Regulation of the Epithelial to Mesenchymal Transition in Osteosarcoma
Abstract
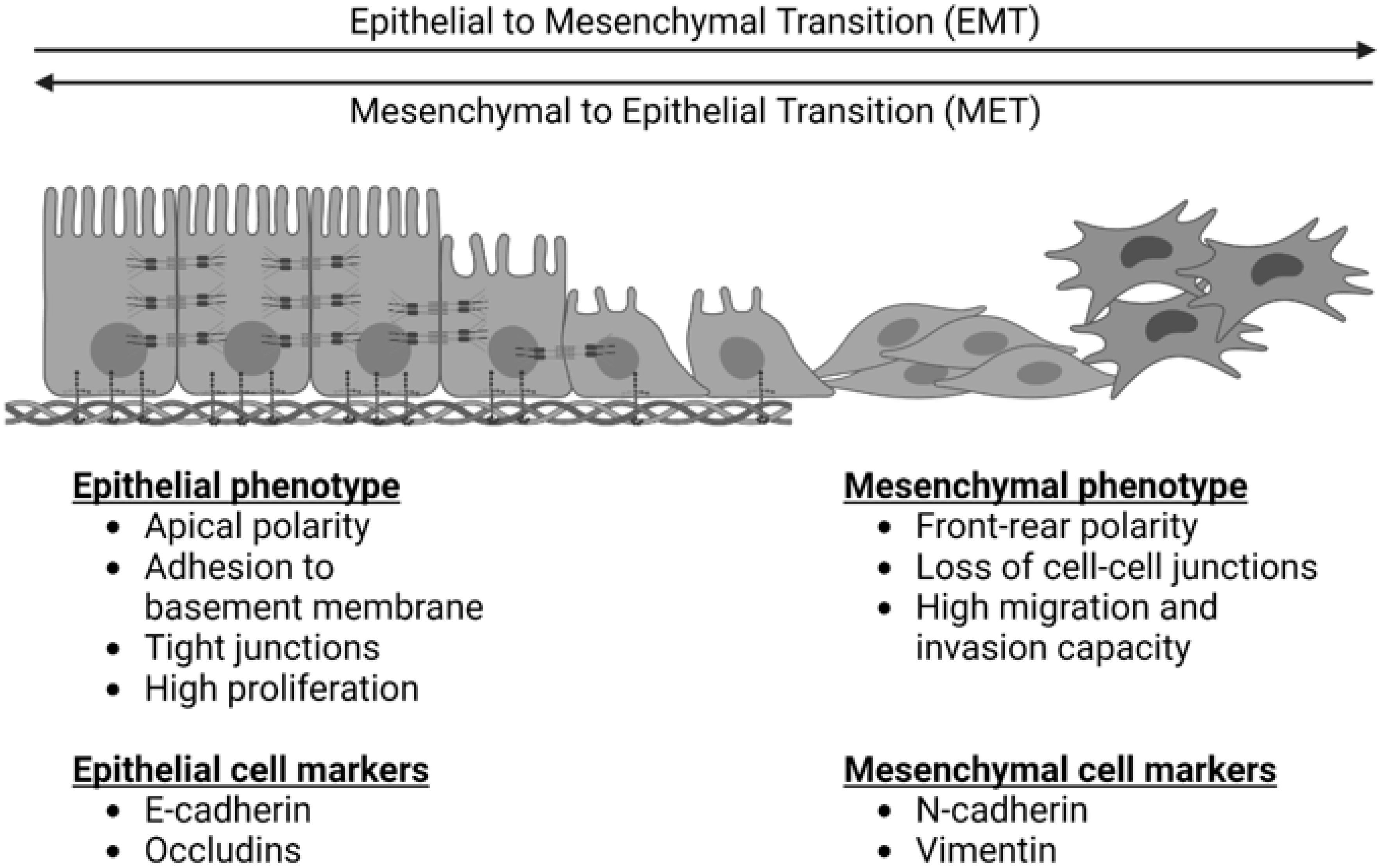
1. Introduction
2. EMT in Cancer
3. EMT Signaling Pathways
3.1. TGFβ/SMAD Pathway
3.2. Canonical wnt Pathway
3.3. Notch Pathway
3.4. Tyrosine Kinase Pathways
4. EMT in OS
5. Regulation of EMT in OS—Proteins
| Protein | Increased Levels in Clinical Sample | Promoted EMT | Promoted Cell Migration/Invasion | Promoted In Vivo Tumor Growth | Promoted In Vivo Metastasis | Endogenous Function |
|---|---|---|---|---|---|---|
| ACTL6A [121] | Yes | Structure | ||||
| AIM2 [114] | No | Immune | ||||
| BMP-2 [93] | No | Cell Signaling | ||||
| Calponin 3 [122] | Yes | Structure | ||||
| Cathepsin E [123] | Yes | Immune | ||||
| COPS3 [116] | Yes | ns | Cell Cycle Regulation | |||
| COX2 [107] | No | Inflammation | ||||
| CPE-ΔN [94] | No | Cell Signaling | ||||
| cPLA2a [124] | Yes | Inflammation | ||||
| CPXM2 [125] | Yes | Cell Signaling | ||||
| Cul4A [126] | Yes | Inflammation | ||||
| CXCR6 [127] | Yes | Immune | ||||
| Cyr61 [61,128] | No | Cell Signaling | ||||
| E2F1 [78] | Yes | Cell Cycle Regulation | ||||
| EPB41L3 [129] | Yes | ↓ | ↓ | Structure | ||
| Fibulin-3 [89] | Yes | Structure | ||||
| Fibulin-4 [106] | Yes | Structure | ||||
| HOXB7 [130] | Yes | Cell Cycle Regulation | ||||
| HuR [131] | Yes | Cell Cycle Regulation | ||||
| ICSBP [44] | Yes | Immune | ||||
| IL-33 [109] | No | Inflammation | ||||
| MAGL [132] | Yes | Inflammation | ||||
| Metadherin [133] | No | Cell Signaling | ||||
| NETO2 [113] | Yes | Cell Signaling | ||||
| OLR1 [134] | Yes | Cell Signaling | ||||
| P2X7 [108] | Yes | Cell Signaling | ||||
| PADI4 [135] | Yes | Inflammation | ||||
| PDGFRβ [112] | No | Cell Cycle Regulation | ||||
| PD-L2 [137] | Yes | Inflammation | ||||
| PGI [136] | No | Metabolism | ||||
| RIPK4 [100] | Yes | Cell Cycle Regulation | ||||
| SenP1 [138] | No | Cell Signaling | ||||
| SENP3 [139] | Yes | Cell Cycle Regulation | ||||
| SIRT1 [64] | Yes | Cell Cycle Regulation | ||||
| SOX3 [53] | Yes | Cell Cycle Regulation | ||||
| SOX5 [54] | Yes | Cell Cycle Regulation | ||||
| ST6Gal-1 [140] | No | Cell Cycle Regulation | ||||
| TANK1 [141] | No | Cell Cycle Regulation | ||||
| Tim-3 [48] | Yes | Immune | ||||
| TRIM29 [142] | Yes | Cell Cycle Regulation | ||||
| TRIM66 [143] | Yes | Cell Cycle Regulation | ||||
| UHRF1 [144] | No | Cell Cycle Regulation | ||||
| USP7 [97] | Yes | Cell Cycle Regulation | ||||
| USP17 [145] | Yes | Cell Cycle Regulation | ||||
| USP22 [146] | Yes | Cell Cycle Regulation |
 : Significant association; ns: No significant association; ↓: Inverse association;
: Significant association; ns: No significant association; ↓: Inverse association;  : Not studied/reported.
: Not studied/reported.| Protein | Decreased Levels in Clinical Sample | Inhibited EMT | Inhibited Cell Migration/Invasion | Inhibited In Vivo Tumor Growth | Inhibited In Vivo Metastasis | Endogenous Function |
|---|---|---|---|---|---|---|
| ARID1a [147] | Yes | Cell Signaling | ||||
| Ezrin [148] | No | Structure | ||||
| FTL [149] | Yes | Metabolism | ||||
| GPER [55] | Yes | Cell Signaling | ||||
| LAIR-1 [150] | Yes | Immune | ||||
| RASSF4 [91] | Yes | Cell Cycle Regulation | ||||
| SOX6 [62] | Yes | Cell Cycle Regulation | ||||
| TSSC3 [49] | Yes | Cell Cycle Regulation | ||||
| WWOX [151] | Yes | Cell Cycle Regulation |
 : Significant association;
: Significant association;  : Not studied/reported.
: Not studied/reported.6. Regulation of EMT in OS—Non-Coding Ribonucleic Acids
7. Regulation of EMT in OS—The Tumor Microenvironment
8. Targeting EMT in Osteosarcoma
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rojas, G.A.; Hubbard, A.K.; Diessner, B.J.; Ribeiro, K.B.; Spector, L.G. International Trends in Incidence of Osteosarcoma (1988–2012). Int. J. Cancer 2021, 149, 1044–1053. [Google Scholar] [CrossRef]
- American Cancer Society Key Statistics for Osteosarcoma. Available online: https://www.cancer.org/cancer/osteosarcoma/about/key-statistics.html (accessed on 12 September 2021).
- Sadykova, L.R.; Ntekim, A.I.; Muyangwa-Semenova, M.; Rutland, C.S.; Jeyapalan, J.N.; Blatt, N.; Rizvanov, A.A. Epidemiology and Risk Factors of Osteosarcoma. Cancer Investig. 2020, 38, 259–269. [Google Scholar] [CrossRef]
- American Cancer Society Survival Rates for Osteosarcoma. Available online: https://www.cancer.org/cancer/osteosarcoma/detection-diagnosis-staging/survival-rates.html (accessed on 11 September 2021).
