Lnc-uc.147 Is Associated with Disease Stage of Liver, Gastric, and Renal Cancer
Abstract
1. Introduction
2. Materials and Methods
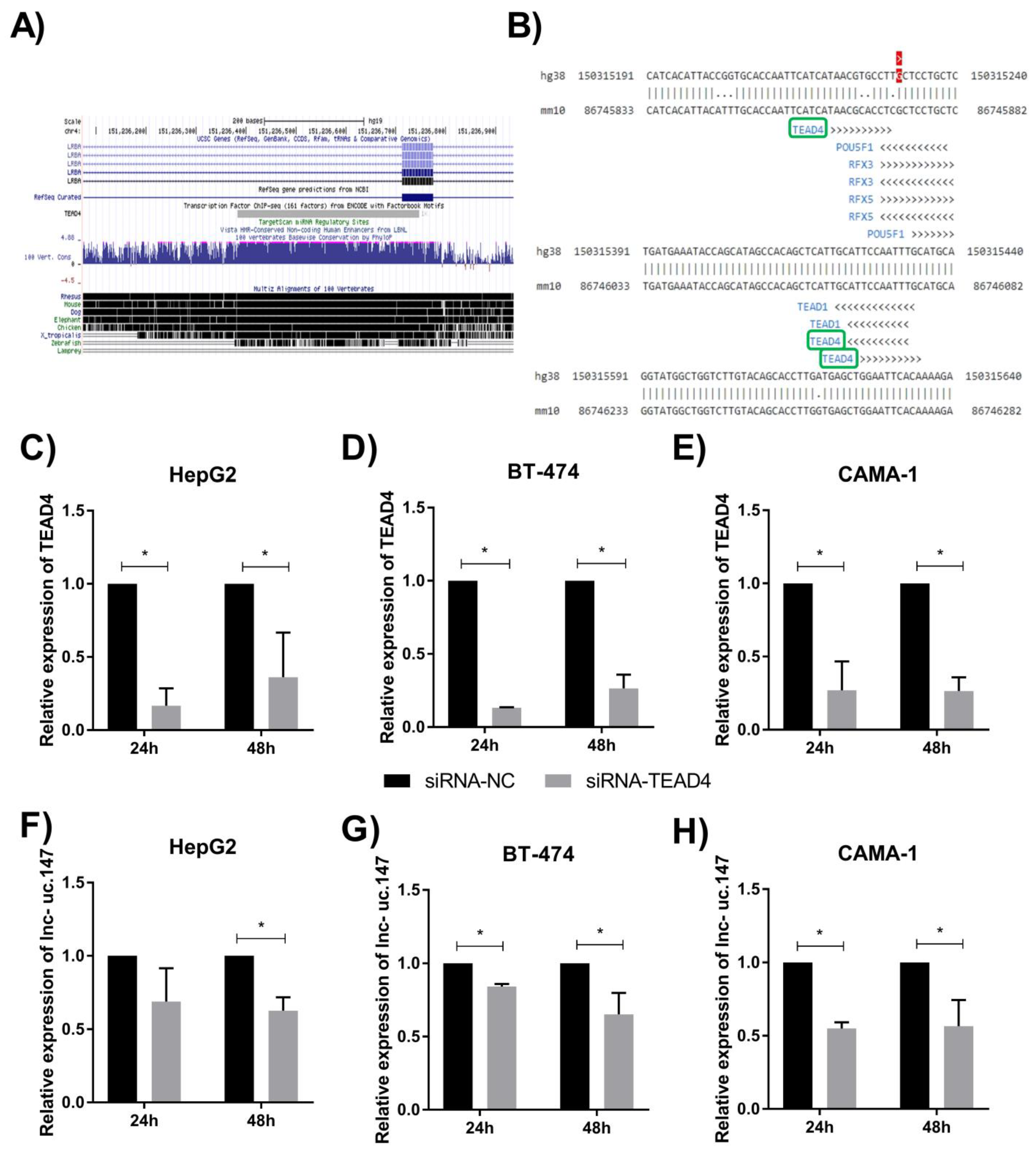
2.1. In Silico Analysis
2.2. Cell Culture and Growth Conditions
2.3. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR Analysis
2.4. siRNA Treatment
2.5. Cell Viability Assay
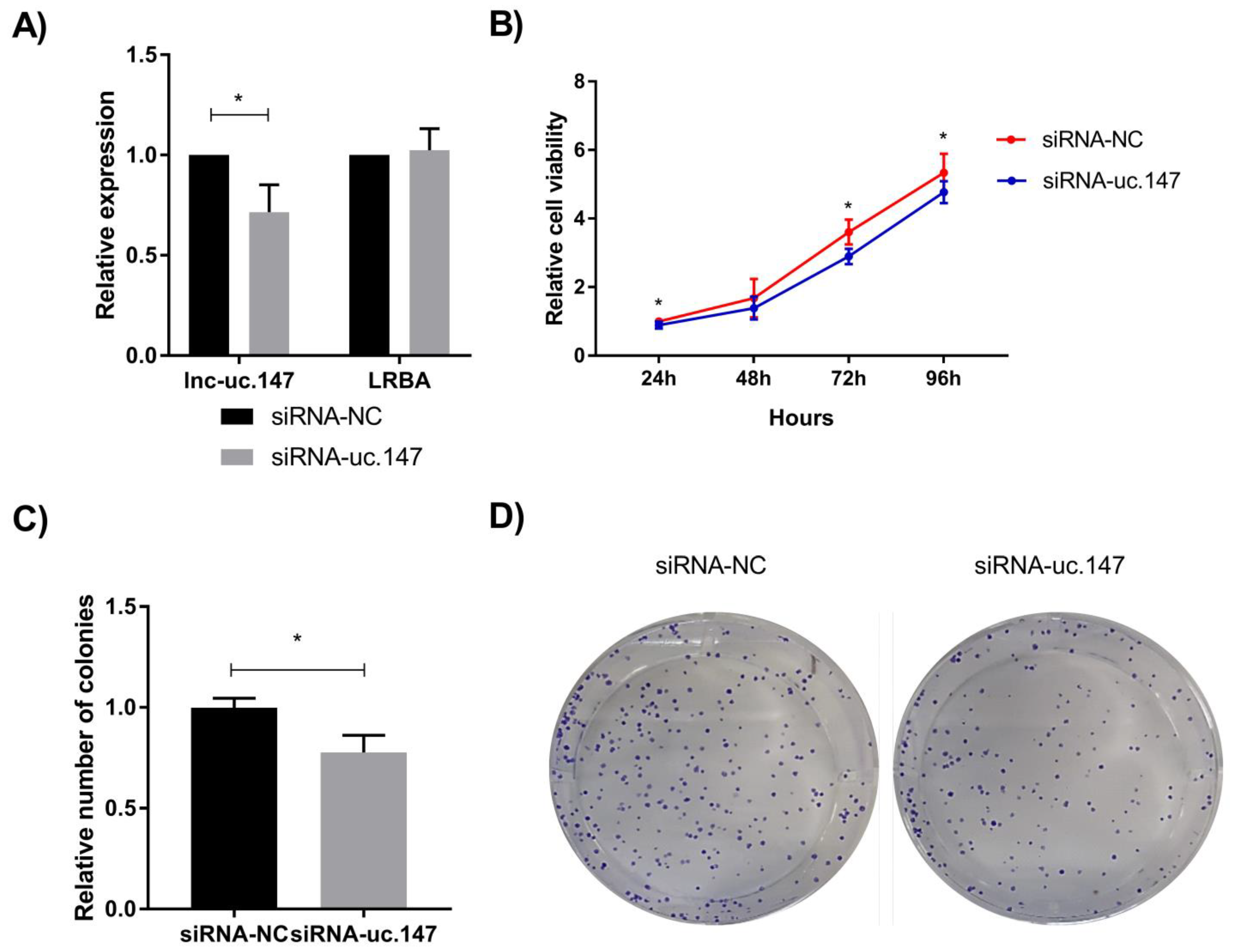
2.6. Cell Proliferation Assay
2.7. Colony Formation Assay
2.8. Apoptosis and Cell Cycle
2.9. Scratch Assay
2.10. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved Elements in the Human Genome. Science 2004, 304, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Fredlund, E.; Pattyn, F.; Rihani, A.; Van Maerken, T.; Vermeulen, J.; Kumps, C.; Menten, B.; De Preter, K.; Schramm, A.; et al. An Integrative Genomics Screen Uncovers NcRNA T-UCR Functions in Neuroblastoma Tumours. Oncogene 2010, 29, 3583–3592. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved Regions Encoding NcRNAs Are Altered in Human Leukemias and Carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef]
- Pereira Zambalde, E.; Mathias, C.; Rodrigues, A.C.; de Souza Fonseca Ribeiro, E.M.; Fiori Gradia, D.; Calin, G.A.; Carvalho de Oliveira, J. Highlighting Transcribed Ultraconserved Regions in Human Diseases. WIREs RNA 2020, 11, e1567. [Google Scholar] [CrossRef] [PubMed]
- Pereira Zambalde, E.; Bayraktar, R.; Schultz Jucoski, T.; Ivan, C.; Rodrigues, A.C.; Mathias, C.; Knutsen, E.; Silveira de Lima, R.; Fiori Gradia, D.; de Souza Fonseca Ribeiro, E.M.; et al. A Novel LncRNA Derived from an Ultraconserved Region: Lnc-Uc.147, a Potential Biomarker in Luminal A Breast Cancer. RNA Biol. 2021, 8 (Suppl. 1), 416–429. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of LncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef] [PubMed]
