The Juvenile-Hormone-Responsive Factor AmKr-h1 Regulates Caste Differentiation in Honey Bees
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Sequence Analysis of AmKr-h1 Gene
2.3. Expression Analysis Using qRT-PCR
2.4. JH Treatment
2.5. RNAi Treatment
2.6. cDNA Library Construction and Sequencing
2.7. Screening of DEGs and DEASEs
3. Results
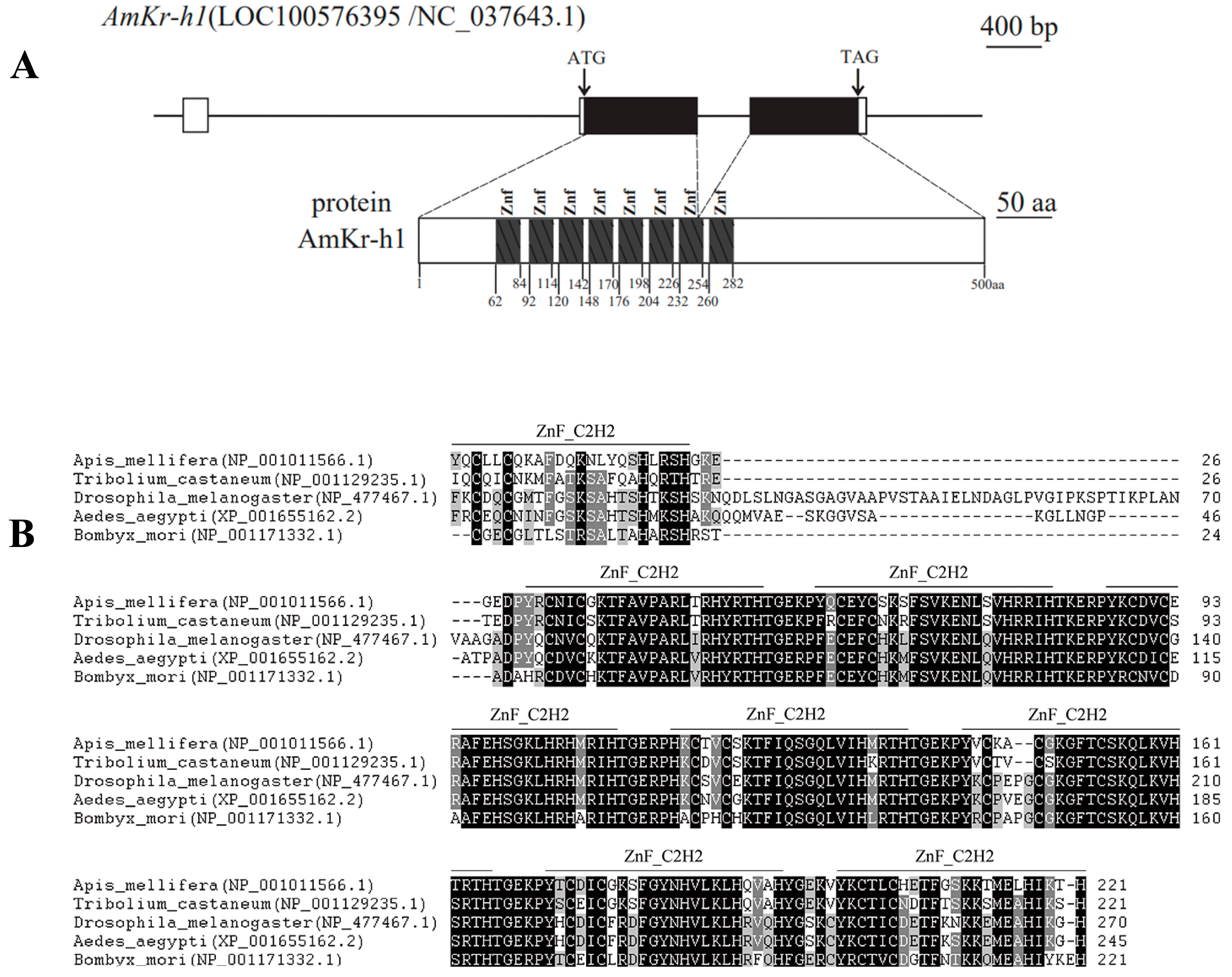
3.1. AmKr-h1 Encodes a Zinc Finger Protein
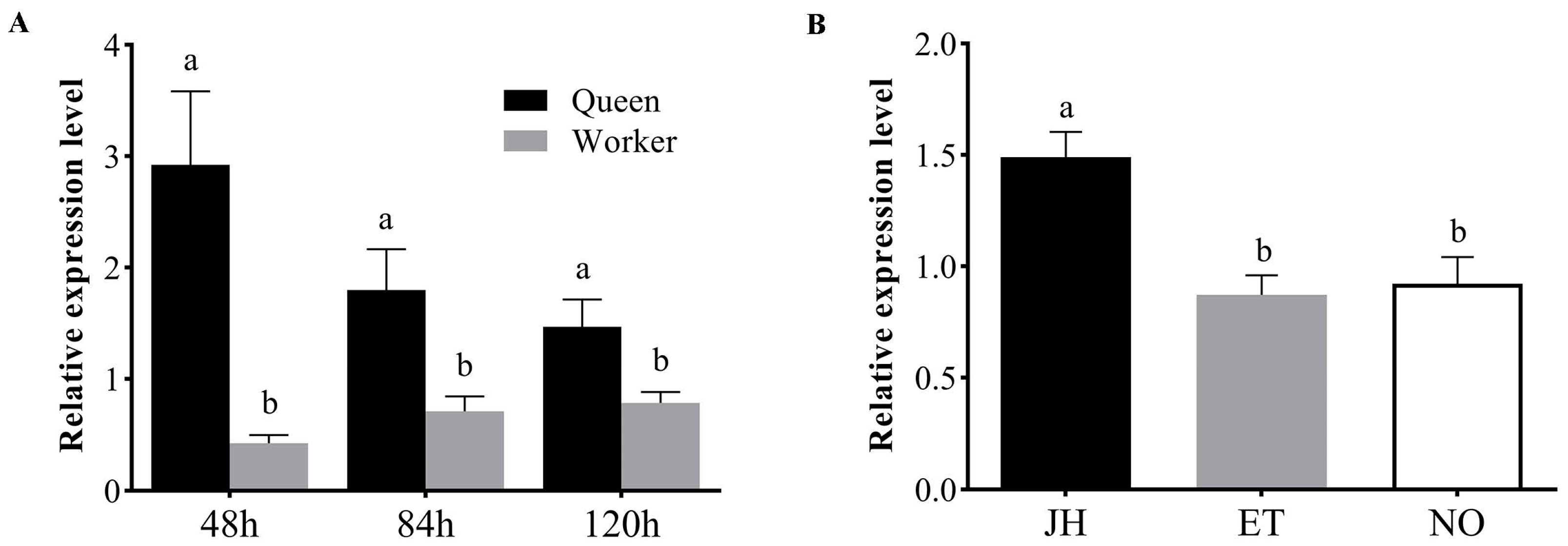
3.2. AmKr-h1 Has Higher Expression in Queen Larvae and Is Regulated by JH
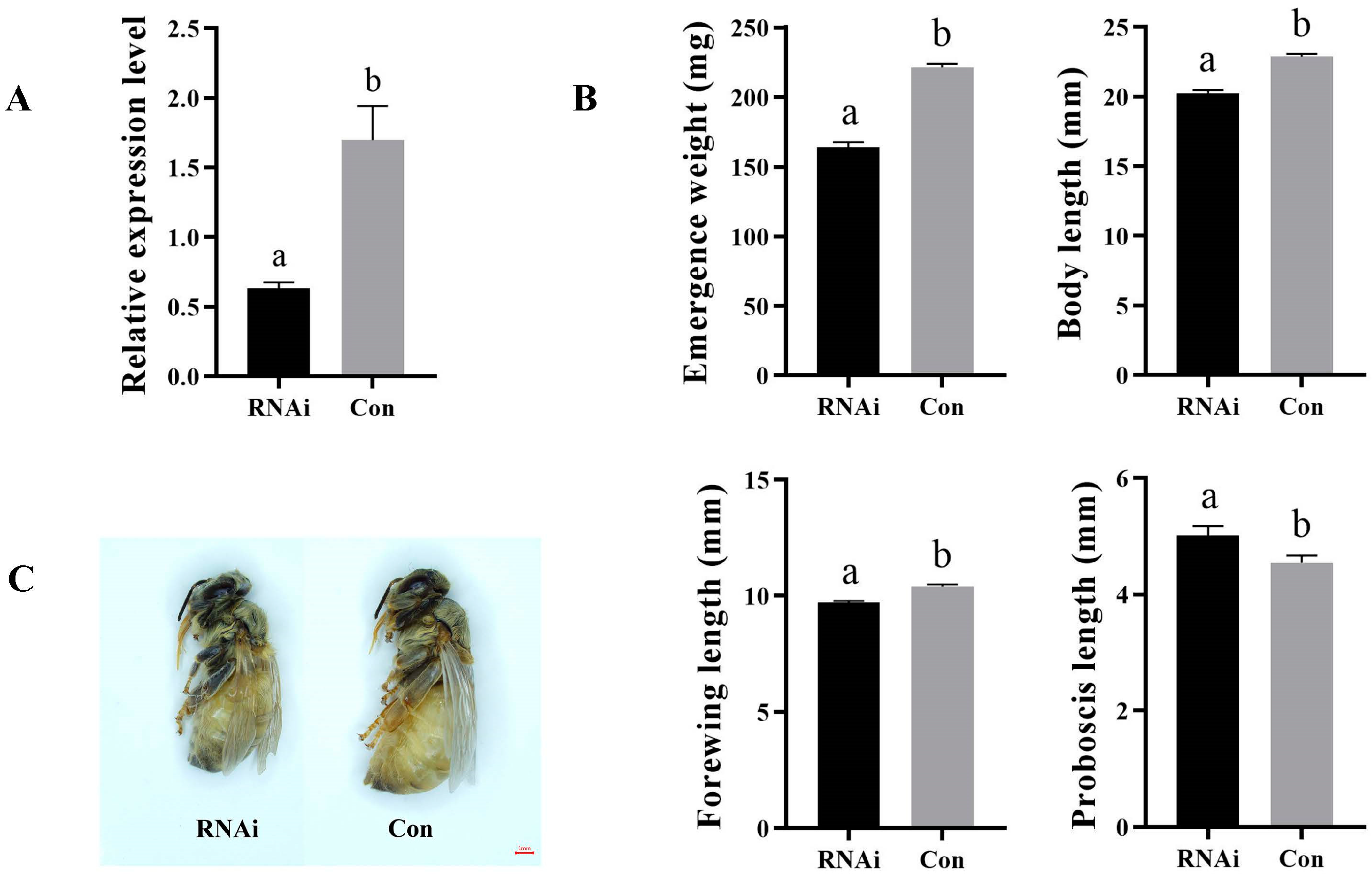
3.3. AmKr-h1 Affects Castes Differentiation of Queen–Worker
3.4. Summary of Transcriptome Sequencing Data
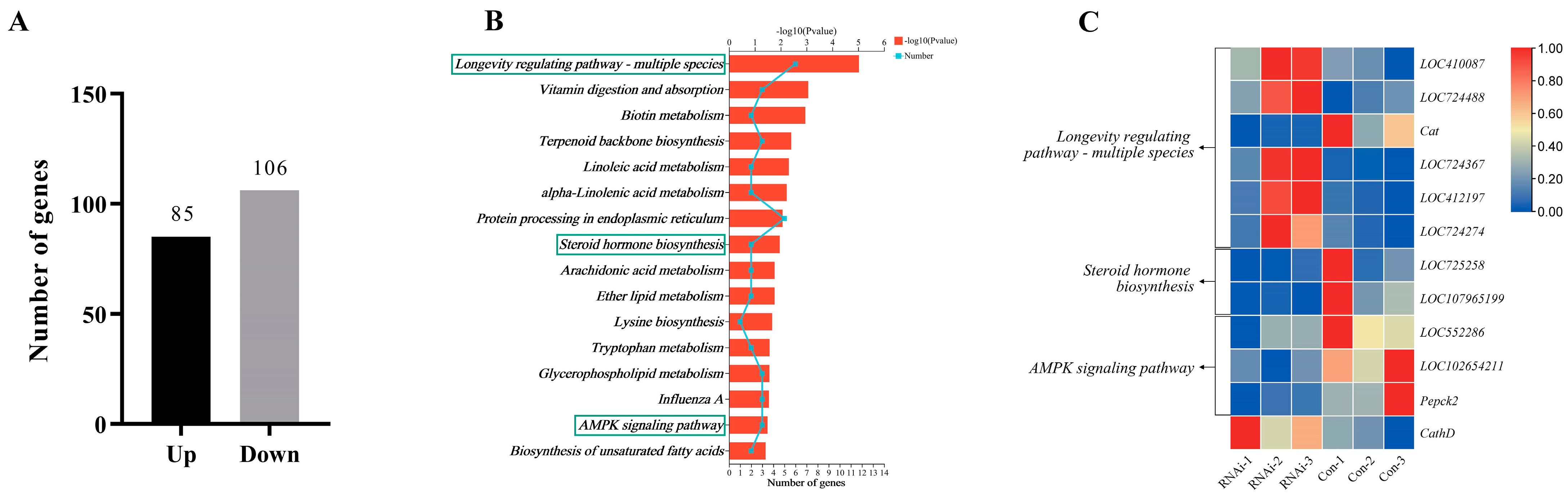
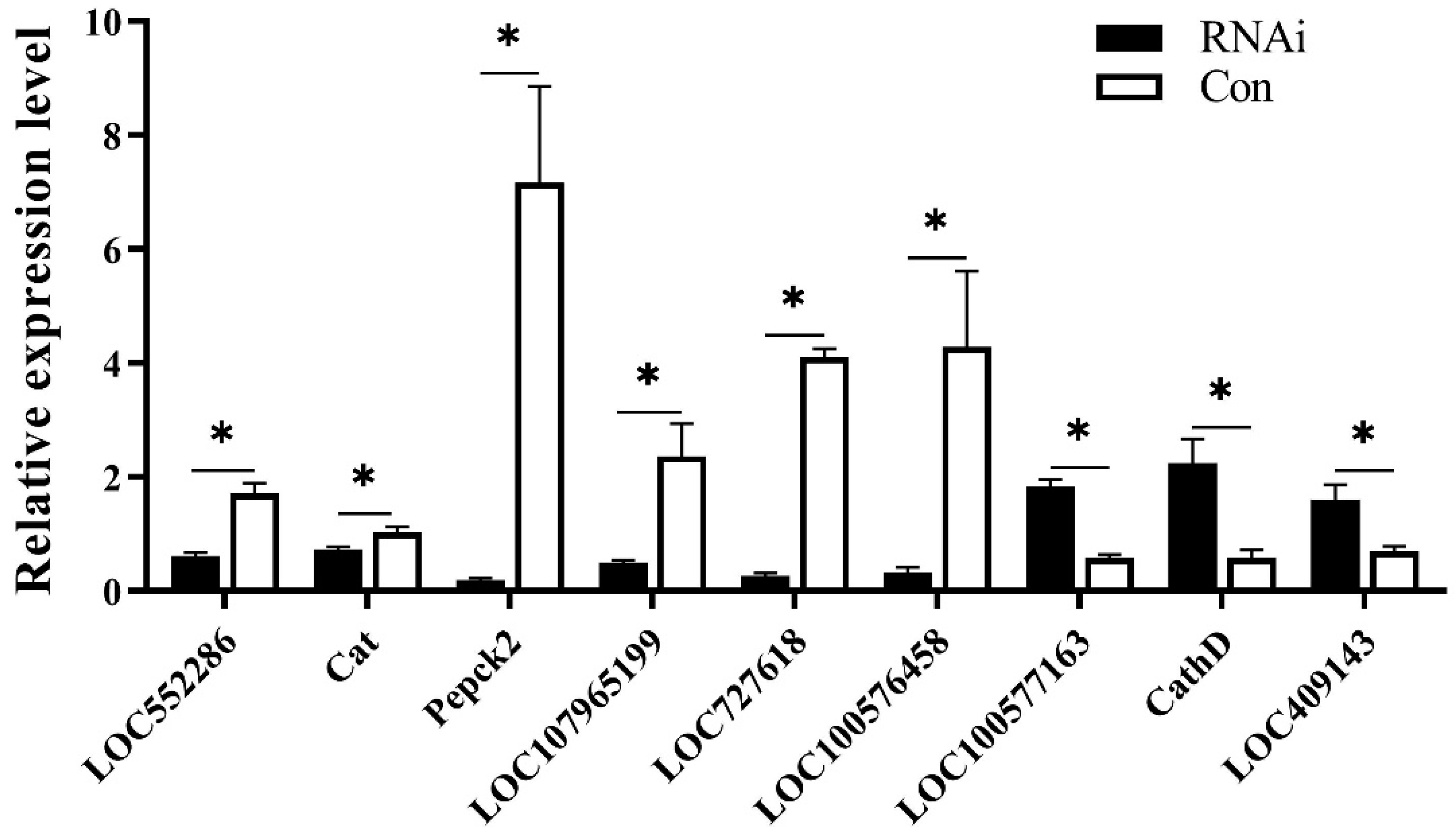
3.5. DEGs between the RNAi and Con Groups
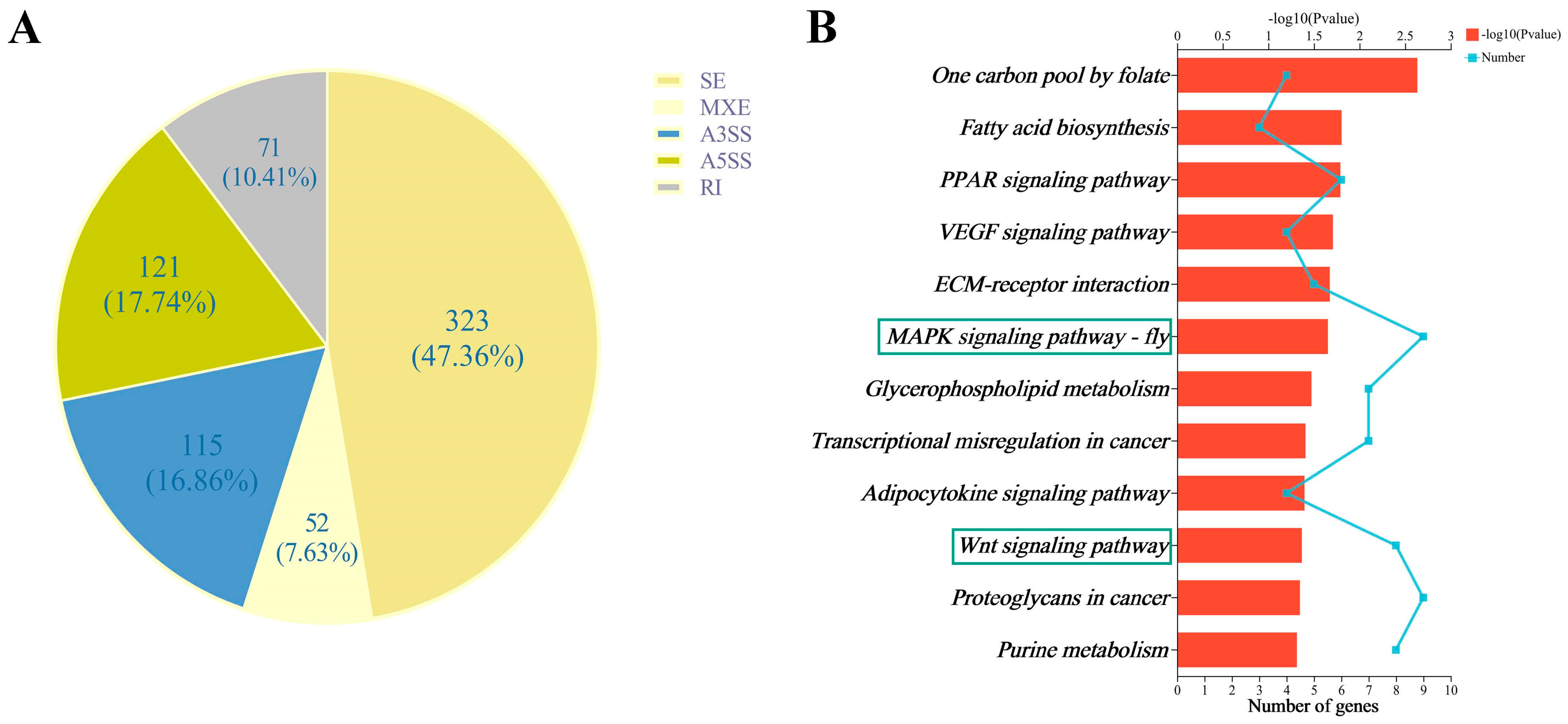
3.6. DEASEs between the RNAi and Control Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barchuk, A.R.; Cristino, A.S.; Kucharski, R.; Costa, L.F.; Simões, Z.L.; Maleszka, R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev. Biol. 2007, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Kamakura, M. Royalactin induces queen differentiation in honeybees. Nature 2011, 473, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Kucharski, R.; Maleszka, J.; Foret, S.; Maleszka, R. Nutritional Control of Reproductive Status in Honeybees via DNA Methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef] [PubMed]
- Shuel, R.W.; Dixon, S.E. The early establishment of dimorphism in the female honeybee, Apis mellifera L. Insect. Soc. 1960, 7, 265–282. [Google Scholar] [CrossRef]
