scholar] [PubMed]
- Ye, H.; Zhou, Q.; Zheng, S.; Li, G.; Lin, Q.; Ye, L.; Wang, Y.; Wei, L.; Zhao, X.; Li, W.; et al. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Hanley, S.J.B.; Yue, J.; Watari, H. Musashi-2, a novel oncoprotein promoting cervical cancer cell growth and invasion, is negatively regulated by p53-induced miR-143 and miR-107 activation. J. Exp. Clin. Cancer Res. 2017, 36, 150. [Google Scholar] [CrossRef]
- Humphreys, K.J.; Cobiac, L.; Le Leu, R.K.; Van der Hoek, M.B.; Michael, M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol. Carcinog. 2013, 52, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Wu, D.; Li, P.; Xu, B.; Lu, Q.; Zhang, W. microRNA-18a, a member of the oncogenic miR-17-92 cluster, targets Dicer and suppresses cell proliferation in bladder cancer T24 cells. Mol. Med. Rep. 2012, 5, 167–172. [Google Scholar]
- Tsuchida, A.; Ohno, S.; Wu, W.; Borjigin, N.; Fujita, K.; Aoki, T.; Ueda, S.; Takanashi, M.; Kuroda, M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011, 102, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Chadalapaka, G.; Lee, S.O.; Yamada, D.; Sastre-Garau, X.; Defossez, P.A.; Park, Y.Y.; Lee, J.S.; Safe, S. Identification of oncogenic microRNA-17-92/ZBTB4/specificity protein axis in breast cancer. Oncogene 2012, 31, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Osada, H.; Takahashi, T. let-7 and miR-17-92: Small-sized major players in lung cancer development. Cancer Sci. 2011, 102, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef]
- Li, S.; Zeng, X.; Ma, R.; Wang, L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp. Ther. Med. 2018, 16, 2038–2045. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Kacimi, S.E.O.; Nguyen, T.L.; Suman, K.H.; Lemus-Martin, R.; Saleem, H.; Do, D.N. MiR-21 in the Cancers of the Digestive System and Its Potential Role as a Diagnostic, Predictive, and Therapeutic Biomarker. Biology 2021, 10, 417. [Google Scholar] [CrossRef]
- Wu, Y.; Song, Y.; Xiong, Y.; Wang, X.; Xu, K.; Han, B.; Bai, Y.; Li, L.; Zhang, Y.; Zhou, L. MicroRNA-21 (Mir-21) Promotes Cell Growth and Invasion by Repressing Tumor Suppressor PTEN in Colorectal Cancer. Cell Physiol. Biochem. 2017, 43, 945–958. [Google Scholar] [CrossRef]
- Sun, Z.; Li, S.; Kaufmann, A.M.; Albers, A.E. miR-21 increases the programmed cell death 4 gene-regulated cell proliferation in head and neck squamous carcinoma cell lines. Oncol. Rep. 2014, 32, 2283–2289. [Google Scholar] [CrossRef]
- Kong, X.; Liu, F.; Gao, J. MiR-155 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells through the activation of PI3K/SGK3/beta-catenin signaling pathways. Oncotarget 2016, 7, 66051–66060. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Guo, J.; Li, S.; Jiang, D. miR-155 inhibitor reduces the proliferation and migration in osteosarcoma MG-63 cells. Exp. Ther. Med. 2014, 8, 1575–1580. [Google Scholar] [CrossRef]
- Pedersen, I.M.; Otero, D.; Kao, E.; Miletic, A.V.; Hother, C.; Ralfkiaer, E.; Rickert, R.C.; Gronbaek, K.; David, M. Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas. EMBO Mol. Med. 2009, 1, 288–295. [Google Scholar] [CrossRef]
- Sagar, S.K. miR-106b as an emerging therapeutic target in cancer. Genes. Dis. 2022, 9, 889–899. [Google Scholar] [CrossRef] [PubMed]
- Mehlich, D.; Garbicz, F.; Wlodarski, P.K. The emerging roles of the polycistronic miR-106b approximately 25 cluster in cancer—A comprehensive review. Biomed. Pharmacother. 2018, 107, 1183–1195. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jung, E.J.; Kim, J.M.; Son, Y.; Lee, H.S.; Kwag, S.J.; Park, J.H.; Cho, J.K.; Kim, H.G.; Park, T.; et al. MiR-221 and miR-222 regulate cell cycle progression and affect chemosensitivity in breast cancer by targeting ANXA3. Exp. Ther. Med. 2023, 25, 127. [Google Scholar] [CrossRef]
- Sun, L.L.; Li, W.D.; Lei, F.R.; Li, X.Q. The regulatory role of microRNAs in angiogenesis-related diseases. J. Cell Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Arbitrio, M.; Caracciolo, D.; Cordua, A.; Cuomo, O.; Grillone, K.; Riillo, C.; Carida, G.; Scionti, F.; Labanca, C.; et al. miR-221/222 as biomarkers and targets for therapeutic intervention on cancer and other diseases: A systematic review. Mol. Ther. Nucleic Acids 2022, 27, 1191–1224. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar]
- Song, Q.; An, Q.; Niu, B.; Lu, X.; Zhang, N.; Cao, X. Role of miR-221/222 in Tumor Development and the Underlying Mechanism. J. Oncol. 2019, 2019, 7252013. [Google Scholar] [CrossRef]
- Zhao, J.J.; Chu, Z.B.; Hu, Y.; Lin, J.; Wang, Z.; Jiang, M.; Chen, M.; Wang, X.; Kang, Y.; Zhou, Y.; et al. Targeting the miR-221-222/PUMA/BAK/BAX Pathway Abrogates Dexamethasone Resistance in Multiple Myeloma. Cancer Res. 2015, 75, 4384–4397. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.T.; Gulla, A.; Cantafio, M.E.; Lionetti, M.; Leone, E.; Amodio, N.; Guzzi, P.H.; Foresta, U.; Conforti, F.; Cannataro, M.; et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget 2013, 4, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci. Signal 2011, 4, ra41. [Google Scholar] [CrossRef] [PubMed]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal 2011, 4, ra41. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Zhang, A.; Wang, Y.; Han, L.; You, Y.; Pu, P.; Kang, C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int. J. Oncol. 2010, 37, 1621–1626. [Google Scholar]
- Chang, H.Y.; Lee, C.H.; Li, Y.S.; Huang, J.T.; Lan, S.H.; Wang, Y.F.; Lai, W.W.; Wang, Y.C.; Lin, Y.J.; Liu, H.S.; et al. MicroRNA-146a suppresses tumor malignancy via targeting vimentin in esophageal squamous cell carcinoma cells with lower fibronectin membrane assembly. J. Biomed. Sci. 2020, 27, 102. [Google Scholar] [CrossRef]
- Cabello, P.; Torres-Ruiz, S.; Adam-Artigues, A.; Fores-Martos, J.; Martinez, M.T.; Hernando, C.; Zazo, S.; Madoz-Gurpide, J.; Rovira, A.; Burgues, O.; et al. miR-146a-5p Promotes Angiogenesis and Confers Trastuzumab Resistance in HER2+ Breast Cancer. Cancers 2023, 15, 2138. [Google Scholar] [CrossRef]
- Do, Y.; Cho, J.G.; Park, J.Y.; Oh, S.; Park, D.; Yoo, K.H.; Lee, M.S.; Kwon, B.S.; Kim, J.; Yang, Y. MiR-146a Regulates Migration and Invasion by Targeting NRP2 in Circulating-Tumor Cell Mimicking Suspension Cells. Genes 2020, 12, 45. [Google Scholar] [CrossRef]
- Shahriar, A.; Ghaleh-Aziz Shiva, G.; Ghader, B.; Farhad, J.; Hosein, A.; Parsa, H. The dual role of mir-146a in metastasis and disease progression. Biomed. Pharmacother. 2020, 126, 110099. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; Yu, Q.; Yao, Y. Effect of miR-146a-5p on proliferation and metastasis of triple-negative breast cancer via regulation of SOX5. Exp. Ther. Med. 2018, 15, 4515–4521. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, W.; Zhang, L.; Chen, X.; Liu, F.; Zhang, J.; Guan, S.; Sun, Y.; Chen, P.; Wang, D.; et al. miR-146a-5p mediates epithelial-mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br. J. Cancer 2016, 115, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhao, X.; Liu, X.; Wang, Y.; Huang, J.; Jiang, B.; Chen, Q.; Yu, J. miR-146a functions as a tumor suppressor in prostate cancer by targeting Rac1. Prostate 2014, 74, 1613–1621. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. Causes and consequences of microRNA dysregulation. Cancer J. 2012, 18, 215–222. [Google Scholar] [CrossRef]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V.; Kuznetsova, E.B. Histone modifications and non-coding rRNAs: Mutual epigenetic regulation and role in pathogenesis. Int. J. Mol. Sci. 2022, 23, 5801. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, D.; Sohrabi, B.; Mosallaei, M.; Nariman-Saleh-Fam, Z.; Bastami, M.; Mansoori, Y.; Daraei, A.; Zununi Vahed, S.; Navid, S.; Saadatian, Z.; et al. Epi-miRNAs: Regulators of the Histone Modification Machinery in Human Cancer. J. Oncol. 2022, 2022, 4889807. [Google Scholar] [CrossRef]
- Bianchi, M.; Renzini, A.; Adamo, S.; Moresi, V. Coordinated Actions of MicroRNAs with other Epigenetic Factors Regulate Skeletal Muscle Development and Adaptation. Int. J. Mol. Sci. 2017, 18, 840. [Google Scholar] [CrossRef]
- Ramzan, F.; Vickers, M.H.; Mithen, R.F. Epigenetics, microRNA and Metabolic Syndrome: A Comprehensive Review. Int. J. Mol. Sci. 2021, 22, 5047. [Google Scholar] [CrossRef]
- Zhang, P.; Torres, K.; Liu, X.; Liu, C.G.; Pollock, R.E. An Overview of Chromatin-Regulating Proteins in Cells. Curr. Protein Pept. Sci. 2016, 17, 401–410. [Google Scholar] [CrossRef]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [PubMed]
- Yang, Y.; Zhang, M.; Wang, Y. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J. Natl. Cancer Cent. 2022, 2, 277–290. [Google Scholar]
- Kondo, Y.; Shen, L.; Issa, J.P. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell Biol. 2003, 23, 206–215. [Google Scholar]
- Khan, S.A.; Reddy, D.; Gupta, S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment? World J. Biol. Chem. 2015, 6, 333–345. [Google Scholar] [PubMed]
- Chervona, Y.; Costa, M. Histone modifications and cancer: Biomarkers of prognosis? Am. J. Cancer Res. 2012, 2, 589–597. [Google Scholar]
- Kristeleit, R.; Stimson, L.; Workman, P.; Aherne, W. Histone modification enzymes: Novel targets for cancer drugs. Expert. Opin. Emerg. Drugs 2005, 9, 135–154. [Google Scholar]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar]
- Torres, I.O.; Fujimori, D.G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015, 35, 68–75. [Google Scholar]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [PubMed]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep 2015, 16, 1467–1481. [Google Scholar]