- Zogopoulos, V.L.; Spaho, K.; Ntouka, C.; Lappas, G.A.; Kyranis, I.; Bagos, P.G.; Spandidos, D.A.; Michalopoulos, I. TFBSPred: A Functional Transcription Factor Binding Site Prediction Webtool for Humans and Mice. Int. J. Epigenetics 2021, 1, 9. [Google Scholar] [CrossRef]
- Ahmed, S.A.; Gogal, R.M.; Walsh, J.E. A New Rapid and Simple Non-Radioactive Assay to Monitor and Determine the Proliferation of Lymphocytes: An Alternative to [3H]Thymidine Incorporation Assay. J. Immunol. Methods 1994, 170, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Didier, N.; Denton, M. Determination of Cell Number in Monolayer Cultures. Anal. Biochem. 1986, 159, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Althoff, K.N.; Jing, Y.; Horberg, M.A.; Buchacz, K.; Gill, M.J.; Justice, A.C.; Rabkin, C.S.; Goedert, J.J.; Sigel, K.; et al. Trends in Hepatocellular Carcinoma Incidence and Risk Among Persons with HIV in the US and Canada, 1996-2015. JAMA Netw. Open 2021, 4, e2037512. [Google Scholar] [CrossRef]
- Anbanandam, A.; Albarado, D.C.; Nguyen, C.T.; Halder, G.; Gao, X.; Veeraraghavan, S. Insights into Transcription Enhancer Factor 1 (TEF-1) Activity from the Solution Structure of the TEA Domain. Proc. Natl. Acad. Sci. USA 2006, 103, 17225–17230. [Google Scholar] [CrossRef]
- Jiang, S.W.; Desai, D.; Khan, S.; Eberhardt, N.L. Cooperative Binding of TEF-1 to Repeated GGAATG-Related Consensus Elements with Restricted Spatial Separation and Orientation. DNA Cell Biol. 2000, 19, 507–514. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo Pathway and Human Cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Chen, M.; Huang, B.; Zhu, L.; Chen, K.; Liu, M.; Zhong, C. Structural and Functional Overview of TEAD4 in Cancer Biology. OncoTargets Ther. 2020, 13, 9865–9874. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, M.; Lin, J.; Hu, C. Hippo/TEAD4 Signaling Pathway as a Potential Target for the Treatment of Breast Cancer. Oncol. Lett. 2021, 21, 313. [Google Scholar] [CrossRef] [PubMed]
- Coto-Llerena, M.; Tosti, N.; Taha-Mehlitz, S.; Kancherla, V.; Paradiso, V.; Gallon, J.; Bianco, G.; Garofoli, A.; Ghosh, S.; Tang, F.; et al. Transcriptional Enhancer Factor Domain Family Member 4 Exerts an Oncogenic Role in Hepatocellular Carcinoma by Hippo-Independent Regulation of Heat Shock Protein 70 Family Members. Hepatol. Commun. 2021, 5, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, P.; Tao, J.; Wang, P.; Zhang, Y.; Song, X.; Che, L.; Sumazin, P.; Ribback, S.; Kiss, A.; et al. TEA Domain Transcription Factor 4 Is the Major Mediator of Yes-Associated Protein Oncogenic Activity in Mouse and Human Hepatoblastoma. Am. J. Pathol. 2019, 189, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, L.; Zhang, Z.; Bi, M.; Wang, H.; Su, W.; Hernandez, K.; Liu, P.; Chen, J.; Chen, M.; et al. A Non-Canonical Role of YAP/TEAD Is Required for Activation of Estrogen-Regulated Enhancers in Breast Cancer. Mol. Cell 2019, 75, 791–806.e8. [Google Scholar] [CrossRef] [PubMed]

| Genes | Sequence (5′-3′) | |
|---|---|---|
| RNU6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
| TBP | Forward | TCAAACCCAGAATTGTTCTCCTTAT |
| Reverse | CCTGAATCCCTTTAGAATAGGGTAGA | |
| GAPDH | Forward | GGATTTGGTCGTATTGGG |
| Reverse | GGAAGATGGTGATGGGATT | |
| uc.147 | Forward | CGTCCTAGGCCTGTTCAAAT |
| Reverse | TGGGAATGGAATTTTCCTGA | |
| LRBA | Forward | CCAACTTCAGAGATTTGTCCAAGC |
| Reverse | ATGCTGCTCTTTTTGGGTTCAG | |
