Single-Cell RNA-Seq Analysis Reveals Macrophages Are Involved in the Pathogenesis of Human Sporadic Acute Type A Aortic Dissection
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval and Consent
2.2. Acquisition of Donor Samples
2.3. Preparation of Single-Cell Samples from the Donor Samples
2.4. Single-Cell RNA Sequencing
2.5. Single-Cell RNA-Seq Data Processing
2.6. Dimension–Reduction and Cell Clustering
2.7. Differentially Expressed Gene Analysis
2.8. Marker Gene Analysis and Cell-Type Annotation
2.9. GO and KEGG Enrichment Analysis
2.10. Pseudotime Trajectory Analysis
2.11. Cell-to-Cell Communication Analysis
2.12. Transcription Factor Analysis
2.13. Immunohistology and Western Blot Analyses
2.14. Statistical Analysis
3. Results
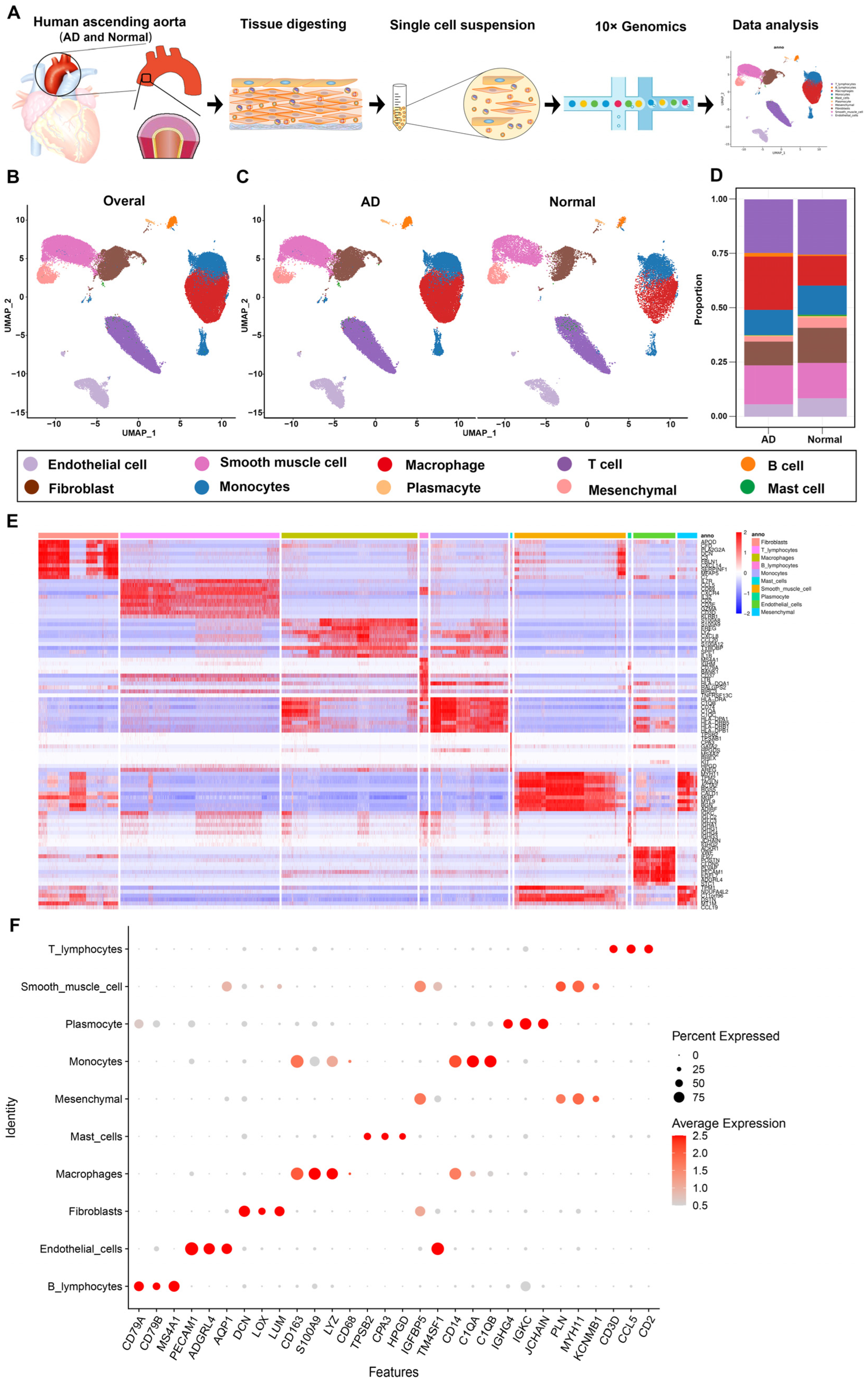
3.1. Overview of Single-Cell Transcriptomic Data from AD and Normal Ascending Aorta Tissues
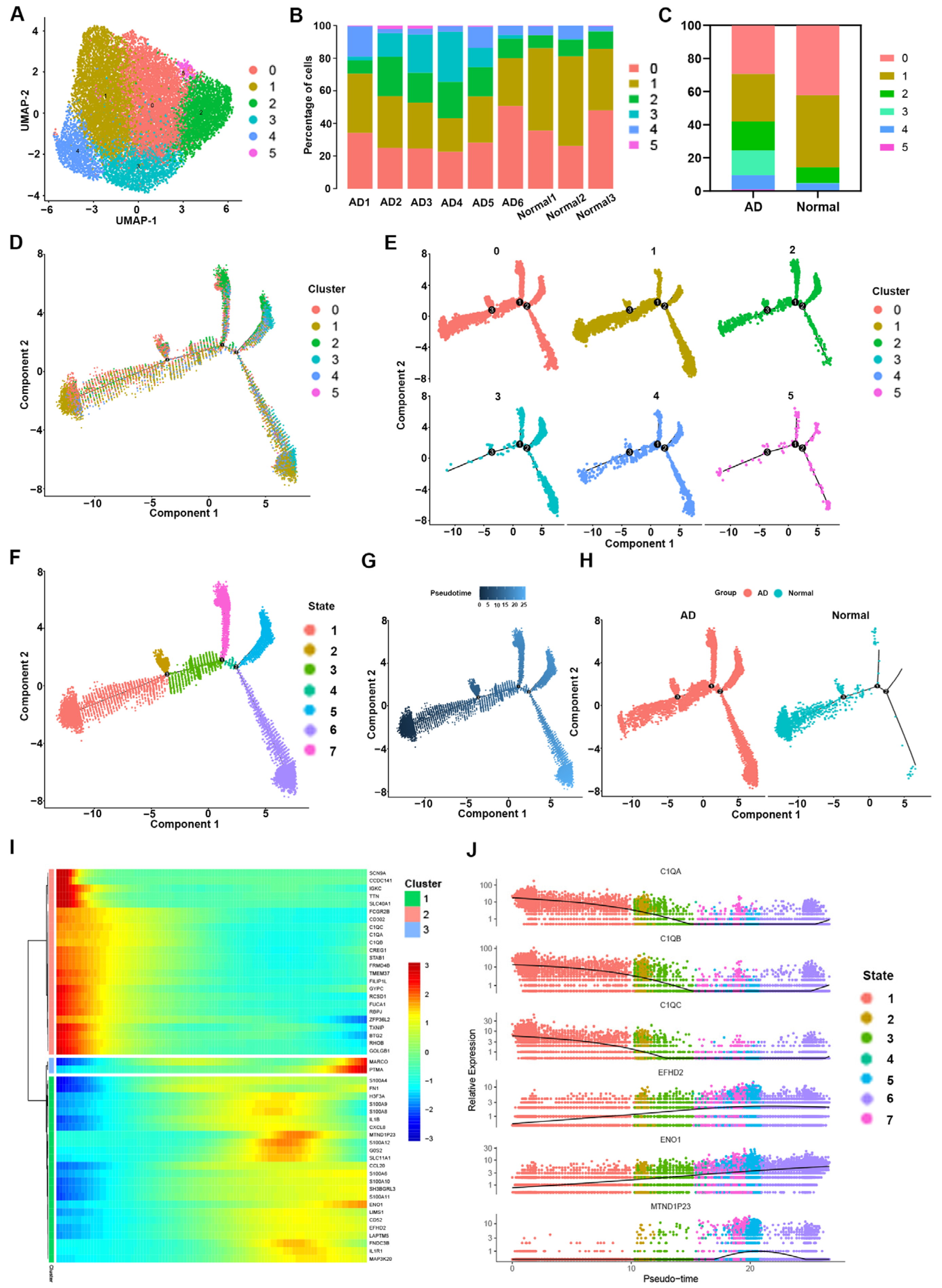
3.2. Heterogeneity of Macrophage Phenotypes and Functions between the AD and Normal Groups
3.3. Transcriptome Differences between Macrophages in the AD Group vs. the Normal Group
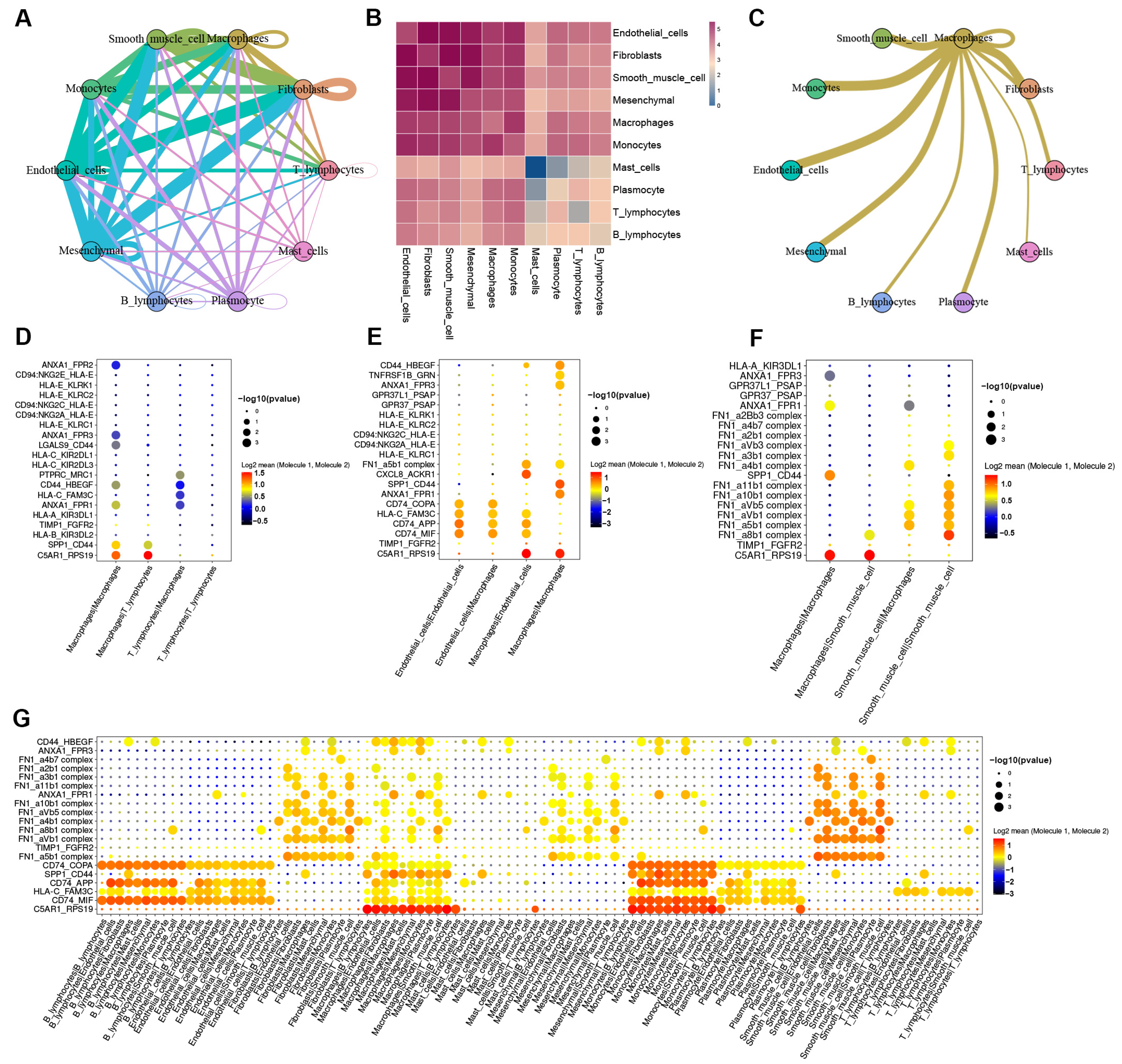
3.4. Cell-to-Cell Communication between Macrophages and Other Cells in AD tissue
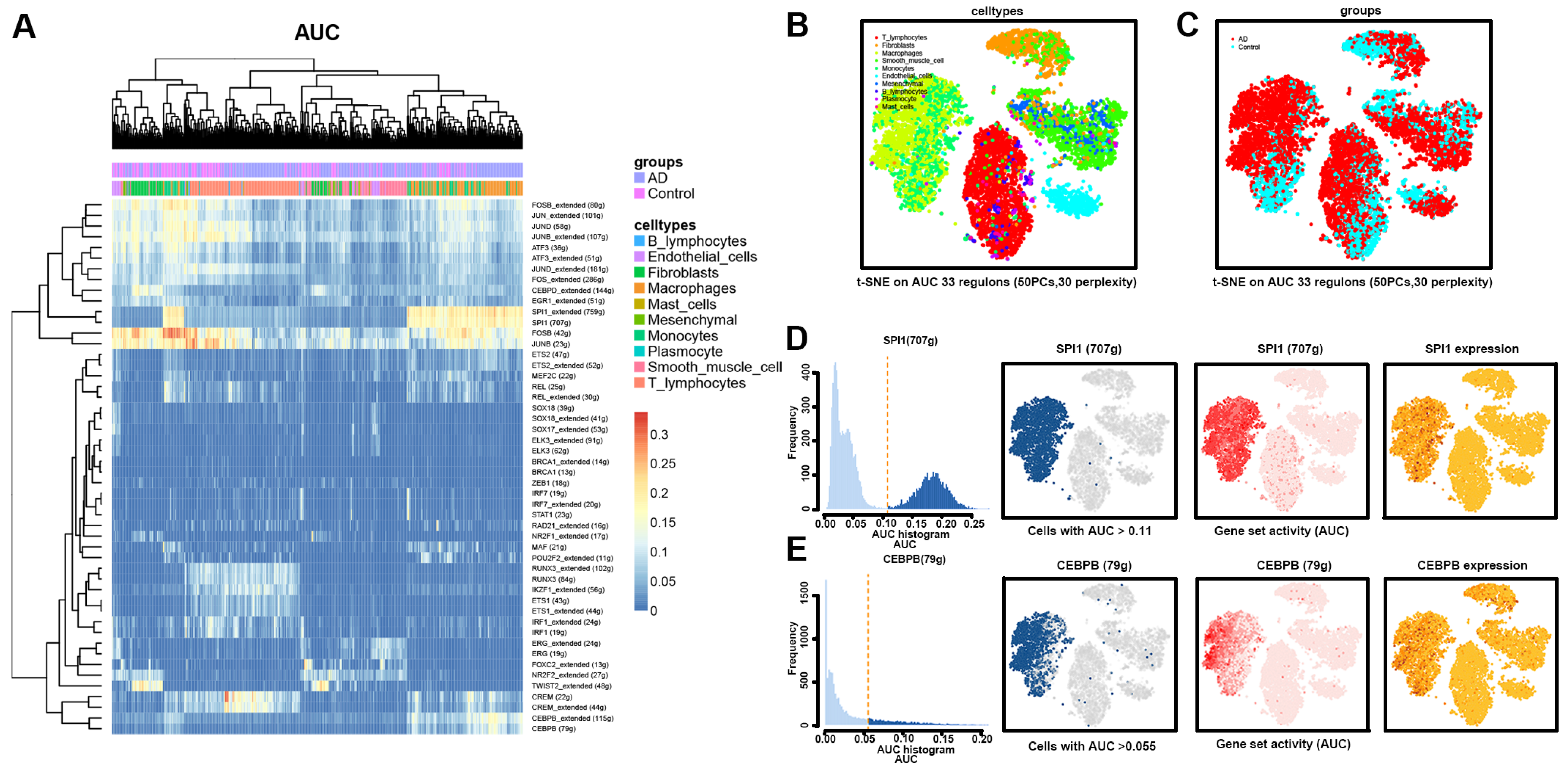
3.5. Transcriptional Regulation of Macrophages in AD Tissues
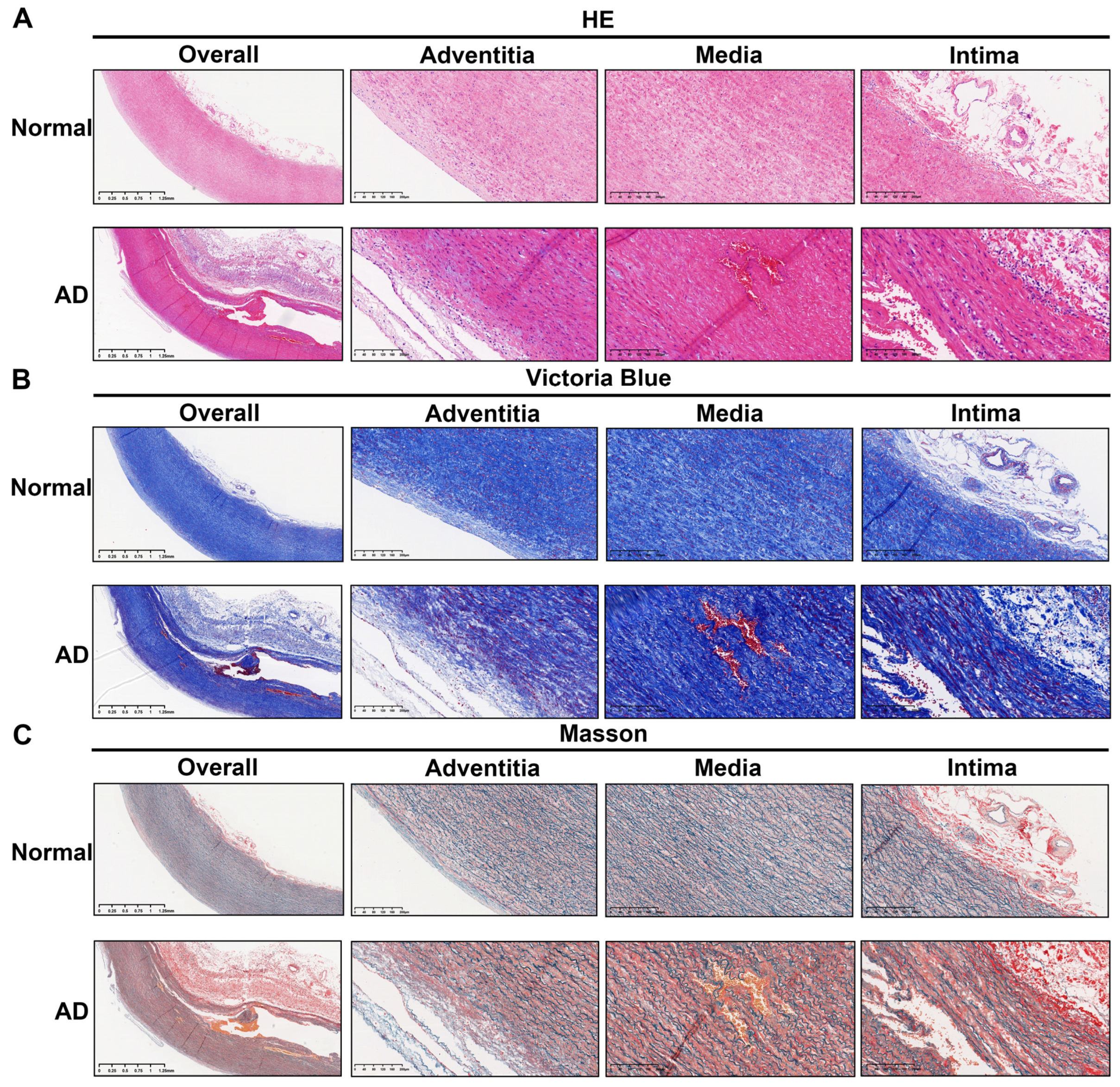
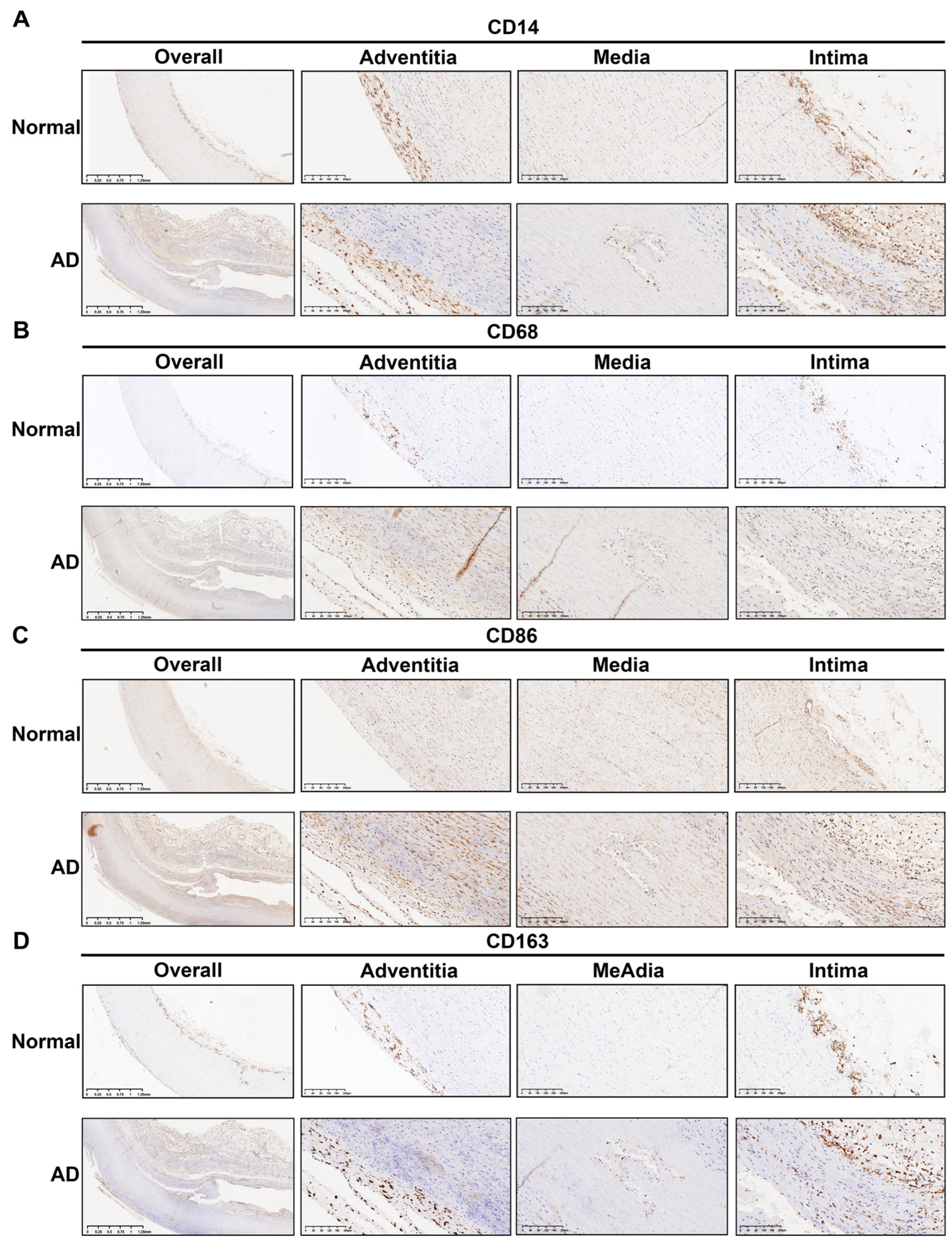
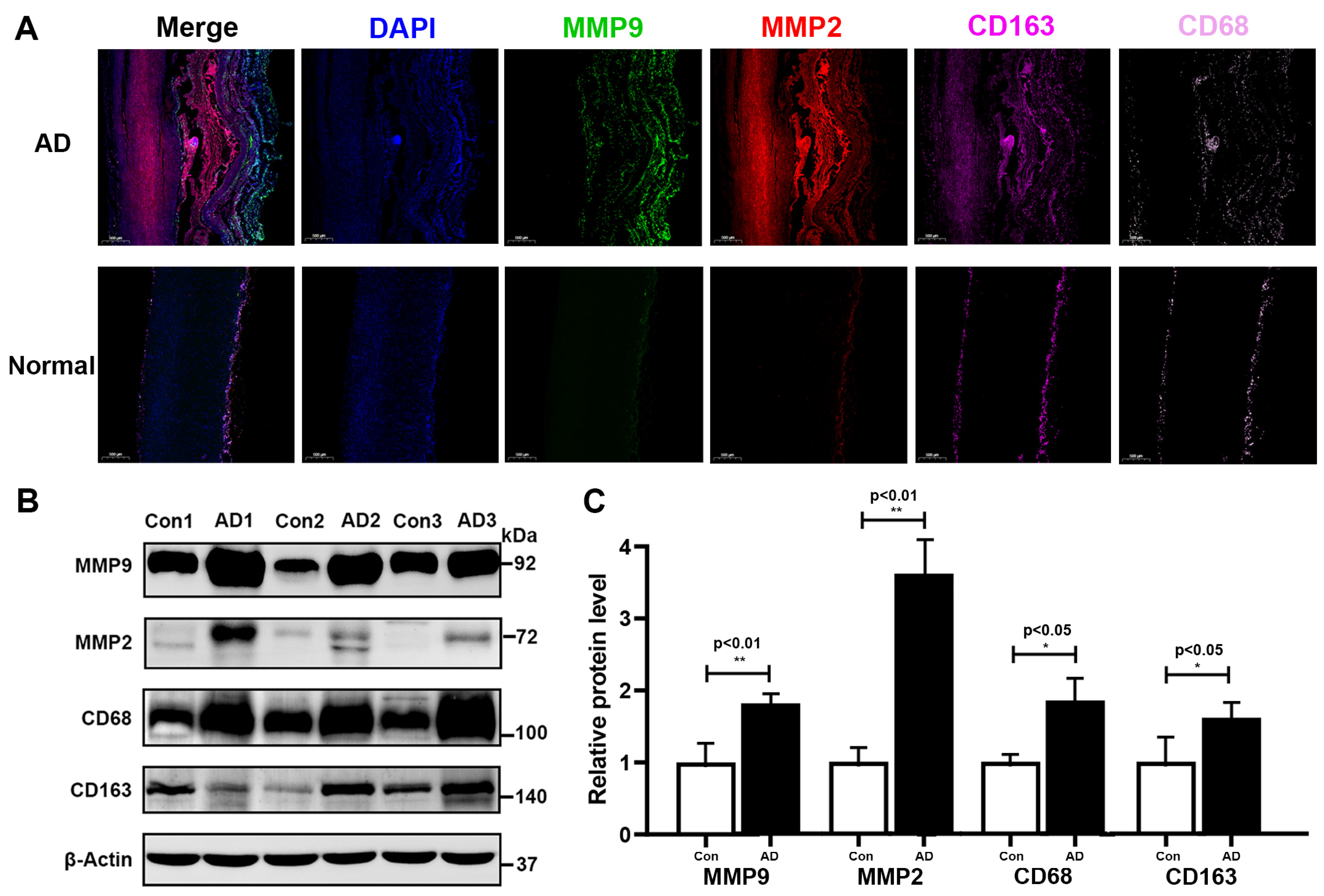
3.6. Histopathological Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harris, K.M.; Nienaber, C.A.; Peterson, M.D.; Woznicki, E.M.; Braverman, A.C.; Trimarchi, S.; Myrmel, T.; Pyeritz, R.; Hutchison, S.; Strauss, C.; et al. Early Mortality in Type A Acute Aortic Dissection: Insights From the International Registry of Acute Aortic Dissection. JAMA Cardiol. 2022, 7, 1009–1015. [Google Scholar] [CrossRef]
- Nienaber, C.A.; Clough, R.E. Management of acute aortic dissection. Lancet 2015, 385, 800–811. [Google Scholar] [CrossRef]
- Zhu, Y.; Lingala, B.; Baiocchi, M.; Tao, J.J.; Toro Arana, V.; Khoo, J.W.; Williams, K.M.; Traboulsi, A.A.; Hammond, H.C.; Lee, A.M.; et al. Type A Aortic Dissection-Experience Over 5 Decades: JACC Historical Breakthroughs in Perspective. J. Am. Coll. Cardiol. 2020, 76, 1703–1713. [Google Scholar] [CrossRef]
- Guo, D.C.; Hostetler, E.M.; Fan, Y.; Kulmacz, R.J.; Zhang, D.; Nickerson, D.A.; Leal, S.M.; LeMaire, S.A.; Regalado, E.S.; Milewicz, D.M. Heritable Thoracic Aortic Disease Genes in Sporadic Aortic Dissection. J. Am. Coll. Cardiol. 2017, 70, 2728–2730. [Google Scholar] [CrossRef]
- Ye, J.; Wang, M.; Jiang, H.; Ji, Q.; Huang, Y.; Liu, J.; Zeng, T.; Xu, Y.; Wang, Z.; Lin, Y.; et al. Increased levels of interleukin-22 in thoracic aorta and plasma from patients with acute thoracic aortic dissection. Clin. Chim. Acta Int. J. Clin. Chem. 2018, 486, 395–401. [Google Scholar] [CrossRef]