- Hartfelder, K.; Engels, W. 2 Social Insect Polymorphism: Hormonal Regulation of Plasticity in Development and Reproduction in the Honeybee. In Current Topics in Developmental Biology; Pedersen, R.A., Schatten, G.P., Eds.; Academic Press: Cambridge, MA, USA, 1998; Volume 40, pp. 45–77. ISBN 0070-2153. [Google Scholar]
- Kotaki, T.; Shinada, T.; Kaihara, K.; Ohfune, Y.; Numata, H. Structure Determination of a New Juvenile Hormone from a Heteropteran Insect. Org. Lett. 2009, 11, 5234–5237. [Google Scholar] [CrossRef]
- Belles, X.; Santos, C.G. The MEKRE93 (Methoprene tolerant-Krüppel homolog 1-E93) pathway in the regulation of insect metamorphosis, and the homology of the pupal stage. Insect Biochem. Mol. Biol. 2014, 52, 60–68. [Google Scholar] [CrossRef]
- Jindra, M.; Bellés, X.; Shinoda, T. Molecular basis of juvenile hormone signaling. Curr. Opin. Insect Sci. 2015, 11, 39–46. [Google Scholar] [CrossRef]
- Li, K.; Jia, Q.Q.; Li, S. Juvenile hormone signaling—A mini review. Insect Sci. 2018, 26, 600–606. [Google Scholar] [CrossRef]
- Liu, S.; Li, K.; Gao, Y.; Liu, X.; Chen, W.; Ge, W.; Feng, Q.; Palli, S.R.; Li, S. Antagonistic actions of juvenile hormone and 20-hydroxyecdysone within the ring gland determine developmental transitions in Drosophila. Proc. Natl. Acad. Sci. USA 2018, 115, 139–144. [Google Scholar] [CrossRef]