- Berndsen, C.E.; Denu, J.M. Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 2008, 18, 682–689. [Google Scholar]
- Liu, R.; Wu, J.; Guo, H.; Yao, W.; Li, S.; Lu, Y.; Jia, Y.; Liang, X.; Tang, J.; Zhang, H. Post-translational modifications of histones: Mechanisms, biological functions, and therapeutic targets. MedComm 2023, 4, e292. [Google Scholar]
- Wapenaar, H.; Dekker, F.J. Histone acetyltransferases: Challenges in targeting bi-substrate enzymes. Clin. Epigenetics 2016, 8, 59. [Google Scholar]
- Milazzo, G.; Mercatelli, D.; Di Muzio, G.; Triboli, L.; De Rosa, P.; Perini, G.; Giorgi, F.M. Histone Deacetylases (HDACs): Evolution, Specificity, Role in Transcriptional Complexes, and Pharmacological Actionability. Genes 2020, 11, 556. [Google Scholar] [PubMed]
- Parbin, S.; Kar, S.; Shilpi, A.; Sengupta, D.; Deb, M.; Rath, S.K.; Patra, S.K. Histone deacetylases: A saga of perturbed acetylation homeostasis in cancer. J. Histochem. Cytochem. 2014, 62, 11–33. [Google Scholar]
- Seto, E.; Yoshida, M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef] [PubMed]
- Rajan, P.K.; Udoh, U.A.; Sanabria, J.D.; Banerjee, M.; Smith, G.; Schade, M.S.; Sanabria, J.; Sodhi, K.; Pierre, S.; Xie, Z.; et al. The Role of Histone Acetylation-/Methylation-Mediated Apoptotic Gene Regulation in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 8894. [Google Scholar]
- Zhang, Y.; Reinberg, D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes. Dev. 2001, 15, 2343–2360. [Google Scholar] [PubMed]
- Cloos, P.A.; Christensen, J.; Agger, K.; Helin, K. Erasing the methyl mark: Histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008, 22, 1115–1140. [Google Scholar]
- Dimitrova, E.; Turberfield, A.H.; Klose, R.J. Histone demethylases in chromatin biology and beyond. EMBO Rep. 2015, 16, 1620–1639. [Google Scholar]
- Zaware, N.; Zhou, M.M. Bromodomain biology and drug discovery. Nat. Struct. Mol. Biol. 2019, 26, 870–879. [Google Scholar] [PubMed]
- Josling, G.A.; Selvarajah, S.A.; Petter, M.; Duffy, M.F. The role of bromodomain proteins in regulating gene expression. Genes 2012, 3, 320–343. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Barton, M.C. Bromodomain Histone Readers and Cancer. J. Mol. Biol. 2017, 429, 2003–2010. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.S.; Wang, W. The recognition specificity of the CHD1 chromodomain with modified histone H3 peptides. J. Mol. Biol. 2011, 406, 527–541. [Google Scholar] [CrossRef]
- Yap, K.L.; Zhou, M.M. Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 2011, 50, 1966–1980. [Google Scholar]
- Chen, Q.; Yang, B.; Liu, X.; Zhang, X.D.; Zhang, L.; Liu, T. Histone acetyltransferases CBP/p300 in tumorigenesis and CBP/p300 inhibitors as promising novel anticancer agents. Theranostics 2022, 12, 4935–4948. [Google Scholar] [CrossRef]
- Welti, J.; Sharp, A.; Brooks, N.; Yuan, W.; McNair, C.; Chand, S.N.; Pal, A.; Figueiredo, I.; Riisnaes, R.; Gurel, B.; et al. Targeting the p300/CBP Axis in Lethal Prostate Cancer. Cancer Discov. 2021, 11, 1118–1137. [Google Scholar]
- Di Cerbo, V.; Schneider, R. Cancers with wrong HATs: The impact of acetylation. Brief. Funct. Genom. 2013, 12, 231–243. [Google Scholar]
- Kittler, R.; Zhou, J.; Hua, S.; Ma, L.; Liu, Y.; Pendleton, E.; Cheng, C.; Gerstein, M.; White, K.P. A comprehensive nuclear receptor network for breast cancer cells. Cell Rep. 2013, 3, 538–551. [Google Scholar]
- Sun, G.; Wang, C.; Wang, S.; Sun, H.; Zeng, K.; Zou, R.; Lin, L.; Liu, W.; Sun, N.; Song, H.; et al. An H3K4me3 reader, BAP18 as an adaptor of COMPASS-like core subunits co-activates ERalpha action and associates with the sensitivity of antiestrogen in breast cancer. Nucleic Acids Res. 2020, 48, 10768–10784. [Google Scholar] [CrossRef]
- Wang, J.; Hevi, S.; Kurash, J.K.; Lei, H.; Gay, F.; Bajko, J.; Su, H.; Sun, W.; Chang, H.; Xu, G.; et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2008, 41, 125–129. [Google Scholar] [CrossRef]
- Takeshima, H.; Wakabayashi, M.; Hattori, N.; Yamashita, S.; Ushijima, T. Identification of coexistence of DNA methylation and H3K27me3 specifically in cancer cells as a promising target for epigenetic therapy. Carcinogenesis 2015, 36, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Kazanets, A.; Shorstova, T.; Hilmi, K.; Marques, M.; Witcher, M. Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim. Biophys. Acta 2016, 1865, 275–288. [Google Scholar]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 245. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, Y.; Jia, J.; Fang, Y.; Tang, Y.; Wu, H.; Fang, D. H3K36me3, message from chromatin to DNA damage repair. Cell Biosci. 2020, 10, 9. [Google Scholar] [CrossRef]
- Sturm, D.; Bender, S.; Jones, D.T.; Lichter, P.; Grill, J.; Becher, O.; Hawkins, C.; Majewski, J.; Jones, C.; Costello, J.F.; et al. Paediatric and adult glioblastoma: Multiform (epi)genomic culprits emerge. Nat. Rev. Cancer 2014, 14, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Komar, D.; Juszczynski, P. Rebelled epigenome: Histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clin. Epigenetics 2020, 12, 147. [Google Scholar] [CrossRef]
- Peng, Y.; Liao, Q.; Tan, W.; Peng, C.; Hu, Z.; Chen, Y.; Li, Z.; Li, J.; Zhen, B.; Zhu, W.; et al. The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat. Commun. 2019, 10, 1224. [Google Scholar] [CrossRef]
- Sekiguchi, M.; Matsushita, N. DNA Damage Response Regulation by Histone Ubiquitination. Int. J. Mol. Sci. 2022, 23, 8187. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.Y.; Hochstrasser, M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res. 2021, 49, 6043–6052. [Google Scholar] [CrossRef]
- Zhu, Q.; Liang, P.; Chu, C.; Zhang, A.; Zhou, W. Protein sumoylation in normal and cancer stem cells. Front. Mol. Biosci. 2022, 9, 1095142. [Google Scholar] [CrossRef]
- Zong, W.; Gong, Y.; Sun, W.; Li, T.; Wang, Z.Q. PARP1: Liaison of Chromatin Remodeling and Transcription. Cancers 2022, 14, 4162. [Google Scholar] [CrossRef]
- Ummarino, S.; Hausman, C.; Di Ruscio, A. The PARP Way to Epigenetic Changes. Genes 2021, 12, 446. [Google Scholar] [CrossRef]
- Ntorla, A.; Burgoyne, J.R. The Regulation and Function of Histone Crotonylation. Front. Cell Dev. Biol. 2021, 9, 624914. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Wu, X.; Wu, Q.; Chatoff, A.; Megill, E.; Gao, J.; Huang, T.; Duan, T.; Yang, K.; Jin, C.; et al. Lysine catabolism reprograms tumour immunity through histone crotonylation. Nature 2023, 617, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Baird, A.-M.; Richard, D.; O’Byrne, K.J.; Gray, S.G. Epigenetic Therapy in Lung Cancer and Mesothelioma. In Epigenetic Cancer Therapy; Academic Press: Cambridge, MA, USA, 2015; pp. 189–213. [Google Scholar] [CrossRef]
- Gray, S.G. Epigenetics of Cisplatin Resistance. In Epigenetic Cancer Therapy; Academic Press: Cambridge, MA, USA, 2015; pp. 613–637. [Google Scholar] [CrossRef]
- Jin, Q.; Yu, L.R.; Wang, L.; Zhang, Z.; Kasper, L.H.; Lee, J.E.; Wang, C.; Brindle, P.K.; Dent, S.Y.; Ge, K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011, 30, 249–262. [Google Scholar] [CrossRef]
- Farago, A.; Zsindely, N.; Farkas, A.; Neller, A.; Siagi, F.; Szabo, M.R.; Csont, T.; Bodai, L. Acetylation State of Lysine 14 of Histone H3.3 Affects Mutant Huntingtin Induced Pathogenesis. Int. J. Mol. Sci. 2022, 23, 15173. [Google Scholar] [CrossRef]
- Weirich, S.; Khella, M.S.; Jeltsch, A. Structure, Activity and Function of the Suv39h1 and Suv39h2 Protein Lysine Methyltransferases. Life 2021, 11, 703. [Google Scholar] [CrossRef]
- Fritsch, L.; Robin, P.; Mathieu, J.R.; Souidi, M.; Hinaux, H.; Rougeulle, C.; Harel-Bellan, A.; Ameyar-Zazoua, M.; Ait-Si-Ali, S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell 2010, 37, 46–56. [Google Scholar] [CrossRef]
- Kaniskan, H.U.; Martini, M.L.; Jin, J. Inhibitors of Protein Methyltransferases and Demethylases. Chem. Rev. 2018, 118, 989–1068. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.; Yu, X.-Y. Histone lysine demethylase inhibitor (HDMi) as chemo-sensitizing agent. In Epigenetic Regulation in Overcoming Chemoresistance; Academic Press: Cambridge, MA, USA, 2021; pp. 41–55. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Li, C. Evolutionarily conserved protein arginine methyltransferases in non-mammalian animal systems. FEBS J. 2012, 279, 932–945. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Huang, K.; Kondengaden, S.; Wooten, J.; Reyhanfard, H.; Qing, Z.; Zhai, B.C.; Wang, P.G. Histone Methyltransferase Inhibitors for Cancer Therapy. In Epigenetic Technological Applications; Academic Press: Cambridge, MA, USA, 2015; pp. 363–395. [Google Scholar] [CrossRef]
- Huang, S.; Litt, M.; Felsenfeld, G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes. Dev. 2005, 19, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Litt, M.; Qiu, Y.; Huang, S. Histone arginine methylations: Their roles in chromatin dynamics and transcriptional regulation. Biosci. Rep. 2009, 29, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation. Epigenetics 2014, 7, 1098–1108. [Google Scholar] [CrossRef]
- Wang, J.; Tian, X.; Feng, C.; Song, C.; Yu, B.; Wang, Y.; Ji, X.; Zhang, X. Histone H3 phospho-regulation by KimH3 in both interphase and mitosis. iScience 2023, 26, 106372. [Google Scholar] [CrossRef]
- Mattiroli, F.; Penengo, L. Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability. Trends Genet. 2021, 37, 566–581. [Google Scholar] [CrossRef]