- Shen, Y.H.; LeMaire, S.A.; Webb, N.R.; Cassis, L.A.; Daugherty, A.; Lu, H.S. Aortic Aneurysms and Dissections Series. Arterioscler. Thromb. Vasc. Biol. 2020, 40, e37–e46. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, L.; Tang, H.; Li, J.; Liu, H.; Jiang, X.; Jiang, B.; Dong, Z.; Fu, W. Single-Cell Sequencing of Immune Cells in Human Aortic Dissection Tissue Provides Insights Into Immune Cell Heterogeneity. Front. Cardiovasc. Med. 2022, 9, 791875. [Google Scholar] [CrossRef]
- Grün, D.; van Oudenaarden, A. Design and Analysis of Single-Cell Sequencing Experiments. Cell 2015, 163, 799–810. [Google Scholar] [CrossRef]
- Potter, S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018, 14, 479–492. [Google Scholar] [CrossRef]
- Kalluri, A.S.; Vellarikkal, S.K.; Edelman, E.R.; Nguyen, L.; Subramanian, A.; Ellinor, P.T.; Regev, A.; Kathiresan, S.; Gupta, R.M. Single-Cell Analysis of the Normal Mouse Aorta Reveals Functionally Distinct Endothelial Cell Populations. Circulation 2019, 140, 147–163. [Google Scholar] [CrossRef]
- Luo, W.; Wang, Y.; Zhang, L.; Ren, P.; Zhang, C.; Li, Y.; Azares, A.R.; Zhang, M.; Guo, J.; Ghaghada, K.B.; et al. Critical Role of Cytosolic DNA and Its Sensing Adaptor STING in Aortic Degeneration, Dissection, and Rupture. Circulation 2020, 141, 42–66. [Google Scholar] [CrossRef]
- Li, Y.; Ren, P.; Dawson, A.; Vasquez, H.G.; Ageedi, W.; Zhang, C.; Luo, W.; Chen, R.; Li, Y.; Kim, S.; et al. Single-Cell Transcriptome Analysis Reveals Dynamic Cell Populations and Differential Gene Expression Patterns in Control and Aneurysmal Human Aortic Tissue. Circulation 2020, 142, 1374–1388. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Shaath, H.; Vishnubalaji, R.; Elkord, E.; Alajez, N.M. Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19. Cells 2020, 9, 2374. [Google Scholar] [CrossRef]
- Shuken, S.R.; McNerney, M.W. Costs and Benefits of Popular P-Value Correction Methods in Three Models of Quantitative Omic Experiments. Anal. Chem. 2023, 95, 2732–2740. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef]
- Nahrendorf, M.; Swirski, F.K. Abandoning M1/M2 for a Network Model of Macrophage Function. Circ. Res. 2016, 119, 414–417. [Google Scholar] [CrossRef]
- Mould, K.J.; Jackson, N.D.; Henson, P.M.; Seibold, M.; Janssen, W.J. Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Cochain, C.; Vafadarnejad, E.; Arampatzi, P.; Pelisek, J.; Winkels, H.; Ley, K.; Wolf, D.; Saliba, A.E.; Zernecke, A. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ. Res. 2018, 122, 1661–1674. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Paavola, C.D.; Hemmerich, S.; Grunberger, D.; Polsky, I.; Bloom, A.; Freedman, R.; Mulkins, M.; Bhakta, S.; McCarley, D.; Wiesent, L.; et al. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 1998, 273, 33157–33165. [Google Scholar] [CrossRef]