- Jindra, M.; Palli, S.R.; Riddiford, L.M. The Juvenile Hormone Signaling Pathway in Insect Development. Annu. Rev. Entomol. 2013, 58, 181–204. [Google Scholar] [CrossRef]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory Pathways Controlling Female Insect Reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, P.; Beetsma, J. Induction of caste differentiation in the honeybee (Apis mellifera) by juvenile hormone. Entomol. Exp. Appl. 1972, 15, 517–520. [Google Scholar] [CrossRef]
- Chen, S.; Lin, C.; Lu, K. cDNA isolation, expression, and hormonal regulation of yolk protein genes in the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae). J. Insect Physiol. 2012, 58, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Kayukawa, T.; Tateishi, K.; Shinoda, T. Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 2013, 3, 1570. [Google Scholar] [CrossRef]
- He, Q.; Wen, D.; Jia, Q.; Cui, C.; Wang, J.; Palli, S.R.; Li, S. Heat Shock Protein 83 (Hsp83) Facilitates Methoprene-tolerant (Met) Nuclear Import to Modulate Juvenile Hormone Signaling. J. Biol. Chem. 2014, 289, 27874–27885. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Y.; Zheng, H.; Cao, L.; Niu, D.; Yu, D.; Sun, Y.; Hu, S.; Hu, F. Transcriptome comparison between honey bee queen- and worker-destined larvae. Insect Biochem. Mol. Biol. 2012, 42, 665–673. [Google Scholar] [CrossRef]
- Li, M.; Liu, P.; Wiley, J.D.; Ojani, R.; Bevan, D.R.; Li, J.; Zhu, J. A steroid receptor coactivator acts as the DNA-binding partner of the methoprene-tolerant protein in regulating juvenile hormone response genes. Mol. Cell. Endocrinol. 2014, 394, 47–58. [Google Scholar] [CrossRef]