| TEAD4 | Forward | GGACACTACTCTTACCGCATCC |
| Reverse | TCAAAGACATAGGCAATGCACA |
| Type of Cancer | Association | p-Value |
|---|---|---|
| Breast invasive carcinoma (BRCA) | PAM50 | 2.72 × 10−12 |
| ER-positive | 2.74 × 10−6 | |
| Therapy | 5.16 × 10−2 | |
| Liver hepatocellular carcinoma (LIHC) | Histologic grade | 0.0181 |
| Stomach adenocarcinoma (STAD) | Disease stage | 0.0228 |
| Kidney renal clear cell carcinoma (KIRC) | Disease stage | 0.0489 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, A.C.; Zambalde, E.P.; Bellan, D.d.L.; Trindade, E.d.S.; Ribeiro, E.M.d.S.F.; Calin, G.; Gradia, D.F.; Carvalho de Oliveira, J. Lnc-uc.147 Is Associated with Disease Stage of Liver, Gastric, and Renal Cancer. Biomolecules 2023, 13, 265. https://doi.org/10.3390/biom13020265
Rodrigues AC, Zambalde EP, Bellan DdL, Trindade EdS, Ribeiro EMdSF, Calin G, Gradia DF, Carvalho de Oliveira J. Lnc-uc.147 Is Associated with Disease Stage of Liver, Gastric, and Renal Cancer. Biomolecules. 2023; 13(2):265. https://doi.org/10.3390/biom13020265
Chicago/Turabian StyleRodrigues, Ana Carolina, Erika Pereira Zambalde, Daniel de Lima Bellan, Edvaldo da Silva Trindade, Enilze Maria de Souza Fonseca Ribeiro, George Calin, Daniela Fiori Gradia, and Jaqueline Carvalho de Oliveira. 2023. "Lnc-uc.147 Is Associated with Disease Stage of Liver, Gastric, and Renal Cancer" Biomolecules 13, no. 2: 265. https://doi.org/10.3390/biom13020265
APA StyleRodrigues, A. C., Zambalde, E. P., Bellan, D. d. L., Trindade, E. d. S., Ribeiro, E. M. d. S. F., Calin, G., Gradia, D. F., & Carvalho de Oliveira, J. (2023). Lnc-uc.147 Is Associated with Disease Stage of Liver, Gastric, and Renal Cancer. Biomolecules, 13(2), 265. https://doi.org/10.3390/biom13020265