- Dasgupta, A.; Mondal, P.; Dalui, S.; Das, C.; Roy, S. Molecular characterization of substrate-induced ubiquitin transfer by UBR7-PHD finger, a newly identified histone H2BK120 ubiquitin ligase. FEBS J. 2021, 289, 1842–1857. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liao, L.; Balaji, K.S.; Wei, C.; Kim, J.; Peng, J. Epigenetic modification and a role for the E3 ligase RNF40 in cancer development and metastasis. Oncogene 2020, 40, 465–474. [Google Scholar] [CrossRef]
- Chernikova, S.B.; Razorenova, O.V.; Higgins, J.P.; Sishc, B.J.; Nicolau, M.; Dorth, J.A.; Chernikova, D.A.; Kwok, S.; Brooks, J.D.; Bailey, S.M.; et al. Deficiency in Mammalian Histone H2B Ubiquitin Ligase Bre1 (Rnf20/Rnf40) Leads to Replication Stress and Chromosomal Instability. Cancer Res. 2012, 72, 2111–2119. [Google Scholar] [CrossRef]
- So, C.C.; Ramachandran, S.; Martin, A. E3 Ubiquitin Ligases RNF20 and RNF40 Are Required for Double-Stranded Break (DSB) Repair: Evidence for Monoubiquitination of Histone H2B Lysine 120 as a Novel Axis of DSB Signaling and Repair. Mol. Cell. Biol. 2023, 39, e00488-18. [Google Scholar] [CrossRef]
- Delcuve, G.P.; Khan, D.H.; Davie, J.R. Roles of histone deacetylases in epigenetic regulation: Emerging paradigms from studies with inhibitors. Clin. Epigenetics 2012, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Kim, J.-S. A short guide to histone deacetylases including recent progress on class II enzymes. Exp. Mol. Med. 2020, 52, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Yruela, C.; Zhang, D.; Wei, W.; Bæk, M.; Liu, W.; Gao, J.; Danková, D.; Nielsen, A.L.; Bolding, J.E.; Yang, L.; et al. Class I histone deacetylases (HDAC1–3) are histone lysine delactylases. Sci. Adv. 2022, 8, eabi6696. [Google Scholar] [CrossRef]
- Li, G.; Tian, Y.; Zhu, W.-G. The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy. Front. Cell Dev. Biol. 2020, 8, 576946. [Google Scholar] [CrossRef]
- Arifuzzaman, S.; Khatun, M.R.; Khatun, R. Emerging of lysine demethylases (KDMs): From pathophysiological insights to novel therapeutic opportunities. Biomed. Pharmacother. 2020, 129, 110392. [Google Scholar] [CrossRef]
- Accari, S.L.; Fisher, P.R. Emerging roles of JmjC domain-containing proteins. Int. Rev. Cell Mol. Biol. 2015, 319, 165–220. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Nottke, A.C.; Shi, Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008, 20, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Soloaga, A. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003, 22, 2788–2797. [Google Scholar] [CrossRef]
- Barber, C.M.; Turner, F.B.; Wang, Y.; Hagstrom, K.; Taverna, S.D.; Mollah, S.; Ueberheide, B.; Meyer, B.J.; Hunt, D.F.; Cheung, P.; et al. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 2004, 112, 360–371. [Google Scholar] [CrossRef]
- Zhang, G.; Pradhan, S. Mammalian epigenetic mechanisms. IUBMB Life 2014, 66, 240–256. [Google Scholar] [CrossRef]
- Hare, A.E.; Parvin, J.D. Processes that Regulate the Ubiquitination of Chromatin and Chromatin-Associated Proteins. In Ubiquitin Proteasome System—Current Insights into Mechanism Cellular Regulation and Disease; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Ma, T.; Keller, J.A.; Yu, X. RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta Biochim. Biophys. Sin. 2011, 43, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, R.M.; Kupai, A.; Rothbart, S.B. Chromatin Regulation through Ubiquitin and Ubiquitin-like Histone Modifications. Trends Biochem. Sci. 2021, 46, 258–269. [Google Scholar] [CrossRef]
- Fang, Y.-Z.; Jiang, L.; He, Q.; Cao, J.; Yang, B. Deubiquitination complex platform: A plausible mechanism for regulating the substrate specificity of deubiquitinating enzymes. Acta Pharm. Sin. B 2023, 13, 2955–2962. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-Y.; Jones, A.; Yang, C.; Zhai, L.; Smith, A.D.; Zhang, Z.; Chandrasekharan, M.B.; Sun, Z.-w.; Renfrow, M.B.; Wang, Y.; et al. Regulation of Histone H2A and H2B Deubiquitination and Xenopus Development by USP12 and USP46. J. Biol. Chem. 2011, 286, 7190–7201. [Google Scholar] [CrossRef]
- Ting, X.; Xia, L.; Yang, J.; He, L.; Si, W.; Shang, Y.; Sun, L. USP11 acts as a histone deubiquitinase functioning in chromatin reorganization during DNA repair. Nucleic Acids Res. 2019, 47, 9721–9740. [Google Scholar] [CrossRef]
- Fontán-Lozano, Á.; Suárez-Pereira, I.; Horrillo, A.; del-Pozo-Martín, Y.; Hmadcha, A.; Carrión, Á.M. Histone H1 Poly[ADP]-Ribosylation Regulates the Chromatin Alterations Required for Learning Consolidation. J. Neurosci. 2010, 30, 13305–13313. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Kraus, W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012, 26, 417–432. [Google Scholar] [CrossRef]
- Kmiecik, S.W.; Drzewicka, K.; Melchior, F.; Mayer, M.P. Heat shock transcription factor 1 is SUMOylated in the activated trimeric state. J. Biol. Chem. 2021, 296, 100324. [Google Scholar] [CrossRef]
- Ramachandran, H.; Herfurth, K.; Grosschedl, R.; Schafer, T.; Walz, G. SUMOylation Blocks the Ubiquitin-Mediated Degradation of the Nephronophthisis Gene Product Glis2/NPHP7. PLoS ONE 2015, 10, e0130275. [Google Scholar] [CrossRef]
- Sun, Y.; Miller Jenkins, L.M.; Su, Y.P.; Nitiss, K.C.; Nitiss, J.L.; Pommier, Y. A conserved SUMO pathway repairs topoisomerase DNA-protein cross-links by engaging ubiquitin-mediated proteasomal degradation. Sci. Adv. 2020, 6, eaba6290. [Google Scholar] [CrossRef]
- Sharma, P.; Yamada, S.; Lualdi, M.; Dasso, M.; Kuehn, M.R. Senp1 is essential for desumoylating Sumo1-modified proteins but dispensable for Sumo2 and Sumo3 deconjugation in the mouse embryo. Cell Rep. 2013, 3, 1640–1650. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Sharma, P.; Athanasiou, M.; Kumar, A.; Yamada, S.; Kuehn, M.R. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell Biol. 2005, 25, 5171–5182. [Google Scholar] [CrossRef] [PubMed]
- Downey, M. Non-histone protein acetylation by the evolutionarily conserved GCN5 and PCAF acetyltransferases. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194608. [Google Scholar] [CrossRef]
- Kollenstart, L.; de Groot, A.J.L.; Janssen, G.M.C.; Cheng, X.; Vreeken, K.; Martino, F.; Cote, J.; van Veelen, P.A.; van Attikum, H. Gcn5 and Esa1 function as histone crotonyltransferases to regulate crotonylation-dependent transcription. J. Biol. Chem. 2019, 294, 20122–20134. [Google Scholar] [CrossRef]
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 2015, 58, 203–215. [Google Scholar] [CrossRef]
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Esteller, M.; Rountree, M.R.; Bachman, K.E.; Schuebel, K.; Herman, J.G. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 2001, 10, 687–692. [Google Scholar] [CrossRef]
- Okabe, A.; Kaneda, A. Transcriptional dysregulation by aberrant enhancer activation and rewiring in cancer. Cancer Sci. 2021, 112, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, S. Epigenetic modifications in cancer. Clin. Genet. 2012, 81, 303–311. [Google Scholar] [CrossRef]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef]
- Sun, X.J.; Man, N.; Tan, Y.; Nimer, S.D.; Wang, L. The Role of Histone Acetyltransferases in Normal and Malignant Hematopoiesis. Front. Oncol. 2015, 5, 108. [Google Scholar] [CrossRef] [PubMed]
- Pathak, S.; Tomar, S.; Pathak, A. Epigenetics and Cancer: A Comprehensive Review. Asian Pac. J. Cancer Biol. 2023, 8, 75–89. [Google Scholar] [CrossRef]
- Flavahan, W.A.; Gaskell, E.; Bernstein, B.E. Epigenetic plasticity and the hallmarks of cancer. Science 2017, 357, eaal2380. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, S.; Bhattacharya, A.; Adhikary, S.; Singh, V.; Gadad, S.S.; Roy, S.; Das, C. The paradigm of drug resistance in cancer: An epigenetic perspective. Biosci. Rep. 2022, 42, BSR20211812. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.L.; Jeong, K.W. Histone modifications in drug-resistant cancers: From a cancer stem cell and immune evasion perspective. Exp. Mol. Med. 2023, 55, 1333–1347. [Google Scholar] [CrossRef]
- Feng, J.; Meng, X. Histone modification and histone modification-targeted anti-cancer drugs in breast cancer: Fundamentals and beyond. Front. Pharmacol. 2022, 13, 946811. [Google Scholar] [CrossRef]
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef]
- Memari, F.; Joneidi, Z.; Taheri, B.; Aval, S.F.; Roointan, A.; Zarghami, N. Epigenetics and Epi-miRNAs: Potential markers/therapeutics in leukemia. Biomed. Pharmacother. 2018, 106, 1668–1677. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A.; Filip, M. A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs. Neurotox. Res. 2015, 27, 172–197. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Sadakierska-Chudy, A. MicroRNAs: Diverse Mechanisms of Action and Their Potential Applications as Cancer Epi-Therapeutics. Biomolecules 2020, 10, 1285. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V. Mutual regulation of ncRNAs and chromatin remodeling complexes in normal and pathological conditions. Int. J. Mol. Sci. 2023, 24, 7848. [Google Scholar]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Chen, B.F.; Lu, Z.; Huo, X.S.; Zhou, W.P.; Wang, F.; Sun, S.H. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology 2011, 54, 2025–2035. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Toh, H.C.; Chow, P.; Chung, A.Y.; Meyers, D.J.; Cole, P.A.; Ooi, L.L.; Lee, C.G. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012, 26, 3032–3041. [Google Scholar] [CrossRef]
- Incoronato, M.; Urso, L.; Portela, A.; Laukkanen, M.O.; Soini, Y.; Quintavalle, C.; Keller, S.; Esteller, M.; Condorelli, G. Epigenetic regulation of miR-212 expression in lung cancer. PLoS ONE 2011, 6, e27722. [Google Scholar]