- Kewley, R.J.; Whitelaw, M.L.; Chapman-Smith, A. The mammalian basic helix–loop–helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004, 36, 189–204. [Google Scholar] [CrossRef]
- Martín, D.; Chafino, S.; Franch-Marro, X. How stage identity is established in insects: The role of the Metamorphic Gene Network. Curr. Opin. Insect Sci. 2021, 43, 29–38. [Google Scholar] [CrossRef]
- Truman, J.W. The Evolution of Insect Metamorphosis. Curr. Biol. 2019, 29, R1252–R1268. [Google Scholar] [CrossRef]
- Jindra, M. Where did the pupa come from? The timing of juvenile hormone signalling supports homology between stages of hemimetabolous and holometabolous insects. Philos. Trans. R. Soc. B-Biol. Sci. 2019, 374, 20190064. [Google Scholar] [CrossRef] [PubMed]
- Pecasse, F.; Beck, Y.; Ruiz, C.; Richards, G. Krüppel-homolog, a Stage-Specific Modulator of the Prepupal Ecdysone Response, Is Essential for Drosophila Metamorphosis. Dev. Biol. 2000, 221, 53–67. [Google Scholar] [CrossRef]
- Minakuchi, C.; Namiki, T.; Shinoda, T. Krüppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 2009, 325, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Lozano, J.; Belles, X. Conserved repressive function of Krüppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci. Rep. 2011, 1, 163. [Google Scholar] [CrossRef]
- Song, J.; Wu, Z.; Wang, Z.; Deng, S.; Zhou, S. Krüppel-homolog 1 mediates juvenile hormone action to promote vitellogenesis and oocyte maturation in the migratory locust. Insect Biochem. Mol. Biol. 2014, 52, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Sun, Z.; Bai, H.; Palli, S.R. Juvenile hormone regulation of vitellogenin synthesis in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 2010, 40, 405–414. [Google Scholar] [CrossRef]