- Nelson, R.T.; Boyd, J.; Gladue, R.P.; Paradis, T.; Thomas, R.; Cunningham, A.C.; Lira, P.; Brissette, W.H.; Hayes, L.; Hames, L.M.; et al. Genomic organization of the CC chemokine mip-3alpha/CCL20/larc/exodus/SCYA20, showing gene structure, splice variants, and chromosome localization. Genomics 2001, 73, 28–37. [Google Scholar] [CrossRef]
- Ryckman, C.; Vandal, K.; Rouleau, P.; Talbot, M.; Tessier, P.A. Proinflammatory activities of S100: Proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. J. Immunol. 2003, 170, 3233–3242. [Google Scholar] [CrossRef]
- Simard, J.C.; Simon, M.M.; Tessier, P.A.; Girard, D. Damage-associated molecular pattern S100A9 increases bactericidal activity of human neutrophils by enhancing phagocytosis. J. Immunol. 2011, 186, 3622–3631. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.; Yang, Z.; Jin, H.; Zou, Q.; Zhan, Q.; Tang, Y.; Tao, Y.; Lei, L.; Jing, Y.; et al. Up-regulation of EMT-related gene VCAN by NPM1 mutant-driven TGF-β/cPML signalling promotes leukemia cell invasion. J. Cancer 2019, 10, 6570–6583. [Google Scholar] [CrossRef]
- Han, Z.L.; Wang, H.Q.; Zhang, T.S.; He, Y.X.; Zhou, H. Up-regulation of exosomal miR-106a may play a significant role in abdominal aortic aneurysm by inducing vascular smooth muscle cell apoptosis and targeting TIMP-2, an inhibitor of metallopeptidases that suppresses extracellular matrix degradation. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8087–8095. [Google Scholar] [CrossRef]
- Yarnazari, A.; Hassanpour, P.; Hosseini-Fard, S.R.; Amirfarhangi, A.; Najafi, M. The sdLDL Reduces MRC1 Expression Level and Secretion of Histamin e in Differentiated M2-macrophages from Patients with Coronary Artery Stenosis. Cardiovasc. Hematol. Disord. Drug Targets 2017, 17, 28–32. [Google Scholar] [CrossRef]
- Purves-Tyson, T.D.; Robinson, K.; Brown, A.M.; Boerrigter, D.; Cai, H.Q.; Weissleder, C.; Owens, S.J.; Rothmond, D.A.; Shannon Weickert, C. Increased Macrophages and C1qA, C3, C4 Transcripts in the Midbrain of People With Schizophrenia. Front. Immunol. 2020, 11, 2002. [Google Scholar] [CrossRef]
- Tong, H.H.; Li, Y.X.; Stahl, G.L.; Thurman, J.M. Enhanced susceptibility to acute pneumococcal otitis media in mice deficient in complement C1qa, factor B, and factor B/C2. Infect. Immun. 2010, 78, 976–983. [Google Scholar] [CrossRef]
- Gerard, N.P.; Gerard, C. The chemotactic receptor for human C5a anaphylatoxin. Nature 1991, 349, 614–617. [Google Scholar] [CrossRef]
- Monk, P.N.; Barker, M.D.; Partridge, L.J.; Pease, J.E. Mutation of glutamate 199 of the human C5a receptor defines a binding site for ligand distinct from the receptor N terminus. J. Biol. Chem. 1995, 270, 16625–16629. [Google Scholar] [CrossRef]