- Saito, Y.; Friedman, J.M.; Chihara, Y.; Egger, G.; Chuang, J.C.; Liang, G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem. Biophys. Res. Commun. 2009, 379, 726–731. [Google Scholar] [CrossRef]
- Li, L.; Yuan, L.; Luo, J.; Gao, J.; Guo, J.; Xie, X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013, 13, 109–117. [Google Scholar] [CrossRef]
- Ma, W.; Xiao, G.G.; Mao, J.; Lu, Y.; Song, B.; Wang, L.; Fan, S.; Fan, P.; Hou, Z.; Li, J.; et al. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget 2015, 6, 10432–10444. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Saini, S.; Shahryari, V.; Arora, S.; Zaman, M.S.; Chang, I.; Yamamura, S.; Tanaka, Y.; Chiyomaru, T.; et al. miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin. Cancer Res. 2013, 19, 73–84. [Google Scholar] [PubMed]
- Thayanithy, V.; Park, C.; Sarver, A.L.; Kartha, R.V.; Korpela, D.M.; Graef, A.J.; Steer, C.J.; Modiano, J.F.; Subramanian, S. Combinatorial treatment of DNA and chromatin-modifying drugs cause cell death in human and canine osteosarcoma cell lines. PLoS ONE 2012, 7, e43720. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Suzuki, H.; Taya, T.; Nishizawa, M.; Tsugawa, H.; Matsuzaki, J.; Hirata, K.; Saito, H.; Hibi, T. Development of a novel microRNA promoter microarray for ChIP-on-chip assay to identify epigenetically regulated microRNAs. Biochem. Biophys. Res. Commun. 2012, 426, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.; Nilsson, J.; Mir, S.E.; van der Stoop, P.M.; Hulleman, E.; Niers, J.M.; de Witt Hamer, P.C.; Marquez, V.E.; Cloos, J.; Krichevsky, A.M.; et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 2010, 1, 710–720. [Google Scholar]
- Pandey, A.K.; Zhang, Y.; Zhang, S.; Li, Y.; Tucker-Kellogg, G.; Yang, H.; Jha, S. TIP60-miR-22 axis as a prognostic marker of breast cancer progression. Oncotarget 2015, 6, 41290–41306. [Google Scholar]
- Wang, B.; Li, D.; Filkowski, J.; Rodriguez-Juarez, R.; Storozynsky, Q.; Malach, M.; Carpenter, E.; Kovalchuk, O. A dual role of miR-22 modulated by RelA/p65 in resensitizing fulvestrant-resistant breast cancer cells to fulvestrant by targeting FOXP1 and HDAC4 and constitutive acetylation of p53 at Lys382. Oncogenesis 2018, 7, 54. [Google Scholar]
- Lv, T.; Song, K.; Zhang, L.; Li, W.; Chen, Y.; Diao, Y.; Yao, Q.; Liu, P. miRNA-34a decreases ovarian cancer cell proliferation and chemoresistance by targeting HDAC1. Biochem. Cell Biol. 2018, 96, 663–671. [Google Scholar] [CrossRef]
- Hsieh, T.H.; Hsu, C.Y.; Tsai, C.F.; Long, C.Y.; Chai, C.Y.; Hou, M.F.; Lee, J.N.; Wu, D.C.; Wang, S.C.; Tsai, E.M. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget 2015, 6, 494–509. [Google Scholar]
- Hsieh, T.H.; Hsu, C.Y.; Tsai, C.F.; Long, C.Y.; Wu, C.H.; Wu, D.C.; Lee, J.N.; Chang, W.C.; Tsai, E.M. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol. Ther. 2015, 23, 656–666. [Google Scholar]
- Trissal, M.C.; Wong, T.N.; Yao, J.C.; Ramaswamy, R.; Kuo, I.; Baty, J.; Sun, Y.; Jih, G.; Parikh, N.; Berrien-Elliott, M.M.; et al. MIR142 Loss-of-Function Mutations Derepress ASH1L to Increase HOXA Gene Expression and Promote Leukemogenesis. Cancer Res. 2018, 78, 3510–3521. [Google Scholar]
- Colamaio, M.; Puca, F.; Ragozzino, E.; Gemei, M.; Decaussin-Petrucci, M.; Aiello, C.; Bastos, A.U.; Federico, A.; Chiappetta, G.; Del Vecchio, L.; et al. miR-142-3p down-regulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J. Clin. Endocrinol. Metab. 2015, 100, E59–E69. [Google Scholar] [CrossRef]
- Yang, Y.; Meng, Q.; Wang, C.; Li, X.; Lu, Y.; Xin, X.; Zheng, Q.; Lu, D. MicroRNA 675 cooperates PKM2 to aggravate progression of human liver cancer stem cells induced from embryonic stem cells. J. Mol. Med. 2018, 96, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Han, C.; Zhang, J.; Lu, D.; Dash, S.; Feitelson, M.; Lim, K.; Wu, T. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology 2013, 58, 1681–1692. [Google Scholar] [CrossRef]
- Zhang, J.G.; Guo, J.F.; Liu, D.L.; Liu, Q.; Wang, J.J. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J. Thorac. Oncol. 2011, 6, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Deng, Z.; Wan, M.; Huang, W.; Cramer, S.D.; Xu, J.; Lei, M.; Sui, G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol. Cancer 2010, 9, 108. [Google Scholar] [CrossRef]
- Sakurai, T.; Bilim, V.N.; Ugolkov, A.V.; Yuuki, K.; Tsukigi, M.; Motoyama, T.; Tomita, Y. The enhancer of zeste homolog 2 (EZH2), a potential therapeutic target, is regulated by miR-101 in renal cancer cells. Biochem. Biophys. Res. Commun. 2012, 422, 607–614. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, J.; Zhou, Z.; He, Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. OncoTargets Ther. 2017, 10, 1027–1038. [Google Scholar] [CrossRef]
- Masucci, M.G.; Du, Z.-M.; Hu, L.-F.; Wang, H.-Y.; Yan, L.-X.; Zeng, Y.-X.; Shao, J.-Y.; Ernberg, I. Upregulation of MiR-155 in Nasopharyngeal Carcinoma is Partly Driven by LMP1 and LMP2A and Downregulates a Negative Prognostic Marker JMJD1A. PLoS ONE 2011, 6, e19137. [Google Scholar]
- Bourassa, M.W.; Ratan, R.R. The interplay between microRNAs and histone deacetylases in neurological diseases. Neurochem. Int. 2014, 77, 33–39. [Google Scholar]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef]
- Noonan, E.J.; Place, R.F.; Pookot, D.; Basak, S.; Whitson, J.M.; Hirata, H.; Giardina, C.; Dahiya, R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene 2009, 28, 1714–1724. [Google Scholar] [CrossRef]
- Singh, P.K.; Campbell, M.J. The Interactions of microRNA and Epigenetic Modifications in Prostate Cancer. Cancers 2013, 5, 998–1019. [Google Scholar] [CrossRef]
- Fan, D.N.Y.; Tsang, F.H.C.; Au, S.L.K.; Wei, L.L.; Tam, A.H.K.; Wong, C.M. Abstract 1059: SUV39H1 promotes HCC tumorigenesis and is targeted by tumor suppressive miRNA-125b. Cancer Res. 2012, 72, 1059. [Google Scholar] [CrossRef]
- Wu, M.; Fan, B.; Guo, Q.; Li, Y.; Chen, R.; Lv, N.; Diao, Y.; Luo, Y. Knockdown of SETDB1 inhibits breast cancer progression by miR-381-3p-related regulation. Biol. Res. 2018, 51, 39. [Google Scholar] [CrossRef]
- Knyazev, E.N.; Samatov, T.R.; Fomicheva, K.A.; Nyushko, K.M.; Alekseev, B.Y.; Shkurnikov, M.Y. MicroRNA hsa-miR-4674 in Hemolysis-Free Blood Plasma Is Associated with Distant Metastases of Prostatic Cancer. Bull. Exp. Biol. Med. 2016, 161, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.-F.; He, S.-M.; Zeng, S.; Zhou, Z.-W.; He, Z. Hsa-microRNA-181a is a regulator of a number of cancer genes and a biomarker for endometrial carcinoma in patients: A bioinformatic and clinical study and the therapeutic implication. Drug Des. Dev. Ther. 2015, 9, 1103–1175. [Google Scholar] [CrossRef]
- Zhu, W.; Qian, J.; Ma, L.; Ma, P.; Yang, F.; Shu, Y. MiR-346 suppresses cell proliferation through SMYD3 dependent approach in hepatocellular carcinoma. Oncotarget 2017, 8, 65218–65229. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Li, Q.; Chen, S.; Zhang, X.; Tao, X.; Tang, X.; Wang, S.; Che, G.; Yu, Y.; He, L. miR-133b suppresses colorectal cancer cell stemness and chemoresistance by targeting methyltransferase DOT1L. Exp. Cell Res. 2019, 385, 111597. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Guo, Z.; Wu, C.; Li, Y.; Kang, S. A polymorphism at the miR-502 binding site in the 3′ untranslated region of the SET8 gene is associated with the risk of epithelial ovarian cancer. Cancer Genet. 2012, 205, 373–376. [Google Scholar] [CrossRef]
- Yang, S.; Guo, H.; Wei, B.; Zhu, S.; Cai, Y.; Jiang, P.; Tang, J. Association of miR-502-binding site single nucleotide polymorphism in the 3′-untranslated region of SET8 and TP53 codon 72 polymorphism with non-small cell lung cancer in Chinese population. Acta Biochim. Biophys. Sin. 2014, 46, 149–154. [Google Scholar]
- Wang, C.; Wu, J.; Zhao, Y.; Guo, Z. miR-502 medaited histone methyltransferase SET8 expression is associated with outcome of esophageal squamous cell carcinoma. Sci. Rep. 2016, 6, 32921. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Z.; Xu, J.; Wang, J.; Zhang, J.; Cui, L.; Zhang, H.; Liu, Y.; Bai, Y. miR-502-mediated histone methyltransferase SET8 expression is associated with clear cell renal cell carcinoma risk. Oncol. Lett. 2017, 14, 7131–7138. [Google Scholar] [CrossRef]
- Yu, N.; Huangyang, P.; Yang, X.; Han, X.; Yan, R.; Jia, H.; Shang, Y.; Sun, L. microRNA-7 Suppresses the Invasive Potential of Breast Cancer Cells and Sensitizes Cells to DNA Damages by Targeting Histone Methyltransferase SET8. J. Biol. Chem. 2013, 288, 19633–19642. [Google Scholar] [CrossRef]
- Zhang, W.; Lin, J.; Wang, P.; Sun, J. miR-17-5p down-regulation contributes to erlotinib resistance in non-small cell lung cancer cells. J. Drug Target. 2016, 25, 125–131. [Google Scholar] [CrossRef]
- Konno, Y.; Dong, P.; Xiong, Y.; Suzuki, F.; Lu, J.; Cai, M.; Watari, H.; Mitamura, T.; Hosaka, M.; Hanley, S.J.; et al. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget 2014, 5, 6049–6062. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Beckebaum, S.; Iacob, S.; Wu, G.; Kaiser, G.M.; Radtke, A.; Liu, C.; Kabar, I.; Schmidt, H.H.; Zhang, X.; et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J. Hepatol. 2014, 60, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; Chadalapaka, G.; Cho, S.G.; Lee, S.O.; Jin, U.H.; Jutooru, I.; Choi, K.; Leung, Y.K.; Ho, S.M.; Safe, S.; et al. The transcriptional repressor ZBTB4 regulates EZH2 through a MicroRNA-ZBTB4-specificity protein signaling axis. Neoplasia 2014, 16, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ni, Z.; Xu, X.; Xiao, J. MiR-32 functions as a tumor suppressor and directly targets EZH2 in human oral squamous cell carcinoma. Med. Sci. Monit. 2014, 20, 2527–2535. [Google Scholar] [PubMed]