- Fichelson, P.; Brigui, A.; Pichaud, F. Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc. Natl. Acad. Sci. USA 2012, 109, 7893–7898. [Google Scholar] [CrossRef]
- Fussnecker, B.; Grozinger, C. Dissecting the role of Kr-h1 brain gene expression in foraging behavior in honey bees (Apis mellifera). Insect Mol. Biol. 2008, 17, 515–522. [Google Scholar] [CrossRef]
- Abdou, M.; Peng, C.; Huang, J.; Zyaan, O.; Wang, S.; Li, S.; Wang, J. Wnt Signaling Cross-Talks with JH Signaling by Suppressing Met and gce Expression. PLoS ONE 2011, 6, e26772. [Google Scholar] [CrossRef]
- Bernardo, T.J.; Dubrovsky, E.B. The Drosophila Juvenile Hormone Receptor Candidates Methoprene-tolerant (MET) and Germ Cell-expressed (GCE) Utilize a Conserved LIXXL Motif to Bind the FTZ-F1 Nuclear Receptor. J. Biol. Chem. 2012, 287, 7821–7833. [Google Scholar] [CrossRef]
- Dubrovsky, E. Hormonal cross talk in insect development. Trends Endocrinol. Metab. 2005, 16, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lei, X.G.; Zhu, J.; Cheng, W.; Bao, Y.; Ho, Y.; Reddi, A.R.; Holmgren, A.; Arnér, E.S.J. Paradoxical Roles of Antioxidant Enzymes: Basic Mechanisms and Health Implications. Physiol. Rev. 2016, 96, 307–364. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.E.; Souza, A.D.O.; Tibério, G.J.; Alberici, L.C.; Hartfelder, K. Differential expression of antioxidant system genes in honey bee (Apis mellifera L.) caste development mitigates ROS-mediated oxidative damage in queen larvae. Genet. Mol. Biol. 2020, 43, e20200173. [Google Scholar] [CrossRef] [PubMed]