- Marko, T.A.; Diessner, B.J.; Spector, L.G. Prevalence of Metastasis at Diagnosis of Osteosarcoma: An International Comparison. Pediatr. Blood Cancer 2016, 63, 1006–1011. [Google Scholar] [CrossRef]
- de Azevedo, J.W.V.; de Medeiros Fernandes, T.A.A.; Fernandes, J.V.; de Azevedo, J.C.V.; Lanza, D.C.F.; Bezerra, C.M.; Andrade, V.S.; de Araújo, J.M.G.; Fernandes, J.V. Biology and Pathogenesis of Human Osteosarcoma (Review). Oncol. Lett. 2020, 19, 1099–1116. [Google Scholar] [CrossRef]
- Strauss, S.J.; Frezza, A.M.; Abecassis, N.; Bajpai, J.; Bauer, S.; Biagini, R.; Bielack, S.; Blay, J.Y.; Bolle, S.; Bonvalot, S.; et al. Bone Sarcomas: ESMO–EURACAN–GENTURIS–ERN PaedCan Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2021, 32, 1520–1536. [Google Scholar] [CrossRef]
- Cole, S.; Gianferante, D.M.; Zhu, B.; Mirabello, L. Osteosarcoma: A Surveillance, Epidemiology, and End Results Program-Based Analysis from 1975 to 2017. Cancer 2022, 128, 2107–2118. [Google Scholar] [CrossRef]
- Brabletz, S.; Schuhwerk, H.; Brabletz, T.; Stemmler, M.P. Dynamic EMT: A Multi-tool for Tumor Progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New Insights into the Mechanisms of Epithelial–Mesenchymal Transition and Implications for Cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; Lebleu, V.S.; Kalluri, R. Epithelial-to-Mesenchymal Transition Is Dispensable for Metastasis but Induces Chemoresistance in Pancreatic Cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.C.; Choi, H.; el Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-Mesenchymal Transition Is Not Required for Lung Metastasis but Contributes to Chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef]
- Lourenco, A.R.; Ban, Y.; Crowley, M.J.; Lee, S.B.; Ramchandani, D.; Du, W.; Elemento, O.; George, J.T.; Jolly, M.K.; Levine, H.; et al. Differential Contributions of Pre- And Post-EMT Tumor Cells in Breast Cancer Metastasis. Cancer Res. 2020, 80, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef] [PubMed]
- Grande, M.T.; Sánchez-Laorden, B.; López-Blau, C.; de Frutos, C.A.; Boutet, A.; Arévalo, M.; Rowe, R.G.; Weiss, S.J.; López-Novoa, J.M.; Nieto, M.A. Snail1-Induced Partial Epithelial-to-Mesenchymal Transition Drives Renal Fibrosis in Mice and Can Be Targeted to Reverse Established Disease. Nat. Med. 2015, 21, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Dhasarathy, A.; Phadke, D.; Mav, D.; Shah, R.R.; Wade, P.A. The Transcription Factors Snail and Slug Activate the Transforming Growth Factor-Beta Signaling Pathway in Breast Cancer. PLoS ONE 2011, 6, e26514. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular Mechanisms of Epithelial-Mesenchymal Transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Wnt/β-Catenin Signaling in Development and Disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Wu, Y.; Ginther, C.; Kim, J.; Mosher, N.; Chung, S.; Slamon, D.; Vadgama, J. v Expression of Wnt3 Activates Wnt/β-Catenin Pathway and Promotes EMT-like Phenotype in Trastuzumab-Resistant HER2-Overexpressing Breast Cancer Cells. Mol. Cancer Res. 2012, 10, 1597–1606. [Google Scholar] [CrossRef]
- Stemmer, V.; de Craene, B.; Berx, G.; Behrens, J. Snail Promotes Wnt Target Gene Expression and Interacts with β-Catenin. Oncogene 2008, 27, 5075–5080. [Google Scholar] [CrossRef]
- Gauger, K.J.; Chenausky, K.L.; Murray, M.E.; Schneider, S.S. SFRP1 Reduction Results in an Increased Sensitivity to TGF-β Signaling. BMC Cancer 2011, 11, 59. [Google Scholar] [CrossRef]
- Balsamo, J.; Arregui, C.; Leung, T.C.; Lilien, J. The Nonreceptor Protein Tyrosine Phosphatase PTP1B Binds to the Cytoplasmic Domain of N-Cadherin and Regulates the Cadherin-Actin Linkage. J. Cell Biol. 1998, 143, 523–532. [Google Scholar] [CrossRef]
- Misiorek, J.O.; Przybyszewska-Podstawka, A.; Kałafut, J.; Paziewska, B.; Rolle, K.; Rivero-Müller, A.; Nees, M. Context Matters: Notch Signatures and Pathway in Cancer Progression and Metastasis. Cells 2021, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Natsuizaka, M.; Whelan, K.A.; Kagawa, S.; Tanaka, K.; Giroux, V.; Chandramouleeswaran, P.M.; Long, A.; Sahu, V.; Darling, D.S.; Que, J.; et al. Interplay between Notch1 and Notch3 Promotes EMT and Tumor Initiation in Squamous Cell Carcinoma. Nat. Commun. 2017, 8, 1758. [Google Scholar] [CrossRef]
- Timmerman, L.A.; Grego-Bessa, J.; Raya, A.; Bertrán, E.; Pérez-Pomares, J.M.; Díez, J.; Aranda, S.; Palomo, S.; McCormick, F.; Izpisúa-Belmonte, J.C.; et al. Notch Promotes Epithelial-Mesenchymal Transition during Cardiac Development and Oncogenic Transformation. Genes Dev. 2004, 18, 99–115. [Google Scholar] [CrossRef]
- Matsuno, Y.; Coelho, A.L.; Jarai, G.; Westwick, J.; Hogaboam, C.M. Notch Signaling Mediates TGF-Β1-Induced Epithelial-Mesenchymal Transition through the Induction of Snai1. Int. J. Biochem. Cell Biol. 2012, 44, 776–789. [Google Scholar] [CrossRef] [PubMed]
- Zavadil, J.; Cermak, L.; Soto-Nieves, N.; Böttinger, E.P. Integration of TGF-β/Smad and Jagged1/Notch Signalling in Epithelial-to-Mesenchymal Transition. EMBO J. 2004, 23, 1155–1165. [Google Scholar] [CrossRef]
- di Domenico, M.; Giordano, A. Signal Transduction Growth Factors: The Effective Governance of Transcription and Cellular Adhesion in Cancer Invasion. Oncotarget 2017, 8, 36869–36884. [Google Scholar] [CrossRef]
- Persad, A.; Venkateswaran, G.; Hao, L.; Garcia, M.E.; Yoon, J.; Sidhu, J.; Persad, S. Active β-Catenin Is Regulated by the PTEN/PI3 Kinase Pathway: A Role for Protein Phosphatase PP2A. Genes Cancer 2016, 7, 368–382. [Google Scholar] [CrossRef]
- Pottier, C.; Fresnais, M.; Gilon, M.; Jérusalem, G.; Longuespée, R.; Sounni, N.E. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers 2020, 12, 731. [Google Scholar] [CrossRef] [PubMed]
- Maehara, O.; Suda, G.; Natsuizaka, M.; Ohnishi, S.; Komatsu, Y.; Sato, F.; Nakai, M.; Sho, T.; Morikawa, K.; Ogawa, K.; et al. Fibroblast Growth Factor-2-Mediated FGFR/Erk Signaling Supports Maintenance of Cancer Stem-like Cells in Esophageal Squamous Cell Carcinoma. Carcinogenesis 2017, 38, 1073–1083. [Google Scholar] [CrossRef]
- Tashiro, E.; Henmi, S.; Odake, H.; Ino, S.; Imoto, M. Involvement of the MEK/ERK Pathway in EGF-Induced E-Cadherin down-Regulation. Biochem. Biophys. Res. Commun. 2016, 477, 801–806. [Google Scholar] [CrossRef]
- Lo, H.-W.; Hsu, S.-C.; Xia, W.; Cao, X.; Shih, J.-Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.-C. Epidermal Growth Factor Receptor Cooperates with Signal Transducer and Activator of Transcription 3 to Induce Epithelial-Mesenchymal Transition in Cancer Cells via Up-Regulation of TWIST Gene Expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef]
- Uttamsingh, S.; Bao, X.; Nguyen, K.T.; Bhanot, M.; Gong, J.; Chan, J.L.K.; Liu, F.; Chu, T.T.; Wang, L.H. Synergistic Effect between EGF and TGF-Β1 in Inducing Oncogenic Properties of Intestinal Epithelial Cells. Oncogene 2008, 27, 2626–2634. [Google Scholar] [CrossRef]
- Kim, J.; Kong, J.; Chang, H.; Kim, H.; Kim, A. EGF Induces Epithelial-Mesenchymal Transition through Phospho-Smad2/3-Snail Signaling Pathway in Breast Cancer Cells. Oncotarget 2016, 7, 85021–85032. [Google Scholar] [CrossRef]
- Vichalkovski, A.; Gresko, E.; Hess, D.; Restuccia, D.F.; Hemmings, B.A. PKB/AKT Phosphorylation of the Transcription Factor Twist-1 at Ser42 Inhibits P53 Activity in Response to DNA Damage. Oncogene 2010, 29, 3554–3565. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.O.; Kim, J.H.; Hong, J.S.; Yoon, H.J.; Lee, J.I.; Hong, S.P.; Hong, S.D. Inhibition of Akt Activity Induces the Mesenchymal-to-Epithelial Reverting Transition with Restoring E-Cadherin Expression in KB and KOSCC-25B Oral Squamous Cell Carcinoma Cells. J. Exp. Clin. Cancer Res. 2009, 28, 28. [Google Scholar] [CrossRef] [PubMed]
- Grotegut, S.; von Schweinitz, D.; Christofori, G.; Lehembre, F. Hepatocyte Growth Factor Induces Cell Scattering through MAPK/Egr-1-Mediated Upregulation of Snail. EMBO J. 2006, 25, 3534–3545. [Google Scholar] [CrossRef] [PubMed]
- Ogunwobi, O.O.; Puszyk, W.; Dong, H.J.; Liu, C. Epigenetic Upregulation of HGF and C-Met Drives Metastasis in Hepatocellular Carcinoma. PLoS ONE 2013, 8, e63765. [Google Scholar] [CrossRef] [PubMed]
- Sannino, G.; Marchetto, A.; Kirchner, T.; Grünewald, T.G.P. Epithelial-to-Mesenchymal and Mesenchymal-to-Epithelial Transition in Mesenchymal Tumors: A Paradox in Sarcomas? Cancer Res. 2017, 77, 4556–4561. [Google Scholar] [CrossRef] [PubMed]
- Kahlert, U.D.; Joseph, J.V.; Kruyt, F.A.E. EMT- and MET-Related Processes in Nonepithelial Tumors: Importance for Disease Progression, Prognosis, and Therapeutic Opportunities. Mol. Oncol. 2017, 11, 860–877. [Google Scholar] [CrossRef]
- Sato, H.; Hasegawa, T.; Abe, Y.; Sakai, H.; Hirohashi, S. Expression of E-Cadherin in Bone and Soft Tissue Sarcomas: A Possible Role in Epithelial Differentiation. Hum. Pathol. 1999, 30, 1344–1349. [Google Scholar] [CrossRef]
- Yin, K.; Liao, Q.; He, H.; Zhong, D. Prognostic Value of Twist and E-Cadherin in Patients with Osteosarcoma. Med. Oncol. 2012, 29, 3449–3455. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.Y.; Park, S.Y.; Kim, J.H.; Kang, H.G.; Yoon, J.H.; Na, Y.S.; Kim, Y.N.; Park, B.K. Interferon Consensus Sequence-Binding Protein (ICSBP) Promotes Epithelial-to-Mesenchymal Transition (EMT)-like Phenomena, Cell-Motility, and Invasion via TGF-β Signaling in U2OS Cells. Cell Death Dis. 2014, 5, e1224. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; Liu, C.; Sun, X.; Zhang, L.; Liu, L.; Ouyang, J.; Li, B. Visfatin Promotes Osteosarcoma Cell Migration and Invasion via Induction of Epithelial-Mesenchymal Transition. Oncol. Rep. 2015, 34, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Guo, Y.; Yang, Y.; Kan, J.; Dai, S.; Helian, M.; Li, B.; Xu, J.; Liu, C. Nitidine Chloride Suppresses Epithelial-to-Mesenchymal Transition in Osteosarcoma Cell Migration and Invasion through Akt/GSK-3β/Snail Signaling Pathway. Oncol. Rep. 2016, 36, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Ru, N.; Liang, J.; Zhang, F.; Wu, W.; Wang, F.; Liu, X.; Du, Y. SPRY4 Intronic Transcript 1 Promotes Epithelial-Mesenchymal Transition Through Association with Snail1 in Osteosarcoma. DNA Cell Biol. 2016, 35, 290–295. [Google Scholar] [CrossRef]