- Zhang, K.; Zhang, Y.; Ren, K.; Zhao, G.; Yan, K.; Ma, B. MicroRNA-101 inhibits the metastasis of osteosarcoma cells by downregulation of EZH2 expression. Oncol. Rep. 2014, 32, 2143–2149. [Google Scholar] [CrossRef]
- Ciarapica, R.; Russo, G.; Verginelli, F.; Raimondi, L.; Donfrancesco, A.; Rota, R.; Giordano, A. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle 2009, 8, 172–175. [Google Scholar] [CrossRef]
- Dang, X.; Ma, A.; Yang, L.; Hu, H.; Zhu, B.; Shang, D.; Chen, T.; Luo, Y. MicroRNA-26a regulates tumorigenic properties of EZH2 in human lung carcinoma cells. Cancer Genet. 2012, 205, 113–123. [Google Scholar] [CrossRef]
- Koumangoye, R.B.; Andl, T.; Taubenslag, K.J.; Zilberman, S.T.; Taylor, C.J.; Loomans, H.A.; Andl, C.D. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol. Cancer 2015, 14, 24. [Google Scholar] [CrossRef]
- Liu, T.; Hou, L.; Huang, Y. EZH2-specific microRNA-98 inhibits human ovarian cancer stem cell proliferation via regulating the pRb-E2F pathway. Tumour Biol. 2014, 35, 7239–7247. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, Y.H.; Liu, T.H.; Chen, S.P.; Bian, X.W.; Guan, X.Y.; Lin, M.C.; Zeng, Y.X.; et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012, 61, 278–289. [Google Scholar] [CrossRef]
- Zeng, Z.; Yang, Y.; Wu, H. MicroRNA-765 alleviates the malignant progression of breast cancer via interacting with EZH1. Am. J. Transl. Res. 2019, 11, 4500–4507. [Google Scholar]
- Liu, S.; Patel, S.H.; Ginestier, C.; Ibarra, I.; Martin-Trevino, R.; Bai, S.; McDermott, S.P.; Shang, L.; Ke, J.; Ou, S.J.; et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012, 8, e1002751. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.; Mir, S.E.; Nilsson, R.J.; van der Stoop, P.M.; Niers, J.M.; Marquez, V.E.; Cloos, J.; Breakefield, X.O.; Krichevsky, A.M.; Noske, D.P.; et al. Down-regulation of miR-101 in endothelial cells promotes blood vessel formation through reduced repression of EZH2. PLoS ONE 2011, 6, e16282. [Google Scholar] [CrossRef] [PubMed]
- Wood, K.; Tellier, M.; Murphy, S. DOT1L and H3K79 Methylation in Transcription and Genomic Stability. Biomolecules 2018, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Ge, S. Targeting the histone H3 lysine 79 methyltransferase DOT1L in MLL-rearranged leukemias. J. Hematol. Oncol. 2022, 15, 35. [Google Scholar] [CrossRef]
- Sander, S.; Bullinger, L.; Klapproth, K.; Fiedler, K.; Kestler, H.A.; Barth, T.F.; Moller, P.; Stilgenbauer, S.; Pollack, J.R.; Wirth, T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood 2008, 112, 4202–4212. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Iwata, T.; Zheng, Q.; Bethel, C.; Yegnasubramanian, S.; De Marzo, A.M. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget 2011, 2, 669–683. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Chen, Z.; Jin, Y.; Wang, Y.; Kolokythas, A.; Dai, Y.; Zhou, X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011, 440, 23–31. [Google Scholar] [CrossRef]
- Castillo-Aguilera, O.; Depreux, P.; Halby, L.; Arimondo, P.B.; Goossens, L. DNA Methylation Targeting: The DNMT/HMT Crosstalk Challenge. Biomolecules 2017, 7, 3. [Google Scholar] [CrossRef]
- Friedman, J.M.; Liang, G.; Liu, C.C.; Wolff, E.M.; Tsai, Y.C.; Ye, W.; Zhou, X.; Jones, P.A. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009, 69, 2623–2629. [Google Scholar] [CrossRef] [PubMed]
- Vella, S.; Pomella, S.; Leoncini, P.P.; Colletti, M.; Conti, B.; Marquez, V.E.; Strillacci, A.; Roma, J.; Gallego, S.; Milano, G.M.; et al. MicroRNA-101 is repressed by EZH2 and its restoration inhibits tumorigenic features in embryonal rhabdomyosarcoma. Clin. Epigenetics 2015, 7, 82. [Google Scholar] [CrossRef]
- Lu, J.; He, M.L.; Wang, L.; Chen, Y.; Liu, X.; Dong, Q.; Chen, Y.C.; Peng, Y.; Yao, K.T.; Kung, H.F.; et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011, 71, 225–233. [Google Scholar] [CrossRef]
- Zhao, W.T.; Lin, X.L.; Liu, Y.; Han, L.X.; Li, J.; Lin, T.Y.; Shi, J.W.; Wang, S.C.; Lian, M.; Chen, H.W.; et al. miR-26a promotes hepatocellular carcinoma invasion and metastasis by inhibiting PTEN and inhibits cell growth by repressing EZH2. Lab. Investig. 2019, 99, 1484–1500. [Google Scholar] [CrossRef]
- Zhuang, C.; Wang, P.; Huang, D.; Xu, L.; Wang, X.; Wang, L.; Hu, L. A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. Int. J. Oncol. 2016, 48, 1195–1204. [Google Scholar] [CrossRef]
- Lu, J.; Zhao, F.P.; Peng, Z.; Zhang, M.W.; Lin, S.X.; Liang, B.J.; Zhang, B.; Liu, X.; Wang, L.; Li, G.; et al. EZH2 promotes angiogenesis through inhibition of miR-1/Endothelin-1 axis in nasopharyngeal carcinoma. Oncotarget 2014, 5, 11319–11332. [Google Scholar] [CrossRef]
- Howe, E.N.; Cochrane, D.R.; Richer, J.K. The miR-200 and miR-221/222 microRNA families: Opposing effects on epithelial identity. J. Mammary Gland. Biol. Neoplasia 2012, 17, 65–77. [Google Scholar] [CrossRef]
- Iliopoulos, D.; Lindahl-Allen, M.; Polytarchou, C.; Hirsch, H.A.; Tsichlis, P.N.; Struhl, K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell 2010, 39, 761–772. [Google Scholar] [CrossRef]
- Lin, T.; Dai, Y.; Guo, X.; Chen, W.; Zhao, J.; Cao, L.; Wu, Z. Silencing Of hsa_circ_0008450 Represses Hepatocellular Carcinoma Progression through Regulation Of microRNA-214-3p/EZH2 Axis. Cancer Manag. Res. 2019, 11, 9133–9143. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Li, G.; Li, L.; Geng, P.; Song, H. microRNA-214 suppresses the growth of cervical cancer cells by targeting EZH2. Oncol. Lett. 2018, 16, 5679–5686. [Google Scholar] [CrossRef]
- Juan, A.H.; Kumar, R.M.; Marx, J.G.; Young, R.A.; Sartorelli, V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 2009, 36, 61–74. [Google Scholar] [CrossRef]
- Yang, C.; Croteau, S.; Hardy, P. Histone deacetylase (HDAC) 9: Versatile biological functions and emerging roles in human cancer. Cell. Oncol. 2021, 44, 997–1017. [Google Scholar] [CrossRef]
- Jin, Q.; He, W.; Chen, L.; Yang, Y.; Shi, K.; You, Z. MicroRNA-101-3p inhibits proliferation in retinoblastoma cells by targeting EZH2 and HDAC9. Exp. Ther. Med. 2018, 16, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yang, C.; Gao, A.; Sun, M.; Lv, D. MiR-101: An Important Regulator of Gene Expression and Tumor Ecosystem. Cancers 2022, 14, 5861. [Google Scholar] [CrossRef] [PubMed]
- Li, C.J.; Cheng, P.; Liang, M.K.; Chen, Y.S.; Lu, Q.; Wang, J.Y.; Xia, Z.Y.; Zhou, H.D.; Cao, X.; Xie, H.; et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Investig. 2015, 125, 1509–1522. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, Z.; Zhang, X.; Liu, J.; Gui, J.; Cui, M.; Li, Y. miR-211-5p is down-regulated and a prognostic marker in bladder cancer. J. Gene Med. 2020, 22, e3270. [Google Scholar] [CrossRef]
- Jeon, H.S.; Lee, S.Y.; Lee, E.J.; Yun, S.C.; Cha, E.J.; Choi, E.; Na, M.J.; Park, J.Y.; Kang, J.; Son, J.W. Combining microRNA-449a/b with a HDAC inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer 2012, 76, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Hou, L.; Zhao, Y.; Huang, Y. Epigenetic silencing of HDAC1 by miR-449a upregulates Runx2 and promotes osteoblast differentiation. Int. J. Mol. Med. 2015, 35, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Puissegur, M.P.; Mazure, N.M.; Bertero, T.; Pradelli, L.; Grosso, S.; Robbe-Sermesant, K.; Maurin, T.; Lebrigand, K.; Cardinaud, B.; Hofman, V.; et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011, 18, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.H.; Ozer, H.G.; Gasparini, P.; Nigita, G.; Distefano, R.; Yu, L.; Ravikrishnan, J.; Yilmaz, S.; Gallegos, J.; Shukla, S.; et al. HDAC1 regulates the chromatin landscape to control transcriptional dependencies in chronic lymphocytic leukemia. Blood Adv. 2023, 7, 2897–2911. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, X.; Wang, C.; Zhang, S.; Wang, Z.; Li, M.; Wang, Y.; Wang, X.; Yang, X. HDAC6, modulated by miR-206, promotes endometrial cancer progression through the PTEN/AKT/mTOR pathway. Sci. Rep. 2020, 10, 3576. [Google Scholar] [CrossRef]
- Cao, J.; Lv, W.; Wang, L.; Xu, J.; Yuan, P.; Huang, S.; He, Z.; Hu, J. Ricolinostat (ACY-1215) suppresses proliferation and promotes apoptosis in esophageal squamous cell carcinoma via miR-30d/PI3K/AKT/mTOR and ERK pathways. Cell Death Dis. 2018, 9, 817. [Google Scholar] [CrossRef]
- Liu, F.; Zhao, X.; Qian, Y.; Zhang, J.; Zhang, Y.; Yin, R. MiR-206 inhibits Head and neck squamous cell carcinoma cell progression by targeting HDAC6 via PTEN/AKT/mTOR pathway. Biomed. Pharmacother. 2017, 96, 229–237. [Google Scholar] [CrossRef]
- Bae, H.J.; Jung, K.H.; Eun, J.W.; Shen, Q.; Kim, H.S.; Park, S.J.; Shin, W.C.; Yang, H.D.; Park, W.S.; Lee, J.Y.; et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J. Hepatol. 2015, 63, 408–419. [Google Scholar] [CrossRef]
- Wang, X.C.; Ma, Y.; Meng, P.S.; Han, J.L.; Yu, H.Y.; Bi, L.J. miR-433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral. Oncol. 2015, 51, 674–682. [Google Scholar] [CrossRef]
- Kiani, M.; Salehi, M.; Mogheiseh, A.; Mohammadi-Yeganeh, S.; Shahidi, S. The Effect of Increased miR-16-1 Levels in Mouse Embryos on Epigenetic Modification, Target Gene Expression, and Developmental Processes. Reprod. Sci. 2020, 27, 2197–2210. [Google Scholar] [CrossRef]