- Raikhel, A.S. Lysosomes in the cessation of vitellogenin secretion by the mosquito fat body; selective degradation of Golgi complexes and secretory granules. Tissue Cell 1986, 18, 125–142. [Google Scholar] [CrossRef]
- Raikhel, A.S. Role of lysosomes in regulating of vitellogenin secretion in the mosquito fat body. J. Insect Physiol. 1986, 32, 597–604. [Google Scholar] [CrossRef]
- Dittmer, N.T.; Raikhel, A.S. Analysis of the mosquito lysosomal aspartic protease gene: An insect housekeeping gene with fat body-enhanced expression. Insect Biochem. Mol. Biol. 1997, 27, 323–335. [Google Scholar] [CrossRef]
- Raikhel, A.S.; Lea, A.O. Previtellogenic development and vitellogenin synthesis in the fat body of a mosquito: An ultrastructural and immunocytochemical study. Tissue Cell 1983, 15, 281–299. [Google Scholar] [CrossRef]
- Nye, C.K.; Hanson, R.W.; Kalhan, S.C. Glyceroneogenesis Is the Dominant Pathway for Triglyceride Glycerol Synthesis in Vivo in the Rat. J. Biol. Chem. 2008, 283, 27565–27574. [Google Scholar] [CrossRef]
- Wang, T.; Geng, S.L.; Guan, Y.M.; Xu, W.H. Deacetylation of metabolic enzymes by Sirt2 modulates pyruvate homeostasis to extend insect lifespan. Aging 2018, 10, 1053–1072. [Google Scholar] [CrossRef] [PubMed]