- Peng, Q.; Wu, W.; Wu, K.Y.; Cao, B.; Qiang, C.; Li, K.; Sacks, S.H.; Zhou, W. The C5a/C5aR1 axis promotes progression of renal tubulointerstitial fibrosis in a mouse model of renal ischemia/reperfusion injury. Kidney Int. 2019, 96, 117–128. [Google Scholar] [CrossRef]
- Lv, J.; Huang, X.R.; Klug, J.; Fröhlich, S.; Lacher, P.; Xu, A.; Meinhardt, A.; Lan, H.Y. Ribosomal protein S19 is a novel therapeutic agent in inflammatory kidney disease. Clin. Sci. 2013, 124, 627–637. [Google Scholar] [CrossRef]
- Fan, C.; Rajasekaran, D.; Syed, M.A.; Leng, L.; Loria, J.P.; Bhandari, V.; Bucala, R.; Lolis, E.J. MIF intersubunit disulfide mutant antagonist supports activation of CD74 by endogenous MIF trimer at physiologic concentrations. Proc. Natl. Acad. Sci. USA 2013, 110, 10994–10999. [Google Scholar] [CrossRef]
- Cao, G.; Lu, Z.; Gu, R.; Xuan, X.; Zhang, R.; Hu, J.; Dong, H. Deciphering the Intercellular Communication Between Immune Cells and Altered Vascular Smooth Muscle Cell Phenotypes in Aortic Aneurysm From Single-Cell Transcriptome Data. Front. Cardiovasc. Med. 2022, 9, 936287. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, X.; Iwama, A.; Yu, C.; Smith, K.A.; Mueller, B.U.; Narravula, S.; Torbett, B.E.; Orkin, S.H.; Tenen, D.G.P.U. 1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood 2000, 96, 2641–2648. [Google Scholar] [CrossRef]
- McIvor, Z.; Hein, S.; Fiegler, H.; Schroeder, T.; Stocking, C.; Just, U.; Cross, M. Transient expression of PU.1 commits multipotent progenitors to a myeloid fate whereas continued expression favors macrophage over granulocyte differentiation. Exp. Hematol. 2003, 31, 39–47. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Q.; Zhou, J.; Wang, Y.; Xu, C.; Tong, F.; Wang, H.; Kang, C. Single-Cell Transcriptomics of Glioblastoma Reveals a Unique Tumor Microenvironment and Potential Immunotherapeutic Target Against Tumor-Associated Macrophage. Front. Oncol. 2021, 11, 710695. [Google Scholar] [CrossRef]
- Kinoshita, S.; Akira, S.; Kishimoto, T. A member of the C/EBP family, NF-IL6 beta, forms a heterodimer and transcriptionally synergizes with NF-IL6. Proc. Natl. Acad. Sci. USA 1992, 89, 1473–1476. [Google Scholar] [CrossRef]
- Chinery, R.; Brockman, J.A.; Dransfield, D.T.; Coffey, R.J. Antioxidant-induced nuclear translocation of CCAAT/enhancer-binding protein beta. A critical role for protein kinase A-mediated phosphorylation of Ser299. J. Biol. Chem. 1997, 272, 30356–30361. [Google Scholar] [CrossRef]
- Roy, S.K.; Hu, J.; Meng, Q.; Xia, Y.; Shapiro, P.S.; Reddy, S.P.; Platanias, L.C.; Lindner, D.J.; Johnson, P.F.; Pritchard, C.; et al. MEKK1 plays a critical role in activating the transcription factor C/EBP-beta-dependent gene expression in response to IFN-gamma. Proc. Natl. Acad. Sci. USA 2002, 99, 7945–7950. [Google Scholar] [CrossRef]