- Pathania, A.S. Crosstalk between Noncoding RNAs and the Epigenetics Machinery in Pediatric Tumors and Their Microenvironment. Cancers 2023, 15, 2833. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Yamamoto, A.; Yin, C.; Ide, S.; Kitajima, T.; Fujikawa, H.; Yasuda, H.; Koike, Y.; et al. Circulating miR-203 derived from metastatic tissues promotes myopenia in colorectal cancer patients. J. Cachexia Sarcopenia Muscle 2019, 10, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Q.; Niu, W.Y.; Li, Y.P.; Huang, H.B.; Zhan, R. miR-203 inhibits cell growth and regulates G1/S transition by targeting Bmi-1 in myeloma cells. Mol. Med. Rep. 2016, 14, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, S.Y.; Yan, H.Z.; Xu, D.D.; Chen, H.X.; Wang, X.Y.; Wang, X.; Liu, Y.T.; Zhang, L.; Wang, S.; et al. miR-203 inhibits proliferation and self-renewal of leukemia stem cells by targeting survivin and Bmi-1. Sci. Rep. 2016, 6, 19995. [Google Scholar] [CrossRef]
- Yang, F.; Lv, L.Z.; Cai, Q.C.; Jiang, Y. Potential roles of EZH2, Bmi-1 and miR-203 in cell proliferation and invasion in hepatocellular carcinoma cell line Hep3B. World J. Gastroenterol. 2015, 21, 13268–13276. [Google Scholar] [CrossRef]
- Chang, X.; Sun, Y.; Han, S.; Zhu, W.; Zhang, H.; Lian, S. MiR-203 inhibits melanoma invasive and proliferative abilities by targeting the polycomb group gene BMI1. Biochem. Biophys. Res. Commun. 2015, 456, 361–366. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, X.; Li, H.; Guo, L.; Jiang, W.; Lu, S.H. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014, 23, 576–585. [Google Scholar] [CrossRef]
- Arif, K.M.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory mechanisms of epigenetic miRNA relationships in human cancer and potential as therapeutic targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef]
- Perri, P.; Ponzoni, M.; Corrias, M.V.; Ceccherini, I.; Candiani, S.; Bachetti, T. A Focus on Regulatory Networks Linking MicroRNAs, Transcription Factors and Target Genes in Neuroblastoma. Cancers 2021, 13, 5528. [Google Scholar] [CrossRef]
- Chiarella, E.; Aloisio, A.; Scicchitano, S.; Bond, H.M.; Mesuraca, M. Regulatory role of microRNAs targeting the transcription co-factor ZNF521 in normal tissues and cancers. Int. J. Mol. Sci. 2021, 22, 8461. [Google Scholar] [CrossRef]
- Park, S.M.; Gaur, A.B.; Lengyel, E.; Peter, M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008, 22, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wu, L.C.; Pang, J.; Santhanam, R.; Schwind, S.; Wu, Y.Z.; Hickey, C.J.; Yu, J.; Becker, H.; Maharry, K.; et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell 2010, 17, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Mortoglou, M.; Wallace, D.; Buha Djordjevic, A.; Djordjevic, V.; Arisan, E.D.; Uysal-Onganer, P. MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma. Stresses 2021, 1, 30–47. [Google Scholar] [CrossRef]
- Sharma, A.; Mir, R.; Galande, S. Epigenetic Regulation of the Wnt/β-Catenin Signaling Pathway in Cancer. Front. Genet. 2021, 12, 681053. [Google Scholar] [CrossRef] [PubMed]
- Taulli, R.; Bersani, F.; Foglizzo, V.; Linari, A.; Vigna, E.; Ladanyi, M.; Tuschl, T.; Ponzetto, C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J. Clin. Investig. 2009, 119, 2366–2378. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Garzon, R.; Sun, H.; Ladner, K.J.; Singh, R.; Dahlman, J.; Cheng, A.; Hall, B.M.; Qualman, S.J.; Chandler, D.S.; et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 2008, 14, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Guo, L.; Dong, L.; Guo, L.; Li, S.; Zhang, J.; Sun, M. TGF-beta1 signal pathway may contribute to rhabdomyosarcoma development by inhibiting differentiation. Cancer Sci. 2010, 101, 1108–1116. [Google Scholar] [CrossRef] [PubMed]
- Ramadan, F.; Saab, R.; Hussein, N.; Clezardin, P.; Cohen, P.A.; Ghayad, S.E. Non-coding RNA in rhabdomyosarcoma progression and metastasis. Front. Oncol. 2022, 12, 971174. [Google Scholar] [CrossRef]
- Sun, M.; Huang, F.; Yu, D.; Zhang, Y.; Xu, H.; Zhang, L.; Li, L.; Dong, L.; Guo, L.; Wang, S. Autoregulatory loop between TGF-beta1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis. 2015, 6, e1859. [Google Scholar] [CrossRef]
- Zhang, X.-Z.; Liu, H.; Chen, S.-R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers 2020, 12, 1245. [Google Scholar] [CrossRef]
- Han, P.; Chang, C.-P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Hajjari, M.; Salavaty, A. HOTAIR: An oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 2015, 12, 1. [Google Scholar]
- Raju, G.S.R.; Pavitra, E.; Bandaru, S.S.; Varaprasad, G.L.; Nagaraju, G.P.; Malla, R.R.; Huh, Y.S.; Han, Y.K. HOTAIR: A potential metastatic, drug-resistant and prognostic regulator of breast cancer. Mol. Cancer 2023, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Saviana, M.; Le, P.; Micalo, L.; Del Valle-Morales, D.; Romano, G.; Acunzo, M.; Li, H.; Nana-Sinkam, P. Crosstalk between miRNAs and DNA Methylation in Cancer. Genes 2023, 14, 1075. [Google Scholar] [CrossRef]
- Wang, S.; Wu, W.; Claret, F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 2017, 12, 187–197. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes 2023, 14, 873. [Google Scholar] [CrossRef]
- Lei, Q.; Liu, X.; Fu, H.; Sun, Y.; Wang, L.; Xu, G.; Wang, W.; Yu, Z.; Liu, C.; Li, P.; et al. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget 2015, 7, 5007–5022. [Google Scholar] [CrossRef]
- Yoo, A.S.; Staahl, B.T.; Chen, L.; Crabtree, G.R. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 2009, 460, 642–646. [Google Scholar] [CrossRef]
- Moi, L.; Braaten, T.; Al-Shibli, K.; Lund, E.; Busund, L.R. Differential expression of the miR-17-92 cluster and miR-17 family in breast cancer according to tumor type; results from the Norwegian Women and Cancer (NOWAC) study. J. Transl. Med. 2019, 17, 334. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Fan, H.; Zhang, Z.; Li, N. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem. Biophys. Res. Commun. 2018, 504, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Shaban, N.Z.; Ibrahim, N.K.; Saada, H.N.; El-Rashidy, F.H.; Shaaban, H.M.; Farrag, M.A.; ElDebaiky, K.; Kodous, A.S. miR-34a and miR-21 as biomarkers in evaluating the response of chemo-radiotherapy in Egyptian breast cancer patients. J. Radiat. Res. Appl. Sci. 2022, 15, 285–292. [Google Scholar] [CrossRef]
- Wang, S.; Huang, J.; Lyu, H.; Lee, C.K.; Tan, J.; Wang, J.; Liu, B. Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis. 2013, 4, e556. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Wang, C.; Wang, M.; Li, Z.; Casimiro, M.C.; Liu, M.; Wu, K.; Whittle, J.; Ju, X.; Hyslop, T.; et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell Biol. 2008, 182, 509–517. [Google Scholar] [CrossRef]
- Hu, Q.; Huang, T. Regulation of the Cell Cycle by ncRNAs Affects the Efficiency of CDK4/6 Inhibition. Int. J. Mol. Sci. 2023, 24, 8939. [Google Scholar] [CrossRef] [PubMed]
- Kalfert, D.; Ludvikova, M.; Pesta, M.; Ludvik, J.; Dostalova, L.; Kholová, I. Multifunctional Roles of miR-34a in Cancer: A Review with the Emphasis on Head and Neck Squamous Cell Carcinoma and Thyroid Cancer with Clinical Implications. Diagnostics 2020, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.-Z.; Sun, P.; Wang, J.-P.; Liu, Y.; Gong, W.; Liu, J. MiR-34a overexpression enhances the inhibitory effect of doxorubicin on HepG2 cells. World J. Gastroenterol. 2019, 25, 2752–2762. [Google Scholar] [CrossRef]
- Fariha, A.; Hami, I.; Tonmoy, M.I.Q.; Akter, S.; Al Reza, H.; Bahadur, N.M.; Rahaman, M.M.; Hossain, M.S. Cell cycle associated miRNAs as target and therapeutics in lung cancer treatment. Heliyon 2022, 8, e11081. [Google Scholar] [CrossRef]
- Pan, W.; Chai, B.; Li, L.; Lu, Z.; Ma, Z. p53/MicroRNA-34 axis in cancer and beyond. Heliyon 2023, 9, e15155. [Google Scholar] [CrossRef]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Gregorova, J.; Vychytilova-Faltejskova, P.; Sevcikova, S. Epigenetic Regulation of MicroRNA Clusters and Families during Tumor Development. Cancers 2021, 13, 1333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, X.; Lin, J.; Lwin, T.; Wright, G.; Moscinski, L.C.; Dalton, W.S.; Seto, E.; Wright, K.; Sotomayor, E.; et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 2012, 31, 3002–3008. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Yang, Q.; Mi, Y.; Wang, F.; Cai, K.; Zhang, Y.; Wang, Y.; Wang, X.; Gui, Y.; Li, Q. miR-29b-3p Inhibitor Alleviates Hypomethylation-Related Aberrations through a Feedback Loop between miR-29b-3p and DNA Methylation in Cardiomyocytes. Front. Cell Dev. Biol. 2022, 10, 788799. [Google Scholar] [CrossRef]
- Vaiopoulos, A.G.; Athanasoula, K.; Papavassiliou, A.G. Epigenetic modifications in colorectal cancer: Molecular insights and therapeutic challenges. Biochim. Biophys. Acta 2014, 1842, 971–980. [Google Scholar] [CrossRef]
- Lan, S.H.; Lin, S.C.; Wang, W.C.; Yang, Y.C.; Lee, J.C.; Lin, P.W.; Chu, M.L.; Lan, K.Y.; Zuchini, R.; Liu, H.S.; et al. Autophagy Upregulates miR-449a Expression to Suppress Progression of Colorectal Cancer. Front. Oncol. 2021, 11, 738144. [Google Scholar] [CrossRef]
- Ishikawa, D.; Takasu, C.; Kashihara, H.; Nishi, M.; Tokunaga, T.; Higashijima, J.; Yoshikawa, K.; Yasutomo, K.; Shimada, M. The Significance of MicroRNA-449a and Its Potential Target HDAC1 in Patients With Colorectal Cancer. Anticancer. Res. 2019, 39, 2855–2860. [Google Scholar] [CrossRef]
- Abu-Hanna, J.; Patel, J.A.; Anastasakis, E.; Cohen, R.; Clapp, L.H.; Loizidou, M.; Eddama, M.M.R. Therapeutic potential of inhibiting histone 3 lysine 27 demethylases: A review of the literature. Clin. Epigenetics 2022, 14, 98. [Google Scholar] [CrossRef]