- He, X.J.; Barron, A.B.; Yang, L.; Chen, H.; He, Y.Z.; Zhang, L.Z.; Huang, Q.; Wang, Z.L.; Wu, X.B.; Yan, W.Y.; et al. Extent and complexity of RNA processing in honey bee queen and worker caste development. iScience 2022, 25, 104301. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tan, A.; Palli, S.R. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J. Insect Physiol. 2010, 56, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Mane-Padros, D.; Cruz, J.; Cheng, A.; Raikhel, A.S. A Critical Role of the Nuclear Receptor HR3 in Regulation of Gonadotrophic Cycles of the Mosquito Aedes aegypti. PLoS ONE 2012, 7, e45019. [Google Scholar] [CrossRef]
- Burmester, T.; Scheller, K. Ligands and receptors: Common theme in insect storage protein transport. Die Naturwissenschaften 1999, 86, 468–474. [Google Scholar] [CrossRef]
- Braun, R.P.; Wyatt, G.R. Sequence of the Hexameric Juvenile Hormone-binding Protein from the Hemolymph of Locusta migratoria. J. Biol. Chem. 1996, 271, 31756–31762. [Google Scholar] [CrossRef]
- Ismail, S.M.U.O.; Gillott, C. Identification, characterization, and developmental profile of a high molecular weight, juvenile hormone-binding protein in the hemolymph of the migratory grasshopper, Melanoplus sanguinipes. Arch. Insect Biochem. Physiol. 1995, 29, 415–430. [Google Scholar] [CrossRef]
- Zhou, X.; Oi, F.M.; Scharf, M.E. Social Exploitation of Hexamerin: RNAi Reveals a Major Caste-Regulatory Factor in Termites. Proc. Natl. Acad. Sci. USA 2006, 103, 4499–4504. [Google Scholar] [CrossRef]
- Martins, J.R.; Nunes, F.M.; Cristino, A.S.; Simoes, Z.L.; Bitondi, M.M. The four hexamerin genes in the honey bee: Structure, molecular evolution and function deduced from expression patterns in queens, workers and drones. BMC Mol. Biol. 2010, 11, 23. [Google Scholar] [CrossRef]