- Feng, Z.M.; Guo, S.M. Tim-3 Facilitates Osteosarcoma Proliferation and Metastasis through the NF-ΚB Pathway and Epithelial-Mesenchymal Transition. Genet. Mol. Res. 2016, 15, gmr7844. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.F.; Dai, H.; Yan, G.; Meng, G.; Zhang, X.; Guo, Q. nan Downregulation of Tumor Suppressing STF CDNA 3 Promotes Epithelial-Mesenchymal Transition and Tumor Metastasis of Osteosarcoma by the Wnt/GSK-3β/β-Catenin/Snail Signaling Pathway. Cancer Lett. 2016, 373, 164–173. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, K.; Li, Y.; He, Q. Estrogen-Related Receptor α Participates Transforming Growth Factor-β (TGF-β) Induced Epithelial-Mesenchymal Transition of Osteosarcoma Cells. Cell Adh. Migr. 2017, 11, 338–346. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, M.; Chen, Q.; Zhang, Q. Downregulation of MicroRNA-145 Promotes Epithelial-Mesenchymal Transition via Regulating Snail in Osteosarcoma. Cancer Gene Ther. 2017, 24, 83–88. [Google Scholar] [CrossRef]
- Kong, G.; Jiang, Y.; Sun, X.; Cao, Z.; Zhang, G.; Zhao, Z.; Zhao, Y.; Yu, Q.; Cheng, G. Irisin Reverses the IL-6 Induced Epithelial-Mesenchymal Transition in Osteosarcoma Cell Migration and Invasion through the STAT3/Snail Signaling Pathway. Oncol. Rep. 2017, 38, 2647–2656. [Google Scholar] [CrossRef]
- Qiu, M.; Chen, D.; Shen, C.; Shen, J.; Zhao, H.; He, Y. Sex-Determining Region Y-Box Protein 3 Induces Epithelial-Mesenchymal Transition in Osteosarcoma Cells via Transcriptional Activation of Snail1. J. Exp. Clin. Cancer Res. 2017, 36, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, S. SOX5 Promotes Epithelial-Mesenchymal Transition in Osteosarcoma via Regulation of Snail. J. Balk. Union Oncol. 2017, 22, 258–264. [Google Scholar]
- Wang, Z.; Chen, X.; Zhao, Y.; Jin, Y.; Zheng, J. G-Protein-Coupled Estrogen Receptor Suppresses the Migration of Osteosarcoma Cells via Post-Translational Regulation of Snail. J. Cancer Res. Clin. Oncol. 2019, 145, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, T.; Liu, X.; Li, Z.; Zhou, D.; Xu, W. Melatonin Suppresses Epithelial-to-Mesenchymal Transition in the MG-63 Cell Line. Mol. Med. Rep. 2020, 21, 1356–1364. [Google Scholar] [CrossRef]
- Wen, J.F.; Jiang, Y.Q.; Li, C.; Dai, X.K.; Wu, T.; Yin, W.Z. LncRNA-XIST Promotes the Oxidative Stress-Induced Migration, Invasion, and Epithelial-to-Mesenchymal Transition of Osteosarcoma Cancer Cells through MiR-153-SNAI1 Axis. Cell Biol. Int. 2020, 44, 1991–2001. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Lu, Y.; Zhu, J.; Yu, L.; Ma, B.; Zhang, Q. Oridonin Prevents Epithelial-Mesenchymal Transition and TGF-Β1-Induced Epithelial-Mesenchymal Transition by Inhibiting TGF-Β1/Smad2/3 in Osteosarcoma. Chem. Biol. Interact. 2018, 296, 57–64. [Google Scholar] [CrossRef]
- Sharili, A.S.; Allen, S.; Smith, K.; Price, J.; McGonnell, I.M. Snail2 Promotes Osteosarcoma Cell Motility through Remodelling of the Actin Cytoskeleton and Regulates Tumor Development. Cancer Lett. 2013, 333, 170–179. [Google Scholar] [CrossRef]
- Ishikawa, T.; Shimizu, T.; Ueki, A.; Yamaguchi, S.I.; Onishi, N.; Sugihara, E.; Kuninaka, S.; Miyamoto, T.; Morioka, H.; Nakayama, R.; et al. Twist2 Functions as a Tumor Suppressor in Murine Osteosarcoma Cells. Cancer Sci. 2013, 104, 880–888. [Google Scholar] [CrossRef]
- Hou, C.H.; Lin, F.L.; Hou, S.M.; Liu, J.F. Cyr61 Promotes Epithelial-Mesenchymal Transition and Tumor Metastasis of Osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 Signaling Pathway. Mol. Cancer 2014, 13, 236. [Google Scholar] [CrossRef]
- Wang, Z.; Li, J.; Li, K.; Xu, J. SOX6 Is Downregulated in Osteosarcoma and Suppresses the Migration, Invasion and Epithelial-Mesenchymal Transition via TWIST1 Regulation. Mol. Med. Rep. 2018, 17, 6803–6811. [Google Scholar] [CrossRef]
- Shi, D.; Wu, F.; Mu, S.; Hu, B.; Zhong, B.; Gao, F.; Qing, X.; Liu, J.; Zhang, Z.; Shao, Z. LncRNA AFAP1-AS1 Promotes Tumorigenesis and Epithelial-Mesenchymal Transition of Osteosarcoma through RhoC/ROCK1/P38MAPK/Twist1 Signaling Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 375. [Google Scholar] [CrossRef]
- Yu, X.J.; Guo, X.Z.; Li, C.; Chong, Y.; Song, T.N.; Pang, J.F.; Shao, M. SIRT1-ZEB1-Positive Feedback Promotes Epithelial-Mesenchymal Transition Process and Metastasis of Osteosarcoma. J. Cell Biochem. 2019, 120, 3727–3735. [Google Scholar] [CrossRef]
- Feng, T.; Zhu, Z.; Jin, Y.; Wang, H.; Mao, X.; Liu, D.; Li, Y.; Lu, L.; Zuo, G. The MicroRNA 708 5p/ZEB1/EMT Axis Mediates the Metastatic Potential of Osteosarcoma. Oncol. Rep. 2020, 43, 491–502. [Google Scholar] [CrossRef]
- Yao, H.; Hou, G.; Wang, Q.Y.; Xu, W.B.; Zhao, H.Q.; Xu, Y.C. LncRNA SPRY4-IT1 Promotes Progression of Osteosarcoma by Regulating ZEB1 and ZEB2 Expression through Sponging of MiR-101 Activity. Int. J. Oncol. 2020, 56, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.; Zhang, Y.; Yang, H.; Xu, R.; Huang, G. Overexpression of ZEB1 Relates to Metastasis and Invasion in Osteosarcoma. J. Surg. Oncol. 2012, 105, 830–834. [Google Scholar] [CrossRef] [PubMed]
- Verrecchia, F.; Rédini, F. Transforming Growth Factor-β Signaling Plays a Pivotal Role in the Interplay between Osteosarcoma Cells and Their Microenvironment. Front. Oncol. 2018, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Hsieh, Y.S.; Chu, S.C.; Hsu, L.S.; Huang, S.C.; Chen, P.N. Reduction of Invasion and Cell Stemness and Induction of Apoptotic Cell Death by Cinnamomum Cassia Extracts on Human Osteosarcoma Cells. Environ. Toxicol. 2022, 37, 1261–1274. [Google Scholar] [CrossRef]
- He, D.; Gao, J.; Zheng, L.; Liu, S.; Ye, L.; Lai, H.; Pan, B.; Pan, W.; Lou, C.; Chen, Z.; et al. TGF-β Inhibitor RepSox Suppresses Osteosarcoma via the JNK/Smad3 Signaling Pathway. Int. J. Oncol. 2021, 59, 84. [Google Scholar] [CrossRef]
- Ma, K.; Zhang, C.; Li, W. Gamabufotalin Suppressed Osteosarcoma Stem Cells through the TGF-β/Periostin/PI3K/AKT Pathway. Chem. Biol. Interact. 2020, 331, 109275. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Z.; Song, C.; Deng, H.; Yang, R.; Zhou, L.; Sun, Y.; Zhang, Q. Glaucocalyxin A Reverses EMT and TGF-Β1-Induced EMT by Inhibiting TGF-Β1/Smad2/3 Signaling Pathway in Osteosarcoma. Chem. Biol. Interact. 2019, 307, 158–166. [Google Scholar] [CrossRef]
- Dong, F.; Liu, T.; Jin, H.; Wang, W. Chimaphilin Inhibits Human Osteosarcoma Cell Invasion and Metastasis through Suppressing the TGF-Β1-Induced Epithelial-to-Mesenchymal Transition Markers via PI-3K/Akt, ERK1/2, and Smad Signaling Pathways. Can. J. Physiol. Pharmacol. 2018, 96, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Zhou, R.; Zhong, W.; Lu, S.; Ma, Z.; Chai, Y. Baicalin Inhibits Human Osteosarcoma Cells Invasion, Metastasis, and Anoikis Resistance by Suppressing the Transforming Growth Factor-Β1-Induced Epithelial-to-Mesenchymal Transition. Anticancer Drugs 2017, 28, 581–587. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Park, B.S.; Kang, H.K.; Park, H.R.; Yu, S.B.; Kim, I.R. Delphinidin Induces Apoptosis and Inhibits Epithelial-to-Mesenchymal Transition via the ERK/P38 MAPK-Signaling Pathway in Human Osteosarcoma Cell Lines. Environ. Toxicol. 2018, 33, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Liu, S.; Wu, B.; Zhao, Y.; Chen, B.; Guo, J.; Qiu, S.H.; Cao, Y.M. MiR-19 Promotes Cell Proliferation, Invasion, Migration, and EMT by Inhibiting SPRED2-Mediated Autophagy in Osteosarcoma Cells. Cell Transplant. 2020, 29, 963689720962460. [Google Scholar] [CrossRef]
- Raimondi, L.; Gallo, A.; Cuscino, N.; de Luca, A.; Costa, V.; Carina, V.; Bellavia, D.; Bulati, M.; Alessandro, R.; Fini, M.; et al. Potential Anti-Metastatic Role of the Novel MiR-CT3 in Tumor Angiogenesis and Osteosarcoma Invasion. Int. J. Mol. Sci. 2022, 23, 705. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, X.; Bao, Y.; Mo, J.; Du, H.; Hu, J.; Zhang, X. E2F1 Silencing Inhibits Migration and Invasion of Osteosarcoma Cells via Regulating DDR1 Expression. Int. J. Oncol. 2017, 51, 1639–1650. [Google Scholar] [CrossRef]
- Zheng, B.; Zhou, C.; Qu, G.; Ren, C.; Yan, P.; Guo, W.; Yue, B. VEGFR2 Promotes Metastasis and PD-L2 Expression of Human Osteosarcoma Cells by Activating the STAT3 and RhoA-ROCK-LIMK2 Pathways. Front. Oncol. 2020, 10, 543562. [Google Scholar] [CrossRef]
- Han, Y.; Guo, W.; Ren, T.; Huang, Y.; Wang, S.; Liu, K.; Zheng, B.; Yang, K.; Zhang, H.; Liang, X. Tumor-Associated Macrophages Promote Lung Metastasis and Induce Epithelial-Mesenchymal Transition in Osteosarcoma by Activating the COX-2/STAT3 Axis. Cancer Lett. 2019, 440–441, 116–125. [Google Scholar] [CrossRef]
- Huang, H.; Han, Y.; Chen, Z.; Pan, X.; Yuan, P.; Zhao, X.; Zhu, H.; Wang, J.; Sun, X.; Shi, P. ML264 Inhibits Osteosarcoma Growth and Metastasis via Inhibition of JAK2/STAT3 and WNT/β-Catenin Signalling Pathways. J. Cell Mol. Med. 2020, 24, 5652–5664. [Google Scholar] [CrossRef]
- Hu, Y.; Luo, X.; Zhou, J.; Chen, S.; Gong, M.; Deng, Y.; Zhang, H. Piperlongumine Inhibits the Progression of Osteosarcoma by Downregulating the SOCS3/JAK2/STAT3 Pathway via MiR-30d-5p. Life Sci. 2021, 277, 119501. [Google Scholar] [CrossRef]
- Zhang, J.; Li, N.; Lu, S.; Chen, Y.; Shan, L.; Zhao, X.; Xu, Y. The Role of Notch Ligand Jagged1 in Osteosarcoma Proliferation, Metastasis, and Recurrence. J. Orthop. Surg. Res. 2021, 16, 226. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xia, K.; Gao, T.; Chen, J.; Zhang, Z.; Sun, X.; Simões, B.M.; Eyre, R.; Fan, Z.; Guo, W.; et al. The Notch Pathway Promotes Osteosarcoma Progression through Activation of Ephrin Reverse Signaling. Mol. Cancer Res. 2019, 17, 2383–2394. [Google Scholar] [CrossRef]