- Mohammed, M.R.S.; Zamzami, M.; Choudhry, H.; Ahmed, F.; Ateeq, B.; Khan, M.I. The Histone H3K27me3 Demethylases KDM6A/B Resist Anoikis and Transcriptionally Regulate Stemness-Related Genes. Front. Cell Dev. Biol. 2022, 10, 780176. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.; Broun, A.; Ge, K. Lysine Demethylase KDM6A in Differentiation, Development, and Cancer. Mol. Cell Biol. 2020, 40, e00341-20. [Google Scholar] [CrossRef]
- Mocavini, I.; Pippa, S.; Licursi, V.; Paci, P.; Trisciuoglio, D.; Mannironi, C.; Presutti, C.; Negri, R. JARID1B expression and its function in DNA damage repair are tightly regulated by miRNAs in breast cancer. Cancer Sci. 2019, 110, 1232–1243. [Google Scholar] [CrossRef]
- Jose, A.; Shenoy, G.G.; Sunil Rodrigues, G.; Kumar, N.A.N.; Munisamy, M.; Thomas, L.; Kolesar, J.; Rai, G.; Rao, P.P.N.; Rao, M. Histone Demethylase KDM5B as a Therapeutic Target for Cancer Therapy. Cancers 2020, 12, 2121. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Xu, W.; Cheng, P.; Jin, H.; Wang, X. Histone demethylase lysine demethylase 5B in development and cancer. Oncotarget 2017, 8, 8980–8991. [Google Scholar] [CrossRef]
- Zhang, J.; An, X.; Han, Y.; Ma, R.; Yang, K.; Zhang, L.; Chi, J.; Li, W.; Llobet-Navas, D.; Xu, Y.; et al. Overexpression of JARID1B promotes differentiation via SHIP1/AKT signaling in human hypopharyngeal squamous cell carcinoma. Cell Death Dis. 2016, 7, e2358. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Liu, D.; Liu, C.; Xu, B.; Chen, S.; Yin, Y.; Ang, L.; Huang, Y.; Zhang, X.; Chen, M. Autoregulatory feedback loop of EZH2/miR-200c/E2F3 as a driving force for prostate cancer development. Biochim. Biophys. Acta 2014, 1839, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Ji, T.; Liu, A.; Weng, Y. ELK4-mediated lncRNA SNHG22 promotes gastric cancer progression through interacting with EZH2 and regulating miR-200c-3p/Notch1 axis. Cell Death Dis. 2021, 12, 957. [Google Scholar] [CrossRef]
- Padi, S.K.; Zhang, Q.; Rustum, Y.M.; Morrison, C.; Guo, B. MicroRNA-627 mediates the epigenetic mechanisms of vitamin D to suppress proliferation of human colorectal cancer cells and growth of xenograft tumors in mice. Gastroenterology 2013, 145, 437–446. [Google Scholar] [CrossRef]
- Olejarz, W.; Kubiak-Tomaszewska, G.; Chrzanowska, A.; Lorenc, T. Exosomes in Angiogenesis and Anti-angiogenic Therapy in Cancers. Int. J. Mol. Sci. 2020, 21, 5840. [Google Scholar] [CrossRef] [PubMed]
- Unterleuthner, D.; Neuhold, P.; Schwarz, K.; Janker, L.; Neuditschko, B.; Nivarthi, H.; Crncec, I.; Kramer, N.; Unger, C.; Hengstschlager, M.; et al. Cancer-associated fibroblast-derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis 2020, 23, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Gholami, M.H.; Hushmandi, K.; Hashemi, F.; Zabolian, A.; Canadas, I.; Zarrabi, A.; Nabavi, N.; Aref, A.R.; Crea, F.; et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J. Hematol. Oncol. 2022, 15, 18. [Google Scholar] [CrossRef]
- Liang, W.; Wu, J.; Qiu, X. LINC01116 facilitates colorectal cancer cell proliferation and angiogenesis through targeting EZH2-regulated TPM1. J. Transl. Med. 2021, 19, 45. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zheng, G.; Gu, Z.; Guo, Z. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J. Neuro-Oncol. 2015, 122, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Tomaselli, D.; Lucidi, A.; Rotili, D.; Mai, A. Epigenetic polypharmacology: A new frontier for epi-drug discovery. Med. Res. Rev. 2020, 40, 190–244. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G.; Yang, H.; Bueso-Ramos, C.; Ferrajoli, A.; Cortes, J.; Wierda, W.G.; Faderl, S.; Koller, C.; Morris, G.; Rosner, G.; et al. Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood 2008, 111, 1060–1066. [Google Scholar] [CrossRef]
- Zagni, C.; Floresta, G.; Monciino, G.; Rescifina, A. The Search for Potent, Small-Molecule HDACIs in Cancer Treatment: A Decade After Vorinostat. Med. Res. Rev. 2017, 37, 1373–1428. [Google Scholar] [CrossRef]
- Duvic, M.; Dimopoulos, M. The safety profile of vorinostat (suberoylanilide hydroxamic acid) in hematologic malignancies: A review of clinical studies. Cancer Treat. Rev. 2016, 43, 58–66. [Google Scholar] [CrossRef]
- Tiffon, C.; Adams, J.; van der Fits, L.; Wen, S.; Townsend, P.; Ganesan, A.; Hodges, E.; Vermeer, M.; Packham, G. The histone deacetylase inhibitors vorinostat and romidepsin downmodulate IL-10 expression in cutaneous T-cell lymphoma cells. Br. J. Pharmacol. 2011, 162, 1590–1602. [Google Scholar] [CrossRef]
- Dedes, K.J.; Dedes, I.; Imesch, P.; von Bueren, A.O.; Fink, D.; Fedier, A. Acquired vorinostat resistance shows partial cross-resistance to ‘second-generation’ HDAC inhibitors and correlates with loss of histone acetylation and apoptosis but not with altered HDAC and HAT activities. Anticancer. Drugs 2009, 20, 321–333. [Google Scholar] [CrossRef]
- Lee, J.K.; Kim, K.C. DZNep, inhibitor of S-adenosylhomocysteine hydrolase, down-regulates expression of SETDB1 H3K9me3 HMTase in human lung cancer cells. Biochem. Biophys. Res. Commun. 2013, 438, 647–652. [Google Scholar] [CrossRef]
- Cheng, L.L.; Itahana, Y.; Lei, Z.D.; Chia, N.Y.; Wu, Y.; Yu, Y.; Zhang, S.L.; Thike, A.A.; Pandey, A.; Rozen, S.; et al. TP53 genomic status regulates sensitivity of gastric cancer cells to the histone methylation inhibitor 3-deazaneplanocin A (DZNep). Clin. Cancer Res. 2012, 18, 4201–4212. [Google Scholar] [CrossRef]
- Miranda, T.B.; Cortez, C.C.; Yoo, C.B.; Liang, G.; Abe, M.; Kelly, T.K.; Marquez, V.E.; Jones, P.A. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009, 8, 1579–1588. [Google Scholar] [CrossRef]
- Hosseinahli, N.; Aghapour, M.; Duijf, P.H.G.; Baradaran, B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell Physiol. 2018, 233, 5574–5588. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.C.; Azevedo-Pouly, A.C.; Schmittgen, T.D. MicroRNA replacement therapy for cancer. Pharm. Res. 2011, 28, 3030–3042. [Google Scholar] [CrossRef]
- Bader, A.G.; Brown, D.; Winkler, M. The promise of microRNA replacement therapy. Cancer Res. 2010, 70, 7027–7030. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Stahl, J.; Joji, A.; Volmar, C.-H.; Wahlestedt, C. Amplifying gene expression with RNA-targeted therapeutics. Nat. Rev. Drug Discov. 2023, 22, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.-W.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, J.; Wang, W.; Zhang, B.; Yu, X.; Shi, S. Epigenetic regulation in the tumor microenvironment: Molecular mechanisms and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 210. [Google Scholar] [CrossRef]
- Brest, P.; Lassalle, S.; Hofman, V.; Bordone, O.; Gavric Tanga, V.; Bonnetaud, C.; Moreilhon, C.; Rios, G.; Santini, J.; Barbry, P.; et al. MiR-129-5p is required for histone deacetylase inhibitor-induced cell death in thyroid cancer cells. Endocr. Relat. Cancer 2011, 18, 711–719. [Google Scholar] [CrossRef]
- Xue, F.; Cheng, Y.; Xu, L.; Tian, C.; Jiao, H.; Wang, R.; Gao, X. LncRNA NEAT1/miR-129/Bcl-2 signaling axis contributes to HDAC inhibitor tolerance in nasopharyngeal cancer. Aging 2020, 12, 14174–14188. [Google Scholar] [CrossRef]


| MicroRNA | Cancer Type | Role of miRNA | Target Gene(s) | Ref. |
|---|---|---|---|---|
| Tumor Suppressor microRNA TSmiRs-underexpressed in cancer cells; inhibit cancer development by negatively suppressing the function of oncogenes and/or mRNAs that modulate the cell proliferation and cycle | ||||
| let-7 family | Breast, nasopharyngeal, and oral cancer | regulates EMT process | RAS family, HMGA2, MYC, OSM | [13,14,15,16,17,18,19,20,21] |
| miR-15/16 | Melanoma, bladder, colorectal, and prostate cancer, pituitary adenomas, CLL | induces apoptosis and inhibits cancer progression; contributes aggressiveness, drug resistance | BCL2, cyclin D1, MCL1, CDC2, ETS1, JUN, ROR1 | [22,23,24,25] |
| miR-140 | Colorectal, lung, breast, and ovarian cancer | inhibits cancer progression and liver metastasis, promotes apoptosis | BCL9, BCL2, PDGFRA, ATP8A1, IGF1R | [26,27,28,29,30] |
| miR-148a | Bladder, gastric, colorectal, pancreatic, and non-small cell lung cancer | regulates growth and promotes apoptosis | BCL2, DNMT1, CCK-BR | [31,32,33,34,35] |
| miR-340 | Colon, ovarian, gastric, and endometrial cancer | triggers apoptosis and inhibits cell proliferation, repression of Wnt pathway | Notch, BCL2, BIM, Bax, RLIP76, REV3L, pro-caspase 3, β-catenin, LGR5, FHL2, NF-κB1 | [36,37,38,39,40] |
| miR-34 family | NSCLC, AML, prostate, colorecta, breast, and lung cancer | increases apoptosis, inhibits oncogene expression, represses proliferation, cell cycle progression, and induces apoptosis | SYT1, PDL-1, CDK4 | [36,41,42,43,44,45,46,47] |
| miR-200 family | Breast, thyroid, bladder, and prostate cancer | regulates EMT mechanism | TGFβ, ZEB1, ZEB2, SNAIL, TWIST, β-catenine, PDGF-D | [48] |
| miR-19 | Gastric cancer | inhibits cell proliferation, repression of Wnt pathway | MEF2D | [49] |
| miR-133a | Gastric, colorectal, cervical, and pancreatic cancer | suppresses proliferation | TCF, IGFF1R, EGFR, FSCN1 | [50,51,52,53,54] |
| miR-29 | Lymphoma, retinoblastoma, leukemia, melanoma, cervical, and lung cancer | regulate multiple oncogenic processes, including epigenetics, proteostasis, metabolism, proliferation, apoptosis, metastasis, fibrosis, angiogenesis, and immunomodulation | CDK6, DNMT3B, TCL1, and MCL1 | [55,56,57,58] |