- Ruffell, D.; Mourkioti, F.; Gambardella, A.; Kirstetter, P.; Lopez, R.G.; Rosenthal, N.; Nerlov, C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA 2009, 106, 17475–17480. [Google Scholar] [CrossRef]
- Schoenen, H.; Huber, A.; Sonda, N.; Zimmermann, S.; Jantsch, J.; Lepenies, B.; Bronte, V.; Lang, R. Differential control of Mincle-dependent cord factor recognition and macrophage responses by the transcription factors C/EBPβ and HIF1α. J. Immunol. 2014, 193, 3664–3675. [Google Scholar] [CrossRef]
- Goldfinger, J.Z.; Halperin, J.L.; Marin, M.L.; Stewart, A.S.; Eagle, K.A.; Fuster, V. Thoracic aortic aneurysm and dissection. J. Am. Coll. Cardiol. 2014, 64, 1725–1739. [Google Scholar] [CrossRef]
- Itoh, Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. J. Int. Soc. Matrix Biol. 2015, 44–46, 207–223. [Google Scholar] [CrossRef]
- Mizrak, D.; Feng, H.; Yang, B. Dissecting the Heterogeneity of Human Thoracic Aortic Aneurysms Using Single-Cell Transcriptomics. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 919–930. [Google Scholar] [CrossRef]
- Pisano, C.; Balistreri, C.R.; Ricasoli, A.; Ruvolo, G. Cardiovascular Disease in Ageing: An Overview on Thoracic Aortic Aneurysm as an Emerging Inflammatory Disease. Mediat. Inflamm. 2017, 2017, 1274034. [Google Scholar] [CrossRef]
- Zhao, G.; Lu, H.; Chang, Z.; Zhao, Y.; Zhu, T.; Chang, L.; Guo, Y.; Garcia-Barrio, M.T.; Chen, Y.E.; Zhang, J. Single-cell RNA sequencing reveals the cellular heterogeneity of aneurysmal infrarenal abdominal aorta. Cardiovasc. Res. 2021, 117, 1402–1416. [Google Scholar] [CrossRef]
- Wu, D.; Choi, J.C.; Sameri, A.; Minard, C.G.; Coselli, J.S.; Shen, Y.H.; LeMaire, S.A. Inflammatory Cell Infiltrates in Acute and Chronic Thoracic Aortic Dissection. Aorta 2013, 1, 259–267. [Google Scholar] [CrossRef]
- Niinimäk, E.; Pynnönen, V.; Kholova, I.; Paavonen, T.; Mennander, A. Neovascularization with chronic inflammation characterizes ascending aortic dissection. Anatol. J. Cardiol. 2018, 20, 289–295. [Google Scholar] [CrossRef]
- Miyabe, C.; Miyabe, Y.; Bricio-Moreno, L.; Lian, J.; Rahimi, R.A.; Miura, N.N.; Ohno, N.; Iwakura, Y.; Kawakami, T.; Luster, A.D. Dectin-2-induced CCL2 production in tissue-resident macrophages ignites cardiac arteritis. J. Clin. Investig. 2019, 129, 3610–3624. [Google Scholar] [CrossRef]
- Epelman, S.; Lavine, K.J.; Beaudin, A.E.; Sojka, D.K.; Carrero, J.A.; Calderon, B.; Brija, T.; Gautier, E.L.; Ivanov, S.; Satpathy, A.T.; et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity 2014, 40, 91–104. [Google Scholar] [CrossRef]