- Li, Z.; Tang, Y.; Xing, W.; Dong, W.; Wang, Z. LncRNA, CRNDE Promotes Osteosarcoma Cell Proliferation, Invasion and Migration by Regulating Notch1 Signaling and Epithelial-Mesenchymal Transition. Exp. Mol. Pathol. 2018, 104, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Zhao, F.; Zhang, Z.; Sun, F.; Wang, M. Long Noncoding RNA SNHG7 Promotes the Tumor Growth and Epithelial-to-Mesenchymal Transition via Regulation of MiR-34a Signals in Osteosarcoma. Cancer Biother Radiopharm. 2018, 33, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Dai, G.; Liu, G.; Zheng, D.; Song, Q. Inhibition of the Notch Signaling Pathway Attenuates Progression of Cell Motility, Metastasis, and Epithelial-to-Mesenchymal Transition-like Phenomena Induced by Low Concentrations of Cisplatin in Osteosarcoma. Eur. J. Pharmacol. 2021, 899, 174058. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ren, Y.; Han, E.Q.; Li, H.; Chen, D.; Jacobs, J.J.; Gitelis, S.; O’Keefe, R.J.; Konttinen, Y.T.; Yin, G.; et al. Inhibition of the Wnt-β-Catenin and Notch Signaling Pathways Sensitizes Osteosarcoma Cells to Chemotherapy. Biochem. Biophys. Res. Commun. 2013, 431, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, D.; Han, S.; Gao, P.; Liu, C.; Li, J.; Pan, X. Fibulin-3 Promotes Osteosarcoma Invasion and Metastasis by Inducing Epithelial to Mesenchymal Transition and Activating the Wnt/β-Catenin Signaling Pathway. Sci. Rep. 2017, 7, 6215. [Google Scholar] [CrossRef]
- Cao, J.; Han, X.; Qi, X.; Jin, X.; Li, X. TUG1 Promotes Osteosarcoma Tumorigenesis by Upregulating EZH2 Expression via MIR-144-3p. Int. J. Oncol. 2017, 51, 1115–1123. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, D.; Zhu, T.; Yin, R. RASSF4 Overexpression Inhibits the Proliferation, Invasion, EMT, and Wnt Signaling Pathway in Osteosarcoma Cells. Oncol. Res. 2017, 25, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Guo, D.; Cao, Z.; Xiao, L.; Wang, G. Inhibitory Effect of MicroRNA-107 on Osteosarcoma Malignancy Through Regulation of Wnt/β-Catenin Signaling in Vitro. Cancer Investig. 2018, 36, 175–184. [Google Scholar] [CrossRef]
- Tian, H.; Zhou, T.; Chen, H.; Li, C.; Jiang, Z.; Lao, L.; Kahn, S.A.; Duarte, M.E.L.; Zhao, J.; Daubs, M.D.; et al. Bone Morphogenetic Protein-2 Promotes Osteosarcoma Growth by Promoting Epithelial-Mesenchymal Transition (EMT) through the Wnt/β-Catenin Signaling Pathway. J. Orthop. Res. 2019, 37, 1638–1648. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Gao, X.; Chen, P.; Li, X. Carboxypeptidase E-ΔN Promotes Migration, Invasiveness, and Epithelial-Mesenchymal Transition of Human Osteosarcoma Cells via the Wnt-β-Catenin Pathway. Biochem. Cell Biol. 2019, 97, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, L.; Li, X.; Liu, W.; Zhao, Y.; Li, J. Down-Regulation of MicroRNA-31-5p Inhibits Proliferation and Invasion of Osteosarcoma Cells through Wnt/β-Catenin Signaling Pathway by Enhancing AXIN1. Exp. Mol. Pathol. 2019, 108, 32–41. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, Z.; Zhou, X.; Tang, M.; Tan, W.; Sun, T.; Deng, Y. MiR-342-5p Inhibits Osteosarcoma Cell Growth, Migration, Invasion, and Sensitivity to Doxorubicin through Targeting Wnt7b. Cell Cycle 2019, 18, 3325–3336. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Li, Z.; Zhao, X.; Guo, L.; Yu, C.; Qin, J.; Zhang, S.; Zhang, Y.; Yang, X. Ubiquitin-Specific Protease 7 Promotes Osteosarcoma Cell Metastasis by Inducing Epithelial-Mesenchymal Transition. Oncol. Rep. 2019, 41, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Mo, F.; Cai, X.; Zhang, W.; Wang, J.; Yang, S.; Liu, X. LncRNA CRNDE Is Activated by SP1 and Promotes Osteosarcoma Proliferation, Invasion, and Epithelial-Mesenchymal Transition via Wnt/β-Catenin Signaling Pathway. J. Cell Biochem. 2020, 121, 3358–3371. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, W.; Fan, W. Long Non-Coding RNA GHET1 Promotes Osteosarcoma Development and Progression via Wnt/β-Catenin Signaling Pathway. Oncol. Rep. 2020, 44, 349–359. [Google Scholar] [CrossRef]
- Yi, Z.; Pu, Y.; Gou, R.; Chen, Y.; Ren, X.; Liu, W.; Dong, P. Silencing of RIPK4 Inhibits Epithelial-Mesenchymal Transition by Inactivating the Wnt/β-Catenin Signaling Pathway in Osteosarcoma. Mol. Med. Rep. 2020, 21, 1154–1162. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, P. LncRNA-CASC15 Promotes Osteosarcoma Proliferation and Metastasis by Regulating Epithelial-Mesenchymal Transition via the Wnt/β-Catenin Signaling Pathway. Oncol. Rep. 2021, 45, 76. [Google Scholar] [CrossRef]
- Liang, K.; Liao, L.; Liu, Q.; Ouyang, Q.; Jia, L.; Wu, G. MicroRNA-377-3p Inhibits Osteosarcoma Progression by Targeting CUL1 and Regulating Wnt/β-Catenin Signaling Pathway. Clin. Transl. Oncol. 2021, 23, 2350–2357. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, Q.; Shen, W. Circ-FOXM1 Promotes the Proliferation, Migration and EMT Process of Osteosarcoma Cells through FOXM1-Mediated Wnt Pathway Activation. J. Orthop. Surg. Res. 2022, 17, 344. [Google Scholar] [CrossRef]
- Bi, W.; Yang, M.; Xing, P.; Huang, T. MicroRNA MiR-331-3p Suppresses Osteosarcoma Progression via the Bcl-2/Bax and Wnt/β-Catenin Signaling Pathways and the Epithelial-Mesenchymal Transition by Targeting N-Acetylglucosaminyltransferase I (MGAT1). Bioengineered 2022, 13, 14159–14174. [Google Scholar] [CrossRef]
- Singla, A.; Wang, J.; Yang, R.; Geller, D.S.; Loeb, D.M.; Hoang, B.H. Wnt Signaling in Osteosarcoma. In Current Advances in Osteosarcoma; Kleinerman, E.S., Ed.; Springer Nature: Cham, Switzerland, 2020; pp. 125–139. ISBN 978-3-319-04843-7. [Google Scholar]
- Zhang, D.; Wang, S.; Chen, J.; Liu, H.; Lu, J.; Hua, J.; Huang, A.; Chen, Y. Fibulin-4 Promotes Osteosarcoma Invasion and Metastasis by Inducing Epithelial to Mesenchymal Transition via the PI3K/Akt/MTOR Pathway. Int. J. Oncol. 2017, 50, 1513–1530. [Google Scholar] [CrossRef]
- Zhang, X.; Qu, P.; Zhao, H.; Zhao, T.; Cao, N. COX-2 Promotes Epithelial-Mesenchymal Transition and Migration in Osteosarcoma MG-63 Cells via PI3K/AKT/NF-ΚB Signaling. Mol. Med. Rep. 2019, 20, 3811–3819. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, H.; Li, W.; Wu, H.; Yang, Y. Highly-Expressed P2X7 Receptor Promotes Growth and Metastasis of Human HOS/MNNG Osteosarcoma Cells via PI3K/Akt/GSK3β/β-Catenin and MTOR/HIF1α/VEGF Signaling. Int. J. Cancer 2019, 145, 1068–1082. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, G.; Zhao, S.; Qiao, Y.; Yang, H. The Effects of Interleukin-33 (IL-33) on Osteosarcoma Cell Viability, Apoptosis, and Epithelial-Mesenchymal Transition Are Mediated through the PI3K/AKT Pathway. Med. Sci. Monit. 2020, 26, e920766-1–e920766-10. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Jiang, D.; Gong, F.; Huang, Y.; Luo, Y.; Rong, Y.; Wang, J.; Ge, X.; Ji, C.; Fan, J.; et al. MiR-210-5p Promotes Epithelial–Mesenchymal Transition by Inhibiting PIK3R5 Thereby Activating Oncogenic Autophagy in Osteosarcoma Cells. Cell Death Dis. 2020, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.-G.; Tang, Q.-L.; Wei, J.-H.; He, F.-Y.; Lu, L.; Tang, Y.-J. Targeting EZH2 by MicroRNA-449a Inhibits Osteosarcoma Cell Proliferation, Invasion and Migration via Regulation of PI3K/AKT Signaling Pathway and Epithelial-Mesenchymal Transition. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1656–1665. [Google Scholar] [CrossRef]
- Xing, S.; Wang, C.; Tang, H.; Guo, J.; Liu, X.; Yi, F.; Liu, G.; Wu, X. Down-Regulation of PDGFRβ Suppresses Invasion and Migration in Osteosarcoma Cells by Influencing Epithelial–Mesenchymal Transition. FEBS Open Bio. 2020, 10, 1748–1757. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bian, Z.; Hou, C.; Li, M.; Jiang, W.; Zhu, L. Neuropilin and Tolloid-like 2 Regulates the Progression of Osteosarcoma. Gene 2021, 768, 145292. [Google Scholar] [CrossRef]
- Zheng, J.; Liu, C.; Shi, J.; Wen, K.; Wang, X. AIM2 Inhibits the Proliferation, Invasion and Migration, and Promotes the Apoptosis of Osteosarcoma Cells by Inactivating the PI3K/AKT/MTOR Signaling Pathway. Mol. Med. Rep. 2022, 25, 53. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.H.; Chen, P.N.; Hsieh, Y.H.; Lin, C.Y.; Cheng, F.Y.; Chiu, P.C.; Chu, S.C.; Hsieh, Y.S. 3-Hydroxyflavone Inhibits Human Osteosarcoma U2OS and 143B Cells Metastasis by Affecting EMT and Repressing u-PA/MMP-2 via FAK-Src to MEK/ERK and RhoA/MLC2 Pathways and Reduces 143B Tumor Growth in Vivo. Food Chem. Toxicol. 2016, 97, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Yan, T.; Guo, W.; Sun, K.; Wang, S.; Bao, X.; Liu, K.; Zheng, B.; Zhang, H.; Ren, T. Novel Oncogene COPS3 Interacts with Beclin1 and Raf-1 to Regulate Metastasis of Osteosarcoma through Autophagy. J. Exp. Clin. Cancer Res. 2018, 37, 135. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.-B.; Zhang, J.-Y.; Gao, K.; Yu, Z.-H.; Sheng, W.-C.; Yang, G.; Gao, Y.-Z. MicroRNA-765 Targets MTUS1 to Promote the Progression of Osteosarcoma via Mediating ERK/EMT Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4618–4628. [Google Scholar] [CrossRef]
- Lin, H.; Hao, Y.; Wan, X.; He, J.; Tong, Y. Baicalein Inhibits Cell Development, Metastasis and EMT and Induces Apoptosis by Regulating ERK Signaling Pathway in Osteosarcoma. J. Recept. Signal. Transduct. 2020, 40, 49–57. [Google Scholar] [CrossRef]
- Greenfield, E.M.; Collier, C.D.; Getty, P.J. Receptor Tyrosine Kinases in Osteosarcoma: 2019 Update. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1258, pp. 141–155. [Google Scholar]
- U.S. National Library of Medicine List of Clinical Trials Studying Tyrosine Kinase Inhibition in Osteosarcoma. Available online: https://clinicaltrials.gov/ct2/results?cond=osteosarcoma&term=tyrosine+kinase (accessed on 5 February 2023).