| Oncogenic microRNA OncoMiRs-overexpressed in cancer cells; promote cancer development and progression by downregulating expression level and function of tumor suppressor gene | ||||
| miR-125 | Pancreatic, colorectal, thyroid and gastric cancer, NSCLC | Stimulates proliferation and invasion | EphA2, TAZ, TEAD2, TRIAP1, FNDC3B | [59,60,61,62,63] |
| miR-103 | Bladder, colorectal, breast and pancreatic cancer | Promotes tumor development and metastasis | DAPK, KLF4, PTEN | [64,65,66,67] |
| miR-107 | Pancreatic and cervical cancer, osteosarcoma | Stimulates proliferation and migration | SALL4, FEZF1-AS1 | [68,69,70] |
| miR-17-92 cluster (including miR106a, miR17-5p, miR19a, miR25, miR93) | Lung, thyroid, breast, colon and bladder cancer | Promotes tumor development through apoptosis inhibition | ZBTB4, BCL2L1 MYCN, GAB1, RBL1, TSG101, p63, STAT3 | [71,72,73,74,75,76] |
| miR-21 | Cervical, colorectal cancer, NSCLC, CML | Promotes proliferation, invasion, and metastasis; Inhibits apoptosis, regulates cell cycle; Increases tumor aggressiveness | PTEN, BCL11B, Ras, KRIT1 | [77,78,79,80] |
| miR-155 | Hepatocellular cancer, osteosarcoma, brain cancer | Enhances inflammatory response, promotes angiogenesis; Controls proliferation, regulates apoptosis; Augments angiogenesis and invasion | TP53, PI3K, SHIP1 | [81,82,83] |
| miR-106b-25 (including miR-106b, miR-93, and miR-25) | Breast, prostate, lung cancer, gastric, colorectal, hepatocellular and esophageal cancer | controls cell proliferation, migration, invasion, and metastases | LARP4B, DAB2, REST-1, ALEX1, FUT6, RUNX3 | [84,85] |
| Dual role microRNA | ||||
| miR-221/222 | Breast, colorectal and epithelial cancers, myeloma, glioma | Enhances cancer cell survival; Inhibits angiogenesis, reduces proliferation; Controls angiogenesis and tumor cell growth | PUMA, TRPS1, PTEN | [86,87,88,89,90,91,92,93,94,95] |
| miR-146a | Prostate, breast, and colon cancer, NSCLC | Modulates inflammation, angiogenesis; Suppresses tumor growth and progression; Regulates tumor microenvironment | Rac1, Notch2, TNFalpha, SOX5, TRAF | [96,97,98,99,100,101,102] |
| Histone Modification Type | Consequences on Gene Expression in Cancer | Examples in Cancer | Ref. |
|---|---|---|---|
| Acetylation (e.g., H3K9ac, H3K27ac) | Enhances transcription: loosens chromatin structure, promoting accessibility of transcription machinery and co-activators. | Overexpression of p300/CBP acetyltransferases in prostate cancer promotes H3K27 acetylation, enhancing androgen receptor-mediated transcription. | [136,137,138] |
| Methylation (e.g., H3K4me3) | Activation marks: enrichment at gene promoters is associated with active transcription initiation. | H3K4me3 marks are found at promoters of genes involved in cell cycle regulation, such as MYC and CCND1, in breast cancer. | [119,139,140,141] |
| Methylation (e.g., H3K9me2/3, H4K20me3, H3K27me3) | Silencing marks: dense methylation at certain sites is linked to gene repression, impacting chromatin compaction. | H3K27me3-mediated silencing of tumor suppressor genes, such as CDKN2A, is observed in various cancer types. | [112,119,141,142,143,144] |
| Methylation (e.g., H3K4me3, H3K36me3) | Transcription activation: associated with active transcription and splicing. | H3K36me3 is enriched in the bodies of actively transcribed genes, including PTEN and TP53, in renal cell carcinoma and glioblastoma. | [112,144,145,146] |
| Phosphorylation (e.g., H3S10ph) | Transcription activation: occurs during gene activation, aids chromatin decondensation and transcription factor recruitment. | Phosphorylation of H3S10 is associated with upregulation of proto-oncogenes, such as MYC, in leukemia. | [147] |
| Ubiquitination (e.g., H2BK120ub) | Transcription regulation: affects gene expression via multiple mechanisms, including recruiting transcriptional regulators. | H2BK120ub facilitates recruitment of DNA damage repair factors at the BRCA1 gene locus in breast cancer cells. | [148,149] |
| SUMOylation (e.g., H4K12su) | Transcriptional repression: can lead to heterochromatin formation, silencing gene expression. | Sumoylation occurs on both oncogenes such as MYC and β-catenin and tumor suppressors such as p53, PTEN, and BRCA1 | [150,151] |
| ADP-ribosylation (e.g., H2B-Glu35) | Gene silencing: impedes access to DNA, contributing to chromatin condensation and gene repression. | PARP-mediated ADP-ribosylation of histone H1 is linked to DNA repair at the BRCA1 gene promoter in breast cancer cells. | [152,153] |
| Crotonylation (e.g., H3K9cr, H3K122cr) | Transcription activation: associated with actively transcribed genes, potentially enhancing transcription. | Increased interferon activation and inhibition of the tumorigenic potential of glioblastoma stem cells, leading to enhanced infiltration of CD8+ T cells and slowed tumor growth. | [154,155] |
| Enzyme Category | Enzyme Types | Substrate Histones | Type of Modification | Ref. |
|---|---|---|---|---|
| Histone Acetyltransferases (HATs) | p300/CBP, GCN5, PCAF, TIP60, hCLOCK | H3, H4 | Acetylation (e.g., H3K9, H3K14, H4K5) | [156,157,158,159] |
| Histone Lysine Methyltransferases (KMTs) | SET1, SET8 SUV39H1, SUV39H2, EZH2, ASH1L, NSD1, SMYD3, DOT1L | H3, H4 | Methylation (e.g., H3K4, H4K5, H3K9, H4K20, H3K27, H3K36, H3K79) | [160,161,162,163] |
| Histone Arginine Methyltransferases (RMTs) | PRMT1,4,5,9 | H3, H4 | Methylation (e.g., H3R8 and H4R3) | [164,165,166,167] |
| Histone Phosphorylating Kinases | MSK1, CDKs | H3 | Phosphorylation (e.g., H3S10) | [147,168,169] |
| Histone Ubiquitin Ligases | RNF20/40, BRCA1 | H2B | Ubiquitination (e.g., H2BK120) | [170,171,172,173,174] |
| Histone Deacetylases (HDACs) | (1) Class I Rpd3-Like Proteins (HDAC1, HDAC2, HDAC3, and HDAC8) (2) Class II Hda1-Like Proteins (HDAC4, HDAC5, HDAC6, HDAC7, and HDAC9) (3) Class III Sir2-Like Proteins (SIRT1, SIRT2, SIRT3, SIRT5, SIRT6, and SIRT7) | H3, H4 | Deacetylation (e.g., H3K9, H3K27) | [124,126,175,176,177,178] |
| Histone Demethylases | (1) Histone lysine demethylases KDM1-8 families (e.g., KDM1A (LSD1), KDM2B (FBXL10), KDM3A (JMJD1A), KDM4B (JMJD2B), KDM5A-D (JARID1A-D), KDM6A (UTX), KDM7 (PHF2), (2) Arginine Demethylase (JMJD6) | H3 | Demethylation (e.g., H3K4, H3K27) | [124,126,129,141,175,176,177,178,179,180,181] |
| Histone Kinases | MSK1,2, PKC, RSK2, JAK2, MAP3K8, LIMK2, NEK6, BUB1, CHEK2, PAK2 | H3, H2A | Phosphorylation (e.g., H3S10, H2AS1) | [147,182,183,184] |
| Ubiquitinating Enzymes | RBX1, RNF8, HUWE1, and UHRF1 | H2A, H2B | Ubiquitinations (e.g., H2BK120ub) | [149,185,186,187] |
| Deubiquitination Enzymes | USP3, USP7, USP11 and USP22 | H2A, H2B | Deubiquitination (e.g., H2AK119, H2BK120) | [188,189,190] |
| Poly(ADP-ribose) Polymerases (PARPs) | PARP1 | H1 | ADP-ribosylation (e.g., H1) | [190,191,192] |
| SUMO Ligases | PIAS1, PIAS4 | H2B | SUMOylation (e.g., H2BK126) | [193,194,195] |
| Desumoylation Proteins | SENP1 | H2B | Desumoylation (e.g., H2BK126) | [196,197] |
| Histone Crotonyltransferases | CAT2A, CAT2B, GCN5 | H3, H4 | Crotonylation (e.g., H3K9) | [198,199,200] |
| MicroRNA | Targeted Histone-Modifying Enzymes | Consequences of Interaction | Impact on Gene Expression | Ref. |
|---|---|---|---|---|
| miR-26a, miR-101, miR-1 | EZH2 | Suppression of EZH2: Reduces trimethylation of H3K27, leading to derepression of silenced genes. | Derepression of target genes, influencing cell differentiation. | [269,280,281,282,283,284,285] |
| miR-200 and miR-221/222 families, miR-206 | SUZ12, BMI1 | Suppression of SUZ12/BMI1: Impedes PRC2 complex function, affecting H3K27me3 marks. | Altered chromatin states and changes in gene expression profiles; modulation of genes associated with cellular differentiation. | [286,287] |
| miR-214 | EZH2 | Suppression of EZH2: Reduces H3K27me3 levels, influencing chromatin accessibility. | Activation of genes associated with tumor suppression. | [286,287,288,289,290] |
| miR-101, miR-188, miR-211, miR-30d | HDAC9 | Suppression of HDAC9: Disrupts deacetylation, leading to altered chromatin structure. | Activation of genes associated with cell cycle regulation. | [291,292,293,294,295] |
| miR-449a, miR-210 | HDAC1 | Suppression of HDAC1: Impedes deacetylation, influencing chromatin compaction. | Impact on genes associated with cellular responses. | [296,297,298,299] |
| miR-22, miR- 27b, miR-206, miR-221, miR-433 | HDAC6 | Suppression of HDAC6: Alters acetylation balance, influencing gene expression. | Modulation of genes associated with cellular processes. | [300,301,302,303,304] |
| miR-16, miR-15b, miR-200 family | SUZ12 | Suppression of SUZ12: Impedes PRC2 activity, affecting H3K27me3 marks. | Altered gene expression patterns and cellular responses. | [287,305,306] |
| miR-203 | BMI1 | Suppression of BMI1: Disrupts PRC1 activity, affecting H2AK119ub marks. | Changes in gene expression profiles and cellular functions. | [285,307,308,309,310,311,312] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szczepanek, J.; Tretyn, A. MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules 2023, 13, 1590. https://doi.org/10.3390/biom13111590
Szczepanek J, Tretyn A. MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules. 2023; 13(11):1590. https://doi.org/10.3390/biom13111590
Chicago/Turabian StyleSzczepanek, Joanna, and Andrzej Tretyn. 2023. "MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications" Biomolecules 13, no. 11: 1590. https://doi.org/10.3390/biom13111590
APA StyleSzczepanek, J., & Tretyn, A. (2023). MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules, 13(11), 1590. https://doi.org/10.3390/biom13111590