- Chen, F.; Han, J.; Tang, B. Patterns of Immune Infiltration and the Key Immune-Related Genes in Acute Type A Aortic Dissection in Bioinformatics Analyses. Int. J. Gen. Med. 2021, 14, 2857–2869. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, H.; Loyd, C.M.; Fu, W.; Diaconu, D.; Liu, S.; Cooper, K.D.; McCormick, T.S.; Simon, D.I.; Ward, N.L. Chronic skin-specific inflammation promotes vascular inflammation and thrombosis. J. Investig. Dermatol. 2012, 132, 2067–2075. [Google Scholar] [CrossRef]
- Farris, S.D.; Hu, J.H.; Krishnan, R.; Emery, I.; Chu, T.; Du, L.; Kremen, M.; Dichek, H.L.; Gold, E.; Ramsey, S.A.; et al. Mechanisms of urokinase plasminogen activator (uPA)-mediated atherosclerosis: Role of the uPA receptor and S100A8/A9 proteins. J. Biol. Chem. 2011, 286, 22665–22677. [Google Scholar] [CrossRef]
- Ganta, V.C.; Choi, M.; Farber, C.R.; Annex, B.H. Antiangiogenic VEGF(165)b Regulates Macrophage Polarization via S100A8/S100A9 in Peripheral Artery Disease. Circulation 2019, 139, 226–242. [Google Scholar] [CrossRef]
- Yunna, C.; Mengru, H.; Lei, W.; Weidong, C. Macrophage M1/M2 polarization. Eur. J. Pharmacol. 2020, 877, 173090. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Cao, L.; He, Y.; Ma, A.; Guo, W. The Role of Macrophages in Aortic Dissection. Front. Physiol. 2020, 11, 54. [Google Scholar] [CrossRef]
- Niyonzima, N.; Rahman, J.; Kunz, N.; West, E.E.; Freiwald, T.; Desai, J.V.; Merle, N.S.; Gidon, A.; Sporsheim, B.; Lionakis, M.S.; et al. Mitochondrial C5aR1 activity in macrophages controls IL-1β production underlying sterile inflammation. Sci. Immunol. 2021, 6, eabf2489. [Google Scholar] [CrossRef]
- Reichhardt, M.P.; Meri, S. Intracellular complement activation-An alarm raising mechanism? Semin. Immunol. 2018, 38, 54–62. [Google Scholar] [CrossRef]
- Von Vietinghoff, S.; Koltsova, E.K.; Mestas, J.; Diehl, C.J.; Witztum, J.L.; Ley, K. Mycophenolate mofetil decreases atherosclerotic lesion size by depression of aortic T-lymphocyte and interleukin-17-mediated macrophage accumulation. J. Am. Coll. Cardiol. 2011, 57, 2194–2204. [Google Scholar] [CrossRef]
- Khawar, M.B.; Abbasi, M.H.; Sheikh, N. IL-32: A Novel Pluripotent Inflammatory Interleukin, towards Gastric Inflammation, Gastric Cancer, and Chronic Rhino Sinusitis. Mediat. Inflamm. 2016, 2016, 8413768. [Google Scholar] [CrossRef]
- Seijkens, T.T.P.; Poels, K.; Meiler, S.; van Tiel, C.M.; Kusters, P.J.H.; Reiche, M.; Atzler, D.; Winkels, H.; Tjwa, M.; Poelman, H.; et al. Deficiency of the T cell regulator Casitas B-cell lymphoma-B aggravates atherosclerosis by inducing CD8+ T cell-mediated macrophage death. Eur. Heart J. 2019, 40, 372–382. [Google Scholar] [CrossRef]
- Pham, L.M.; Kim, E.C.; Ou, W.; Phung, C.D.; Nguyen, T.T.; Pham, T.T.; Poudel, K.; Gautam, M.; Nguyen, H.T.; Jeong, J.H.; et al. Targeting and clearance of senescent foamy macrophages and senescent endothelial cells by antibody-functionalized mesoporous silica nanoparticles for alleviating aorta atherosclerosis. Biomaterials 2021, 269, 120677. [Google Scholar] [CrossRef]