- Xiang, Y.; Liu, Z.; Huang, X. br regulates the expression of the ecdysone biosynthesis gene npc1. Dev. Biol. 2010, 344, 800–808. [Google Scholar] [CrossRef]
- Yamanaka, N.; Rewitz, K.F.; O’Connor, M.B. Ecdysone control of developmental transitions: Lessons from Drosophila research. Annu. Rev. Entomol. 2013, 58, 497–516. [Google Scholar] [CrossRef] [PubMed]
- Moeller, M.E.; Danielsen, E.T.; Herder, R.; O Connor, M.B.; Rewitz, K.F. Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development 2013, 140, 4730–4739. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, Z.-X.; Cheng, F.-P.; Xu, J.-N.; Yan, W.-Y.; Wang, Z.-L. The Juvenile-Hormone-Responsive Factor AmKr-h1 Regulates Caste Differentiation in Honey Bees. Biomolecules 2023, 13, 1657. https://doi.org/10.3390/biom13111657
Gong Z-X, Cheng F-P, Xu J-N, Yan W-Y, Wang Z-L. The Juvenile-Hormone-Responsive Factor AmKr-h1 Regulates Caste Differentiation in Honey Bees. Biomolecules. 2023; 13(11):1657. https://doi.org/10.3390/biom13111657
Chicago/Turabian StyleGong, Zhi-Xian, Fu-Ping Cheng, Jia-Ning Xu, Wei-Yu Yan, and Zi-Long Wang. 2023. "The Juvenile-Hormone-Responsive Factor AmKr-h1 Regulates Caste Differentiation in Honey Bees" Biomolecules 13, no. 11: 1657. https://doi.org/10.3390/biom13111657
APA StyleGong, Z.-X., Cheng, F.-P., Xu, J.-N., Yan, W.-Y., & Wang, Z.-L. (2023). The Juvenile-Hormone-Responsive Factor AmKr-h1 Regulates Caste Differentiation in Honey Bees. Biomolecules, 13(11), 1657. https://doi.org/10.3390/biom13111657