- Sun, W.; Wang, W.; Lei, J.; Li, H.; Wu, Y. Actin-like Protein 6A Is a Novel Prognostic Indicator Promoting Invasion and Metastasis in Osteosarcoma. Oncol. Rep. 2017, 37, 2405–2417. [Google Scholar] [CrossRef]
- Dai, F.; Luo, F.; Zhou, R.; Zhou, Q.; Xu, J.; Zhang, Z.; Xiao, J.; Song, L. Calponin 3 Is Associated with Poor Prognosis and Regulates Proliferation and Metastasis in Osteosarcoma. Aging 2020, 12, 14037–14049. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, W.; Yang, Z.; Li, J.; Jin, Y. MiR-185-5p Represses Cells Growth and Metastasis of Osteosarcoma via Targeting Cathepsin E. Int. J. Toxicol. 2022, 41, 115–125. [Google Scholar] [CrossRef]
- Pang, X.; Yin, P.; Han, J.; Wang, Z.; Zheng, F.; Chen, X. CPLA2a Correlates with Metastasis and Poor Prognosis of Osteosarcoma by Facilitating Epithelial-Mesenchymal Transition. Pathol. Res. Pract. 2019, 215, 152398. [Google Scholar] [CrossRef]
- Zhao, X.; Li, R.; Wang, Q.; Wu, M.; Wang, Y. Overexpression of Carboxypeptidase X M14 Family Member 2 Predicts an Unfavorable Prognosis and Promotes Proliferation and Migration of Osteosarcoma. Diagn. Pathol. 2019, 14, 118. [Google Scholar] [CrossRef]
- Li, X.; Xu, R.; Liu, H.; Fang, K. CUL4A Expression in Pediatric Osteosarcoma Tissues and Its Effect on Cell Growth in Osteosarcoma Cells. Tumor Biol. 2016, 37, 8139–8144. [Google Scholar] [CrossRef]
- Ma, Y.; Xu, X.; Luo, M. CXCR6 Promotes Tumor Cell Proliferation and Metastasis in Osteosarcoma through the Akt Pathway. Cell Immunol. 2017, 311, 80–85. [Google Scholar] [CrossRef]
- Habel, N.; Stefanovska, B.; Carène, D.; Patiño-Garcia, A.; Lecanda, F.; Fromigué, O. CYR61 Triggers Osteosarcoma Metastatic Spreading via an IGF1Rβ-Dependent EMT-like Process. BMC Cancer 2019, 19, 62. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Piao, L.; Wang, L.; Han, X.; Tong, L.; Shao, S.; Xu, X.; Zhuang, M.; Liu, Z. Erythrocyte Membrane Protein Band 4.1-like 3 Inhibits Osteosarcoma Cell Invasion through Regulation of Snai1-Induced Epithelial-to-Mesenchymal Transition. Aging 2020, 13, 1947–1961. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, J.; Chen, Q. Upregulation of HOXB7 Promotes Proliferation and Metastasis of Osteosarcoma Cells. Mol. Med. Rep. 2017, 16, 2773–2778. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Chen, C.; Xu, R.; Li, Y.; Hu, R.; Li, Z.; Zhu, X. Knockdown of HuR Represses Osteosarcoma Cells Migration, Invasion and Stemness through Inhibition of YAP Activation and Increases Susceptibility to Chemotherapeutic Agents. Biomed. Pharmacother. 2018, 102, 587–593. [Google Scholar] [CrossRef]
- Gong, X.; Zheng, X.; Huang, Y.; Song, W.; Chen, G.; Chen, T. Monoacylglycerol Lipase (MAGL) Inhibition Impedes the Osteosarcoma Progression by Regulating Epithelial Mesenchymal Transition. Tohoku J. Exp. Med. 2022, 256, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Shen, L.; Yang, Q.; Zhang, C. Overexpression of Metadherin Mediates Metastasis of Osteosarcoma by Regulating Epithelial-Mesenchymal Transition. Cell Prolif. 2014, 47, 427–434. [Google Scholar] [CrossRef]
- Jiang, L.; Jiang, S.; Zhou, W.; Huang, J.; Lin, Y.; Long, H.; Luo, Q. Oxidized Low Density Lipoprotein Receptor 1 Promotes Lung Metastases of Osteosarcomas through Regulating the Epithelial-Mesenchymal Transition. J. Transl. Med. 2019, 17, 369. [Google Scholar] [CrossRef]
- Zhai, Q.; Qin, J.; Jin, X.; Sun, X.; Wang, L.; Du, W.; Li, T.; Xiang, X. PADI4 Modulates the Invasion and Migration of Osteosarcoma Cells by Down-Regulation of Epithelial-Mesenchymal Transition. Life Sci. 2020, 256, 117968. [Google Scholar] [CrossRef]
- Niinaka, Y.; Harada, K.; Fujimuro, M.; Oda, M.; Haga, A.; Hosoki, M.; Uzawa, N.; Arai, N.; Yamaguchi, S.; Yamashiro, M.; et al. Silencing of Autocrine Motility Factor Induces Mesenchymal-to-Epithelial Transition and Suppression of Osteosarcoma Pulmonary Metastasis. Cancer Res. 2010, 70, 9483–9493. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Zheng, B.; Huang, Y.; Wang, S.; Bao, X.; Liu, K.; Guo, W. Osteosarcoma Cell Intrinsic PD-L2 Signals Promote Invasion and Metastasis via the RhoA-ROCK-LIMK2 and Autophagy Pathways. Cell Death Dis. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, X.; Liang, H.; Wang, B. SENP1/HIF-1α Feedback Loop Modulates Hypoxia-Induced Cell Proliferation, Invasion, and EMT in Human Osteosarcoma Cells. J. Cell Biochem. 2018, 119, 1819–1826. [Google Scholar] [CrossRef]
- Yang, P.; Liu, Y.; Qi, Y.C.; Lian, Z.H. High SENP3 Expression Promotes Cell Migration, Invasion, and Proliferation by Modulating DNA Methylation of E-Cadherin in Osteosarcoma. Technol. Cancer Res. Treat. 2020, 19, 1533033820956988. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Ren, C.; Wang, L.; Zhao, Y.; Wang, S. Knockdown of ST6Gal-I Inhibits the Growth and Invasion of Osteosarcoma MG-63 Cells. Biomed. Pharmacother. 2015, 72, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Jin, Q.; Xiao, W.; Sun, C. Tankyrase1 Antisense Oligodeoxynucleotides Suppress the Proliferation, Migration and Invasion through Hippo/YAP Pathway in Human Osteosarcoma Cells. Pathol. Res. Pract. 2019, 215, 152381. [Google Scholar] [CrossRef]
- Zeng, S.X.; Cai, Q.C.; Guo, C.H.; Zhi, L.Q.; Dai, X.; Zhang, D.F.; Ma, W. High Expression of TRIM29 (ATDC) Contributes to Poor Prognosis and Tumor Metastasis by Inducing Epithelial-Mesenchymal Transition in Osteosarcoma. Oncol. Rep. 2017, 38, 1645–1654. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, Y.; Yang, H.; Shi, G.; Xu, G.; Shi, J.; Yin, N.; Chen, D. TRIM66 Overexpresssion Contributes to Osteosarcoma Carcin¬ Ogenesis and Indicates Poor Survival Outcome. Oncotarget 2015, 6, 23708. [Google Scholar] [CrossRef]
- Liu, W.; Qiao, R.H.; Wang, D.M.; Huang, X.W.; Li, B.; Wang, D. UHRF1 Promotes Human Osteosarcoma Cell Invasion by Downregulating the Expression of E-Cadherin in an Rb1-Dependent Manner. Mol. Med. Rep. 2016, 13, 315–320. [Google Scholar] [CrossRef]
- Song, C.; Liu, W.; Li, J. USP17 Is Upregulated in Osteosarcoma and Promotes Cell Proliferation, Metastasis, and Epithelial-Mesenchymal Transition through Stabilizing SMAD4. Tumor Biol. 2017, 39, 1010428317717138. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, F.; Wang, X.; Li, G. Downregulation of Ubiquitin-Specific Protease 22 Inhibits Proliferation, Invasion, and Epithelial-Mesenchymal Transition in Osteosarcoma Cells. Oncol. Res. 2017, 25, 743–751. [Google Scholar] [CrossRef]
- Xu, N.; Wang, L.; Sun, P.; Xu, S.; Fu, S.; Sun, Z. Low Arid1a Expression Correlates with Poor Prognosis and Promotes Cell Proliferation and Metastasis in Osteosarcoma. Pathol. Oncol. Res. 2019, 25, 875–881. [Google Scholar] [CrossRef]
- Liu, P.; Yang, P.; Zhang, Z.; Liu, M.; Hu, S. Ezrin/NF-ΚB Pathway Regulates EGF-Induced Epithelial-Mesenchymal Transition (EMT), Metastasis, and Progression of Osteosarcoma. Med. Sci. Monit. 2018, 24, 2098–2108. [Google Scholar] [CrossRef]
- Yu, G.-H.; Fu, L.; Chen, J.; Wei, F.; Shi, W.-X. Decreased Expression of Ferritin Light Chain in Osteosarcoma and Its Correlation with Epithelial-Mesenchymal Transition. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2580–2587. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Y.; Cheng, S.; Mu, Y.; Liu, Y.; Yi, X.; Jiang, D.; Ding, Y.; Zhuang, R. LAIR-1 Overexpression Inhibits Epithelial-Mesenchymal Transition in Osteosarcoma via GLUT1-Related Energy Metabolism. World J. Surg. Oncol. 2020, 18, 136. [Google Scholar] [CrossRef]
- Gao, K.; Yin, J.; Dong, J. Deregulated WWOX Is Involved in a Negative Feedback Loop with MicroRNA-214-3p in Osteosarcoma. Int. J. Mol. Med. 2016, 38, 1850–1856. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Fedele, V.; Melisi, D. Permissive State of EMT: The Role of Immune Cell Compartment. Front. Oncol. 2020, 10, 587. [Google Scholar] [CrossRef]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A.K. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Angela Nieto, M. The Transcription Factor Snail Controls Epithelial-Mesenchymal Transitions by Repressing E-Cadherin Expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Cech, T.R.; Steitz, J.A. The Noncoding RNA Revolution—Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef]
- Xu, S.; Gong, Y.; Yin, Y.; Xing, H.; Zhang, N. The Multiple Function of Long Noncoding RNAs in Osteosarcoma Progression, Drug Resistance and Prognosis. Biomed. Pharmacother. 2020, 127, 110141. [Google Scholar] [CrossRef]
- Lietz, C.E.; Garbutt, C.; Barry, W.T.; Deshpande, V.; Chen, Y.L.; Lozano-Calderon, S.A.; Wang, Y.; Lawney, B.; Ebb, D.; Cote, G.M.; et al. MicroRNA-MRNA Networks Define Translatable Molecular Outcome Phenotypes in Osteosarcoma. Sci. Rep. 2020, 10, 4409. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Zhang, Y.; Yu, H. Comprehensive Characterization of Circular RNAs in Osteosarcoma Cell Lines. Cell. Signal. 2020, 71, 109603. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Wang, Y.; Jing, J.; Li, J. The Roles of Circular RNAs in Osteosarcoma. Med. Sci. Monit. 2019, 25, 6378–6382. [Google Scholar] [CrossRef]