- Poddar, R.; Sivasubramanian, N.; DiBello, P.M.; Robinson, K.; Jacobsen, D.W. Homocysteine induces expression and secretion of monocyte chemoattractant protein-1 and interleukin-8 in human aortic endothelial cells: Implications for vascular disease. Circulation 2001, 103, 2717–2723. [Google Scholar] [CrossRef]
- Tellides, G.; Pober, J.S. Inflammatory and immune responses in the arterial media. Circ. Res. 2015, 116, 312–322. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, T.; Yao, F.; Gao, X.; Li, D.; Fu, S.; Mao, L.; Liu, F.; Zhang, X.; Xu, Y.; et al. Dysregulation of interaction between LOX(high) fibroblast and smooth muscle cells contributes to the pathogenesis of aortic dissection. Theranostics 2022, 12, 910–928. [Google Scholar] [CrossRef]
- Ren, K.; Li, B.; Liu, Z.; Xia, L.; Zhai, M.; Wei, X.; Duan, W.; Yu, S. GDF11 prevents the formation of thoracic aortic dissection in mice: Promotion of contractile transition of aortic SMCs. J. Cell. Mol. Med. 2021, 25, 4623–4636. [Google Scholar] [CrossRef]
- Li, G.; Wang, M.; Caulk, A.W.; Cilfone, N.A.; Gujja, S.; Qin, L.; Chen, P.Y.; Chen, Z.; Yousef, S.; Jiao, Y.; et al. Chronic mTOR activation induces a degradative smooth muscle cell phenotype. J. Clin. Investig. 2020, 130, 1233–1251. [Google Scholar] [CrossRef]
- Hu, H.; Zhang, G.; Hu, H.; Liu, W.; Liu, J.; Xin, S.; Zhao, X.; Han, L.; Duan, L.; Huang, X.; et al. Interleukin-18 Expression Increases in the Aorta and Plasma of Patients with Acute Aortic Dissection. Mediat. Inflamm. 2019, 2019, 8691294. [Google Scholar] [CrossRef]
- Jin, J.; Du, X.; Zhou, L.; Yao, D.; Zou, Q. SPI1-related protein inhibits cervical cancer cell progression and prevents macrophage cell migration. J. Obstet. Gynaecol. Res. 2022, 48, 2419–2430. [Google Scholar] [CrossRef]
- Zakrzewska, A.; Cui, C.; Stockhammer, O.W.; Benard, E.L.; Spaink, H.P.; Meijer, A.H. Macrophage-specific gene functions in Spi1-directed innate immunity. Blood 2010, 116, e1–e11. [Google Scholar] [CrossRef]
- Lamkin, D.M.; Srivastava, S.; Bradshaw, K.P.; Betz, J.E.; Muy, K.B.; Wiese, A.M.; Yee, S.K.; Waggoner, R.M.; Arevalo, J.M.G.; Yoon, A.J.; et al. C/EBPβ regulates the M2 transcriptome in β-adrenergic-stimulated macrophages. Brain Behav. Immun. 2019, 80, 839–848. [Google Scholar] [CrossRef]
- Zhao, L.; Lv, F.; Zheng, Y.; Yan, L.; Cao, X. Characterization of an Aging-Based Diagnostic Gene Signature and Molecular Subtypes With Diverse Immune Infiltrations in Atherosclerosis. Front. Mol. Biosci. 2021, 8, 792540. [Google Scholar] [CrossRef]
- Wesolowski, R.; Kowenz-Leutz, E.; Zimmermann, K.; Dörr, D.; Hofstätter, M.; Slany, R.K.; Mildner, A.; Leutz, A. Myeloid transformation by MLL-ENL depends strictly on C/EBP. Life Sci. Alliance 2021, 4. [Google Scholar] [CrossRef]
- Li, X.; Liu, D.; Zhao, L.; Wang, L.; Li, Y.; Cho, K.; Tao, C.; Jiang, B. Targeted depletion of monocyte/macrophage suppresses aortic dissection with the spatial regulation of MMP-9 in the aorta. Life Sci. 2020, 254, 116927. [Google Scholar] [CrossRef]
- Wu, T.C.; Chang, W.H.; Lu, H.Y.; Shih, C.C. Tolvaptan reduces angiotensin II-induced experimental abdominal aortic aneurysm and dissection. Vasc. Pharmacol. 2022, 144, 106973. [Google Scholar] [CrossRef]
- Vandestienne, M.; Zhang, Y.; Santos-Zas, I.; Al-Rifai, R.; Joffre, J.; Giraud, A.; Laurans, L.; Esposito, B.; Pinet, F.; Bruneval, P.; et al. TREM-1 orchestrates angiotensin II-induced monocyte trafficking and promotes experimental abdominal aortic aneurysm. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Liao, M.; Zou, S.; Bao, Y.; Jin, J.; Yang, J.; Liu, Y.; Green, M.; Yang, F.; Qu, L. Matrix metalloproteinases are regulated by MicroRNA 320 in macrophages and are associated with aortic dissection. Exp. Cell Res. 2018, 370, 98–102. [Google Scholar] [CrossRef]
- Barhoumi, T.; Fraulob-Aquino, J.C.; Mian, M.O.R.; Ouerd, S.; Idris-Khodja, N.; Huo, K.G.; Rehman, A.; Caillon, A.; Dancose-Giambattisto, B.; Ebrahimian, T.; et al. Matrix metalloproteinase-2 knockout prevents angiotensin II-induced vascular injury. Cardiovasc. Res. 2017, 113, 1753–1762. [Google Scholar] [CrossRef]
- Cifani, N.; Proietta, M.; Tritapepe, L.; Di Gioia, C.; Ferri, L.; Taurino, M.; Del Porto, F. Stanford-A acute aortic dissection, inflammation, and metalloproteinases: A review. Ann. Med. 2015, 47, 441–446. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, B.; Zeng, K.; Guan, R.-C.; Jiang, H.-Q.; Qiang, Y.-J.; Zhang, Q.; Yang, M.; Deng, B.-P.; Yang, Y.-Q. Single-Cell RNA-Seq Analysis Reveals Macrophages Are Involved in the Pathogenesis of Human Sporadic Acute Type A Aortic Dissection. Biomolecules 2023, 13, 399. https://doi.org/10.3390/biom13020399
Zhang B, Zeng K, Guan R-C, Jiang H-Q, Qiang Y-J, Zhang Q, Yang M, Deng B-P, Yang Y-Q. Single-Cell RNA-Seq Analysis Reveals Macrophages Are Involved in the Pathogenesis of Human Sporadic Acute Type A Aortic Dissection. Biomolecules. 2023; 13(2):399. https://doi.org/10.3390/biom13020399
Chicago/Turabian StyleZhang, Bin, Kuan Zeng, Rui-Cong Guan, Hui-Qi Jiang, Yong-Jia Qiang, Qing Zhang, Mo Yang, Bao-Ping Deng, and Yan-Qi Yang. 2023. "Single-Cell RNA-Seq Analysis Reveals Macrophages Are Involved in the Pathogenesis of Human Sporadic Acute Type A Aortic Dissection" Biomolecules 13, no. 2: 399. https://doi.org/10.3390/biom13020399
APA StyleZhang, B., Zeng, K., Guan, R.-C., Jiang, H.-Q., Qiang, Y.-J., Zhang, Q., Yang, M., Deng, B.-P., & Yang, Y.-Q. (2023). Single-Cell RNA-Seq Analysis Reveals Macrophages Are Involved in the Pathogenesis of Human Sporadic Acute Type A Aortic Dissection. Biomolecules, 13(2), 399. https://doi.org/10.3390/biom13020399