- Liu, F.; Xing, L.; Zhang, X.; Zhang, X. A Four-Pseudogene Classifier Identified by Machine Learning Serves as a Novel Prognostic Marker for Survival of Osteosarcoma. Genes 2019, 10, 414. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Z.; Fu, Q.; Wang, X.; Yang, L. Identification of 9-Gene Epithelial-Mesenchymal Transition Related Signature of Osteosarcoma by Integrating Multi Cohorts. Technol. Cancer Res. Treat. 2020, 19, 1533033820980769. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.H.; Zheng, L.; Yao, T.; Tao, S.; Wei, X.A.; Zheng, Z.Y.; Zheng, B.J.; Zhang, X.Y.; Huang, B.; Liu, J.H.; et al. EIF4A3-Induced Circular RNA PRKAR1B Promotes Osteosarcoma Progression by MiR-361-3p-Mediated Induction of FZD4 Expression. Cell Death Dis. 2021, 12, 1025. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yu, Z.; Wu, B.; Zhang, H.; Ruan, J. LINC00319 Promotes Osteosarcoma Progression by Regulating the MiR-455-3p/NFIB Axis. J. Gene Med. 2020, 22, e3248. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gu, Z.; Wu, Y.; Wu, W.; Mao, B.; Zhao, S. LINC00324 Accelerates the Proliferation and Migration of Osteosarcoma through Regulating WDR66. J. Cell Physiol. 2020, 235, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Lian, H.; Xie, P.; Yin, N.; Zhang, J.; Zhang, X.; Li, J.; Zhang, C. Linc00460 Promotes Osteosarcoma Progression via MiR-1224-5p/FADS1 Axis. Life Sci. 2019, 233, 116757. [Google Scholar] [CrossRef]
- Bian, X.; Sun, Y.M.; Wang, L.M.; Shang, Y.L. ELK1-Induced Upregulation LncRNA LINC02381 Accelerates the Osteosarcoma Tumorigenesis through Targeting CDCA4 via Sponging MiR-503–5p. Biochem. Biophys. Res. Commun. 2021, 548, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Guo, Q.; Ma, N.; Bi, W.; Xu, M.; Jia, J.; Wang, W. LncRNA BCRT1 Facilitates Osteosarcoma Progression via Regulating MiR-1303/FGF7 Axis. Aging 2021, 13, 15501–15510. [Google Scholar] [CrossRef]
- Yan, L.; Wu, X.; Yin, X.; Du, F.; Liu, Y.; Ding, X. LncRNA CCAT2 Promoted Osteosarcoma Cell Proliferation and Invasion. J. Cell Mol. Med. 2018, 22, 2592–2599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lin, J.; Chen, J.; Gu, W.; Mao, Y.; Wang, H.; Zhang, Y.; Liu, W. DDX11-AS1 Contributes to Osteosarcoma Progression via Stabilizing DDX11. Life Sci. 2020, 254, 117392. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Z.; Zhang, S.; Li, Z.; Li, D.; Yang, S.; Zhang, H.; Zeng, X.; Liu, J. LncRNA FAL1 Is a Negative Prognostic Biomarker and Exhibits Pro-Oncogenic Function in Osteosarcoma. J. Cell Biochem. 2018, 119, 8481–8489. [Google Scholar] [CrossRef]
- Yang, W.; Shan, Z.; Zhou, X.; Peng, L.; Zhi, C.; Chai, J.; Liu, H.; Yang, J.; Zhang, Z. Knockdown of LncRNA GHET1 Inhibits Osteosarcoma Cells Proliferation, Invasion, Migration and EMT in Vitro and in Vivo. Cancer Biomark. 2018, 23, 589–601. [Google Scholar] [CrossRef]
- Zhao, W.; Li, L. SP1-Induced Upregulation of Long Non-Coding RNA HCP5 Promotes the Development of Osteosarcoma. Pathol. Res. Pract. 2019, 215, 439–445. [Google Scholar] [CrossRef]
- Cai, L.; Lv, J.; Zhang, Y.; Li, J.; Wang, Y.; Yang, H. The LncRNA HNF1A-AS1 Is a Negative Prognostic Factor and Promotes Tumorigenesis in Osteosarcoma. J. Cell Mol. Med. 2017, 21, 2654–2662. [Google Scholar] [CrossRef]
- Lin, H.; Zhao, Z.; Hao, Y.; He, J.; He, J. Long Noncoding RNA HIF1A-AS2 Facilitates Cell Survival and Migration by Sponging MiR-33b-5p to Modulate SIRT6 Expression in Osteosarcoma. Biochem. Cell Biol. 2020, 98, 284–292. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Cheng, G.; Xu, R.; Han, X. Long Non-Coding RNA HOXA-AS2 Promotes Migration and Invasion by Acting as a CeRNA of MiR-520c-3p in Osteosarcoma Cells. Cell Cycle 2018, 17, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- He, J.W.; Li, D.; Zhou, J.H.; Zhu, Y.L.; Yu, B. qing SP1-Mediated Upregulation of LncRNA LMCD1-AS1 Functions a CeRNA for MiR-106b-5p to Facilitate Osteosarcoma Progression. Biochem. Biophys. Res. Commun. 2020, 526, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, Q.M.; Wang, X.Q.; Zhang, C.Q. Long Noncoding RNA MiR210HG Sponges MiR-503 to Facilitate Osteosarcoma Cell Invasion and Metastasis. DNA Cell Biol. 2017, 36, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhong, Y.; Lang, X.-B.; Tu, Y.-L.; Sun, S.-F. MNX1-AS1 Accelerates the Epithelial-Mesenchymal Transition in Osteosarcoma Cells by Activating MNX1 as a Functional Oncogene. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8194–8202. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Ji, S.; Yuan, Q.; Zhou, H. Downregulated Long Non-Coding RNA MSC-AS1 Inhibits Osteosarcoma Progression and Increases Sensitivity to Cisplatin by Binding to MicroRNA-142. Med. Sci. Monit. 2020, 26, e921594-1–e921594-15. [Google Scholar] [CrossRef]
- Tan, H.; Zhao, L. LncRNA Nuclear-Enriched Abundant Transcript 1 Promotes Cell Proliferation and Invasion by Targeting MiR-186-5p/HIF-1α in Osteosarcoma. J. Cell Biochem. 2019, 120, 6502–6514. [Google Scholar] [CrossRef]
- Liu, W.; Liu, P.; Gao, H.; Wang, X.; Yan, M. Long Non-Coding RNA PGM5-AS1 Promotes Epithelial-Mesenchymal Transition, Invasion and Metastasis of Osteosarcoma Cells by Impairing MiR-140-5p-Mediated FBN1 Inhibition. Mol. Oncol. 2020, 14, 2660–2677. [Google Scholar] [CrossRef]
- Xun, C.; Jiang, D.; Tian, Z.; Yunus, A.; Chen, J. Long Noncoding RNA Plasmacytoma Variant Translocation Gene 1 Promotes Epithelial-Mesenchymal Transition in Osteosarcoma. J. Clin. Lab. Anal. 2021, 35, e23587. [Google Scholar] [CrossRef]
- Tong, C.-J.; Deng, Q.-C.; Ou, D.-J.; Long, X.; Liu, H.; Huang, K. LncRNA RUSC1-AS1 Promotes Osteosarcoma Progression through Regulating the MiR-340-5p and PI3K/AKT Pathway. Aging 2021, 13, 20116–20130. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, J.; Chen, J. LncRNA SNHG1 Negatively Regulates MiRNA-101-3p to Enhance the Expression of ROCK1 and Promote Cell Proliferation, Migration and Invasion in Osteosarcoma. Int. J. Mol. Med. 2019, 43, 1157–1166. [Google Scholar] [CrossRef]
- Huang, Y.F.; Lu, L.; Shen, H.L.; Lu, X.X. LncRNA SNHG4 Promotes Osteosarcoma Proliferation and Migration by Sponging MiR-377-3p. Mol. Genet. Genom. Med. 2020, 8, e1349. [Google Scholar] [CrossRef]
- Zhang, J.; Ju, C.; Zhang, W.; Xie, L. LncRNA SNHG20 Is Associated with Clinical Progression and Enhances Cell Migration and Invasion in Osteosarcoma. IUBMB Life 2018, 70, 1115–1121. [Google Scholar] [CrossRef]
- Yu, X.; Hu, L.; Li, S.; Shen, J.; Wang, D.; Xu, R.; Yang, H. Long Non-Coding RNA Taurine Upregulated Gene 1 Promotes Osteosarcoma Cell Metastasis by Mediating HIF-1α via MiR-143-5p. Cell Death Dis. 2019, 10, 280. [Google Scholar] [CrossRef]
- Zhao, X.; Xu, Y.; Sun, X.; Ma, Y.; Zhang, Y.; Wang, Y.; Guan, H.; Jia, Z.; Li, Y.; Wang, Y. MiR-17-5p Promotes Proliferation and Epithelial-Mesenchymal Transition in Human Osteosarcoma Cells by Targeting SRC Kinase Signaling Inhibitor 1. J. Cell Biochem. 2019, 120, 5495–5504. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Meng, F.; Zhu, H.; Yan, H.; Guo, Y.; Zhang, S. MicroRNA-93 Promotes the Tumorigenesis of Osteosarcoma by Targeting TIMP2. Biosci. Rep. 2019, 39, BSR20191237. [Google Scholar] [CrossRef]
- Chen, J.; Yan, D.; Wu, W.; Zhu, J.; Ye, W.; Shu, Q. Micro RNA-130a Promotes the Metastasis and Epithelialmesenchymal Transition of Osteosarcoma by Targeting PTEN. Oncol. Rep. 2016, 35, 3285–3292. [Google Scholar] [CrossRef]
- Shen, S.; Huang, K.; Wu, Y.; Ma, Y.; Wang, J.; Qin, F.; Ma, J. A MiR-135b-TAZ Positive Feedback Loop Promotes Epithelial–Mesenchymal Transition (EMT) and Tumorigenesis in Osteosarcoma. Cancer Lett. 2017, 407, 32–44. [Google Scholar] [CrossRef]
- Yao, J.; Lin, J.; He, L.; Huang, J.; Liu, Q. TNF-α/MiR-155 Axis Induces the Transformation of Osteosarcoma Cancer Stem Cells Independent of TP53INP1. Gene 2020, 726, 144224. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Zhang, X.; Xing, C.; Liu, R.; Zhang, F. MiR-196a Promoted Cell Migration, Invasion and the Epithelial-Mesenchymal Transition by Targeting HOXA5 in Osteosarcoma. Cancer Biomark. 2020, 29, 291–298. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, G.; Zhang, Y.; Ma, Y.; Ding, Y.; Xu, N. MiR-199b-5p Promotes Malignant Progression of Osteosarcoma by Regulating HER2. J. BUON 2018, 23, 1816–1824. [Google Scholar]
- Shi, C.; Huang, C.M.; Wang, B.; Sun, T.F.; Zhu, A.X.; Zhu, Y.C. Pseudogene MSTO2P Enhances Hypoxia-Induced Osteosarcoma Malignancy by Upregulating PD-L1. Biochem. Biophys. Res. Commun. 2020, 530, 673–679. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, L.; Guo, A.; Liu, L.C.; Yu, F.; Diao, N.; Xu, C.; Wang, D. Overexpression of FER1L4 Promotes the Apoptosis and Suppresses Epithelial-Mesenchymal Transition and Stemness Markers via Activating PI3K/AKT Signaling Pathway in Osteosarcoma Cells. Pathol. Res. Pract. 2019, 215, 152412. [Google Scholar] [CrossRef]
- Ye, F.; Tian, L.; Zhou, Q.; Feng, D. LncRNA FER1L4 Induces Apoptosis and Suppresses EMT and the Activation of PI3K/AKT Pathway in Osteosarcoma Cells via Inhibiting MiR-18a-5p to Promote SOCS5. Gene 2019, 721, 144093. [Google Scholar] [CrossRef]
- Ye, K.; Wang, S.; Zhang, H.; Han, H.; Ma, B.; Nan, W. Long Noncoding RNA GAS5 Suppresses Cell Growth and Epithelial–Mesenchymal Transition in Osteosarcoma by Regulating the MiR-221/ARHI Pathway. J. Cell Biochem. 2017, 118, 4772–4781. [Google Scholar] [CrossRef]
- Shen, B.; Zhou, N.; Hu, T.; Zhao, W.; Wu, D.; Wang, S. LncRNA MEG3 Negatively Modified Osteosarcoma Development through Regulation of MiR-361-5p and FoxM1. J. Cell Physiol. 2019, 234, 13464–13480. [Google Scholar] [CrossRef]
- Zhang, G.; Li, Y.; Liao, G.; Qiu, H. LncRNA NKILA Inhibits Invasion and Migration of Osteosarcoma Cells via NF-ΚB/Snail Signaling Pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4118–4125. [Google Scholar]
- Fan, H.; Liu, T.; Tian, H.; Zhang, S. Tusc8 Inhibits the Development of Osteosarcoma by Sponging Mir-197-3p and Targeting Ehd2. Int. J. Mol. Med. 2020, 46, 1311–1320. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, M.; Wang, G. Upregulation of MicroRNA-7 Contributes to Inhibition of the Growth and Metastasis of Osteosarcoma Cells through the Inhibition of IGF1R. J. Cell Physiol. 2019, 234, 22195–22206. [Google Scholar] [CrossRef]
- Jiao, Z.-H.; Wang, J.-D.; Wang, X.-J. MicroRNA-16 Suppressed the Invasion and Migration of Osteosarcoma by Directly Inhibiting RAB23. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2598–2605. [Google Scholar] [CrossRef]
- Chen, B.; Liu, J.; Qu, J.; Song, Y.; Li, Y.; Pan, S. MicroRNA-25 Suppresses Proliferation, Migration, and Invasion of Osteosarcoma by Targeting SOX4. Tumor Biol. 2017, 39, 1010428317703841. [Google Scholar] [CrossRef]
- Gong, H.L.; Tao, Y.; Mao, X.Z.; Song, D.Y.; You, D.; Ni, J.D. MicroRNA-29a Suppresses the Invasion and Migration of Osteosarcoma Cells by Regulating the SOCS1/NF-ΚB Signalling Pathway through Negatively Targeting DNMT3B. Int. J. Mol. Med. 2019, 44, 1219–1232. [Google Scholar] [CrossRef]
- Waresijiang, N.; Sun, J.; Abuduaini, R.; Jiang, T.; Zhou, W.; Yuan, H. The Downregulation of MIR-125a-5p Functions as a Tumor Suppressor by Directly Targeting MMP-11 in Osteosarcoma. Mol. Med. Rep. 2016, 13, 4859–4864. [Google Scholar] [CrossRef]
- Liu, X.; Liang, Z.; Gao, K.; Li, H.; Zhao, G.; Wang, S.; Fang, J. MicroRNA-128 Inhibits EMT of Human Osteosarcoma Cells by Directly Targeting Integrin A2. Tumor Biol. 2016, 37, 7951–7957. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Y.; Liu, J.; Wu, Y.; Zhu, Q. MicroRNA-132 Inhibits Cell Growth and Metastasis in Osteosarcoma Cell Lines Possibly by Targeting Sox4. Int. J. Oncol. 2015, 47, 1672–1684. [Google Scholar] [CrossRef]
- Shi, Y.K.; Guo, Y.H. MiR-139-5p Suppresses Osteosarcoma Cell Growth and Invasion through Regulating DNMT1. Biochem. Biophys. Res. Commun. 2018, 503, 459–466. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, N.; Liu, S.; Pang, Z.; Chen, Z. By Targeting TRAF6, MiR-140-3p Inhibits TGF-Β1-Induced Human Osteosarcoma Epithelial-to-Mesenchymal Transition, Migration, and Invasion. Biotechnol. Lett. 2020, 42, 2123–2133. [Google Scholar] [CrossRef]
- Fu, Y.; Tang, Y.; Wang, J.; Guo, Z. MicroRNA-181c Suppresses the Biological Progression of Osteosarcoma via Targeting Smad7 and Regulating Transforming Growth Factor-β (TGF-β) Signaling Pathway. Med. Sci. Monit. 2019, 25, 4801–4810. [Google Scholar] [CrossRef]
- Yang, D.; Liu, G.; Wang, K. MiR-203 Acts as a Tumor Suppressor Gene in Osteosarcoma by Regulating RAB22A. PLoS ONE 2015, 10, e0132225. [Google Scholar] [CrossRef]
- He, F.; Fang, L.; Yin, Q. MiR-363 Acts as a Tumor Suppressor in Osteosarcoma Cells by Inhibiting PDZD2. Oncol. Rep. 2019, 41, 2729–2738. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, F.; Wang, L.; Zhang, Q. MiR-363 Suppresses Cell Migration, Invasion, and Epithelial-Mesenchymal Transition of Osteosarcoma by Binding to NOB1. World J. Surg. Oncol. 2020, 18, 83. [Google Scholar] [CrossRef]
- Xu, M.; Jin, H.; Xu, C.X.; Sun, B.; Song, Z.G.; Bi, W.Z.; Wang, Y. MiR-382 Inhibits Osteosarcoma Metastasis and Relapse by Targeting y Box-Binding Protein 1. Mol. Ther. 2015, 23, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Chen, L.; Li, S.; Hao, H.; Zhang, D. MiR-384 Inhibits Malignant Biological Behavior Such as Proliferation and Invasion of Osteosarcoma by Regulating IGFBP3. Technol. Cancer Res. Treat. 2020, 19, 1533033820909125. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J.; Xing, C.; Wei, S.; Guo, N.; Wang, Y. MiR-486 Inhibited Osteosarcoma Cells Invasion and Epithelial-Mesenchymal Transition by Targeting PIM1. Cancer Biomark. 2018, 23, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, Y.; Chen, H.; Guo, Z. MicroRNA-488 Inhibits Proliferation, Invasion and EMT in Osteosarcoma Cell Lines by Targeting Aquaporin 3. Int. J. Oncol. 2018, 53, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, S.; Chen, L.; Qiu, H.; Wang, J. MicroRNA-489 Suppresses Osteosarcoma Invasion, Migration and Epithelial-to-Mesenchymal Transition by Directly Targeting NAA10. Minerva Endocrinol. 2020, 45, 150–153. [Google Scholar] [CrossRef]
- Wang, T.; Wang, D.; Zhang, L.; Yang, P.; Wang, J.; Liu, Q.; Yan, F.; Lin, F. The TGFβ-MiR-499a-SHKBP1 Pathway Induces Resistance to EGFR Inhibitors in Osteosarcoma Cancer Stem Cell-like Cells. J. Exp. Clin. Cancer Res. 2019, 38, 226. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, J.; Pang, J.; He, S.; Li, G.; Chong, Y.; Li, C.; Jiao, Z.; Zhang, S.; Shao, M. MicroRNA-503 Represses Epithelial–Mesenchymal Transition and Inhibits Metastasis of Osteosarcoma by Targeting c-Myb. Tumor Biol. 2016, 37, 9181–9187. [Google Scholar] [CrossRef]
- Wang, D.; Bao, F.; Teng, Y.; Li, Q.; Li, J. MicroRNA-506-3p Initiates Mesenchymal-to-Epithelial Transition and Suppresses Autophagy in Osteosarcoma Cells by Directly Targeting SPHK1. Biosci. Biotechnol. Biochem. 2019, 83, 836–844. [Google Scholar] [CrossRef]
- Wang, X.; Li, C.; Yao, W.; Tian, Z.; Liu, Z.; Ge, H. MicroRNA-761 Suppresses Tumor Progression in Osteosarcoma via Negatively Regulating ALDH1B1. Life Sci. 2020, 262, 118544. [Google Scholar] [CrossRef]
- di Fiore, R.; Drago-Ferrante, R.; Pentimalli, F.; di Marzo, D.; Forte, I.M.; Carlisi, D.; de Blasio, A.; Tesoriere, G.; Giordano, A.; Vento, R. Let-7d MiRNA Shows Both Antioncogenic and Oncogenic Functions in Osteosarcoma-Derived 3AB-OS Cancer Stem Cells. J. Cell Physiol. 2016, 231, 1832–1841. [Google Scholar] [CrossRef]
- Yang, H.; Zhao, L.; Zhang, Y.; Li, F.F. A Comprehensive Analysis of Immune Infiltration in the Tumor Microenvironment of Osteosarcoma. Cancer Med. 2021, 10, 5696–5711. [Google Scholar] [CrossRef]
- Ling, J.; Sun, Y.; Pan, J.; Wang, H.; Ma, Z.; Yin, J.; Bao, Z.; Yang, H.; Liu, L. Feedback Modulation of Endothelial Cells Promotes Epithelial-Mesenchymal Transition and Metastasis of Osteosarcoma Cells by Von Willebrand Factor Release. J. Cell Biochem. 2019, 120, 15971–15979. [Google Scholar] [CrossRef]
- Dai, J.; Qin, L.; Chen, Y.; Wang, H.; Lin, G.; Li, X.; Liao, H.; Fang, H. Matrix Stiffness Regulates Epithelial-Mesenchymal Transition via Cytoskeletal Remodeling and MRTF-A Translocation in Osteosarcoma Cells. J. Mech. Behav. Biomed. Mater. 2019, 90, 226–238. [Google Scholar] [CrossRef]
- Bielack, S.; Kempf-Bielack, B.; Delling, G.; Exner, G.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W.; et al. Prognostic Factors in High-Grade Osteosarcoma of the Extremities of Trunk: An Analysis of 1,702 Patients Treated on Neoadjuvant Coperative Osteosarcoma Study Group Protocols. J. Clin. Oncol. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- de Las Rivas, J.; Brozovic, A.; Izraely, S.; Casas-Pais, A.; Witz, I.P.; Figueroa, A. Cancer Drug Resistance Induced by EMT: Novel Therapeutic Strategies. Arch. Toxicol. 2021, 95, 2279–2297. [Google Scholar] [CrossRef]
- Ding, L.; Wang, C.; Cui, Y.; Han, X.; Zhou, Y.; Bai, J.; Li, R. S-Phase Kinase-Associated Protein 2 Is Involved in Epithelial-Mesenchymal Transition in Methotrexate-Resistant Osteosarcoma Cells. Int. J. Oncol. 2018, 52, 1841–1852. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, J.; Shao, H.; Liu, J.; Jin, M.; Chen, J.; Zhao, H. MiR-33a Mediates the Anti-Tumor Effect of Lovastatin in Osteosarcoma by Targeting CYR61. Cell. Physiol. Biochem. 2018, 51, 938–948. [Google Scholar] [CrossRef]
- Cheng, H.-L.; Lin, C.-W.; Yang, J.-S.; Hsieh, M.-J.; Yang, S.-F.; Lu, K.-H. Zoledronate Blocks Geranylgeranylation Not Farnesylation to Suppress Human Osteosarcoma U2OS Cells Metastasis by EMT via Rho A Activation and FAK-Inhibited JNK and P38 Pathways. Oncotarget 2016, 7, 9742–9758. [Google Scholar] [CrossRef]
- Kim, E.H.; Kim, M.-S.; Lee, K.-H.; Koh, J.-S.; Jung, W.-G.; Kong, C.-B. Zoledronic Acid Is an Effective Radiosensitizer in the Treatment of Osteosarcoma. Oncotarget 2016, 7, 70869–70880. [Google Scholar] [CrossRef]
- Fang, D.; Yang, H.; Lin, J.; Teng, Y.; Jiang, Y.; Chen, J.; Li, Y. 17β-Estradiol Regulates Cell Proliferation, Colony Formation, Migration, Invasion and Promotes Apoptosis by Upregulating MiR-9 and Thus Degrades MALAT-1 in Osteosarcoma Cell MG-63 in an Estrogen Receptor-Independent Manner. Biochem. Biophys. Res. Commun. 2015, 457, 500–506. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, A.M.; Turos-Cabal, M.; Puente-Moncada, N.; Herrera, F.; Rodríguez, C.; Martín, V. Calcium Acts as a Central Player in Melatonin Antitumor Activity in Sarcoma Cells. Cell. Oncol. 2022, 45, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.; Xue, Y.; Lian, W.; Wang, C.; He, J.; Fu, Q.; Zhong, L.; Lin, N.; Lai, L.; Ye, Z.; et al. Melatonin Inhibits Osteosarcoma Stem Cells by Suppressing SOX9-Mediated Signaling. Life Sci. 2018, 207, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Fontanella, R.; Pelagalli, A.; Nardelli, A.; D’Alterio, C.; Ieranò, C.; Cerchia, L.; Lucarelli, E.; Scala, S.; Zannetti, A. A Novel Antagonist of CXCR4 Prevents Bone Marrow-Derived Mesenchymal Stem Cell-Mediated Osteosarcoma and Hepatocellular Carcinoma Cell Migration and Invasion. Cancer Lett. 2016, 370, 100–107. [Google Scholar] [CrossRef]
- Zheng, B.; Ren, T.; Huang, Y.; Guo, W. Apatinib Inhibits Migration and Invasion as Well as PD-L1 Expression in Osteosarcoma by Targeting STAT3. Biochem. Biophys. Res. Commun. 2018, 495, 1695–1701. [Google Scholar] [CrossRef]
- Seba, V.; Silva, G.; dos Santos, M.B.; Baek, S.J.; França, S.d.C.; Fachin, A.L.; Regasini, L.O.; Marins, M. Chalcone Derivatives 4′-Amino-1-Naphthyl-Chalcone (D14) and 4′-Amino-4-Methyl-1-Naphthyl-Chalcone (D15) Suppress Migration and Invasion of Osteosarcoma Cells Mediated by P53 Regulating Emt-Related Genes. Int. J. Mol. Sci. 2018, 19, 2838. [Google Scholar] [CrossRef]
- Mishra, R.; Nathani, S.; Varshney, R.; Sircar, D.; Roy, P. Berberine Reverses Epithelial-Mesenchymal Transition and Modulates Histone Methylation in Osteosarcoma Cells. Mol. Biol. Rep. 2020, 47, 8499–8511. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, K.; Du, G.; Wang, J.; Zhao, J. Berberine Enhances the Radiosensitivity of Osteosarcoma by Targeting Rad51 and Epithelial-Mesenchymal Transition. J. Cancer Res. Ther. 2020, 16, 215–221. [Google Scholar] [CrossRef]
- Liu, X.; Fan, Y.; Xie, J.; Zhang, L.; Li, L.; Wang, Z. Dehydroandrographolide Inhibits Osteosarcoma Cell Growth and Metastasis by Targeting SATB2-Mediated EMT. Anticancer Agents Med. Chem. 2019, 19, 1728–1736. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, G.; Wang, W.; Qiu, E.; Pei, Y.; Zhang, X. Magnoflorine Inhibits the Malignant Phenotypes and Increases Cisplatin Sensitivity of Osteosarcoma Cells via Regulating MiR-410-3p/HMGB1/NF-ΚB Pathway. Life Sci. 2020, 256, 117967. [Google Scholar] [CrossRef]
- Chang, J.; Wang, H.; Wang, X.; Zhao, Y.; Zhao, D.; Wang, C.; Li, Y.; Yang, Z.; Lu, S.; Zeng, Q.; et al. Molecular Mechanisms of Polyphyllin I-Induced Apoptosis and Reversal of the Epithelial-Mesenchymal Transition in Human Osteosarcoma Cells. J. Ethnopharmacol. 2015, 170, 117–127. [Google Scholar] [CrossRef]
- Ma, Z.; Yang, J.; Yang, Y.; Wang, X.; Chen, G.; Shi, A.; Lu, Y.; Jia, S.; Kang, X.; Lu, L. Rosmarinic Acid Exerts an Anticancer Effect on Osteosarcoma Cells by Inhibiting DJ-1 via Regulation of the PTEN-PI3K-Akt Signaling Pathway. Phytomedicine 2020, 68, 153186. [Google Scholar] [CrossRef] [PubMed]
- Muscella, A.; Stefàno, E.; de Bellis, L.; Nutricati, E.; Negro, C.; Marsigliante, S. Antitumor and Antimigration Effects of Salvia Clandestina L. Extract on Osteosarcoma Cells. Ann. N. Y. Acad. Sci. 2021, 1500, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; He, L. Sauchinone Inhibits Hypoxia-Induced Invasion and Epithelial–Mesenchymal Transition in Osteosarcoma Cells via Inactivation of the Sonic Hedgehog Pathway. J. Recept. Signal. Transduct. 2022, 42, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Fang, X.; Zhang, H.; Wang, X.; Li, M.; Jiang, W.; Tian, F.; Zhu, L.; Bian, Z. Triptolide Inhibits the Growth of Osteosarcoma by Regulating MicroRNA-181a via Targeting PTEN Gene in Vivo and Vitro. Tumor Biol. 2017, 39, 1010428317697556. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.Y.; Lee, Y.J.; Kim, E.H. Tumor-Treating Fields Inhibit the Metastatic Potential of Osteosarcoma Cells. Technol. Cancer Res. Treat. 2020, 19, 1533033820947481. [Google Scholar] [CrossRef] [PubMed]

| Ribonucleic Acid | Increased Levels in Clinical Sample | Promoted EMT | Promoted Cell Migration/Invasion | Promoted In Vivo Tumor Growth | Promoted In Vivo Metastasis | Associated Pathways/Targets |
|---|---|---|---|---|---|---|
| circ-FOXM1 [103] | No | miR-320a, wnt | ||||
| circ-PRKAR1B [163] | No | miR-361-3p, FZD4 | ||||
| LINC00319 [164] | Yes | miR-455-3p, NFIB | ||||
| LINC00324 [165] | Yes | WDR66, HuR | ||||
| LINC00460 [166] | No | miR-1224-5p, FADS1 | ||||
| LINC02381 [167] | Yes | miR-503-5p, CDCA4 | ||||
| lncRNA AFAP1-AS1 [63] | Yes | Rho, ROCK, p38 | ||||
| lncRNA BCRT1 [168] | Yes | miR-1303, FGF7 | ||||
| lncRNA CASC15 [101] | Yes | wnt/β-catenin | ||||
| lncRNA CCAT2 [169] | Yes | LATS2, c-Myc | ||||
| lncRNA CRNDE [85,98] | Yes | Notch1, SP1, wnt/β-catenin | ||||
| lncRNA DDX11-AS1 [170] | No | miR-873-5p, IGF2BP2 | ||||
| lncRNA FAL1 [171] | Yes | GSK-3β | ||||
| lncRNA GHET1 [99,172] | Yes | Ki67, wnt/β-catenin | ||||
| lncRNA HCP5 [173] | No | SP1 | ||||
| lncRNA HNF1A-AS1 [174] | Yes | |||||
| lncRNA HIF1A-AS2 [175] | Yes | miR-33b-5p, SIRT6 | ||||
| lncRNA HOXA-AS2 [176] | Yes | miR-502c-3p | ||||
| lncRNA LMCD1-AS1 [177] | Yes | miR-106b-5p, SP1 | ||||
| lncRNA miR210HG [178] | Yes | miR-503 | ||||
| lncRNA MNX1-AS1 [179] | Yes | Snail | ||||
| lncRNA MSC-AS1 [180] | Yes | miR-142, CDK6, PI3K/Akt | ||||
| lncRNA NEAT1 [181] | Yes | miR-186-5p, HIF-1α | ||||
| lncRNA PGM5-AS1 [182] | Yes | miR-140-5p, FBN1 | ||||
| lncRNA PVT1 [183] | Yes | |||||
| lncRNA RUSC1-AS1 [184] | Yes | miR-340-5p, PI3K/Akt | ||||
| lncRNA SNHG1 [185] | Yes | miRNA-101-3p, ROCK1, PI3K/Akt | ||||
| lncRNA SNHG4 [186] | Yes | miR-377-3p | ||||
| lncRNA SNHG7 [86] | Yes | MiR-34a, Notch-1, BCL-2, CDK6, SMAD4 | ||||
| lncRNA SNHG20 [187] | Yes | |||||
| lncRNA SPRY4-IT1 [47,66] | No | miR-101 | ||||
| lncRNA TUG1 [90,188] | Yes | miR-144-3p, miR-143-5p, EZH2, HIF-1α, wnt | ||||
| lncRNA XIST [57] | Yes | miR-153, SNAI1 | ||||
| miR-17-5p [189] | Yes | SRCIN1 | ||||
| miR-19 [76] | Yes | SPRED2, ERK/MAPK | ||||
| miR-31-5p [95] | Yes | AXIN1, wnt/β-catenin | ||||
| miR-93 [190] | Yes | TIMP2 | ||||
| miR-130a [191] | Yes | PTEN | ||||
| miR-135b [192] | No | TAZ | ||||
| miR-155 [193] | No | TNFa, TP53INP1 | ||||
| miR-196a [194] | Yes | HOXA5 | ||||
| miR-199b-5p [195] | Yes | HER2 | ||||
| miR-210-5p [110] | Yes | PIK3R5, Akt | ||||
| Pseudogene MSTO2P [196] | Yes | PD-L1 |
 : Significant association;
: Significant association;  : Not studied/reported.
: Not studied/reported.| Ribonucleic Acid | Decreased Levels in Clinical Samples | Inhibited EMT | Inhibited Cell Migration/Invasion | Inhibited In Vivo Tumor Growth | Inhibited In Vivo Metastasis | Associated Pathways/Targets |
|---|---|---|---|---|---|---|
| lncRNA FER1L4 [197,198] | Yes | miR-18a-5p, PI3K/Akt | ||||
| lncRNA GAS5 [199] | Yes | miR-221, ARHI | ||||
| lncRNA MEG3 [200] | Yes | miR-361-5p, FoxM1 | ||||
| lncRNA NKILA [201] | Yes | ↑ | NFκB, Snail | |||
| lncRNA TUSC8 [202] | Yes | miR-197-3p, EHD2 | ||||
| miR-7 [203] | Yes | IGF1R | ||||
| miR-16 [204] | Yes | RAB23 | ||||
| miR-25 [205] | Yes | SOX4 | ||||
| miR-29a [206] | Yes | SOCS1/NFκB, DNMT3B | ||||
| miR-107 [92] | Yes | wnt/β-catenin | ||||
| miR-125a-5p [207] | Yes | MMP11 | ||||
| miR-128 [208] | Yes | Integrin A2 | ||||
| miR-132 [209] | No | SOX4 | ||||
| miR-139-5p [210] | Yes | DNMT1 | ||||
| miR-140-3p [211] | Yes | TRAF6, TGFB | ||||
| miR-145 [51] | Yes | Snail | ||||
| miR-181c [212] | Yes | SMAD7, TGFB | ||||
| miR-203 [213] | Yes | RAB22A | ||||
| miR-331-3p [104] | Yes | MGAT1, Bcl/Bax, wnt/β-catenin | ||||
| miR-342-5p [96] | Yes | wnt/β-catenin | ||||
| miR-363 [214,215] | Yes | PDZD2, NOB1 | ||||
| miR-377-3p [186] | Yes | CuL1, wnt/β-catenin | ||||
| miR-382 [216] | Yes | YB-1 | ||||
| miR-384 [217] | Yes | MECP2, IGFBP3 | ||||
| miR-449a [111] | Yes | EZH2, PI3K/Akt | ||||
| miR-486 [218] | Yes | PIM1 | ||||
| miR-488 [219] | Yes | AQP3 | ||||
| miR-489 [220] | Yes | NAA10 | ||||
| miR-499a [221] | Yes | TGFβ, EGFR, Akt, SHKBP1 | ||||
| miR-503 [222] | No | c-myc | ||||
| miR-506-3p [223] | No | SPHK1, LC3II/I | ||||
| miR-708-5p [65] | No | ZEB1 | ||||
| miR-761 [224] | Yes | ALDH1B1, TGFB | ||||
| miR-765 [117] | Yes | MTUS, ERK | ||||
| miR-CT3 [77] | Yes | p38/MAPK | ||||
| miR-let-7d [225] | No | ↑ | CCND2, E2F2 |
 : Significant association; ↑: Inverse association;
: Significant association; ↑: Inverse association;  :Not studied/reported.
:Not studied/reported.| Compound | Inhibits EMT | Inhibits Cell Migration/Invasion | Inhibits In Vivo Tumor Growth | Inhibits In Vivo Metastasis | Associated Pathways/Targets |
|---|---|---|---|---|---|
| 3’hydroxyflavone [115] | MEK/ERK | ||||
| Baicalin [74,118] | ERK, TGF-β | ||||
| Berberine [241,242] | EZH2, Rad51 | ||||
| Chimaphilin [73] | PI3K/Akt, ERK, TGF-β | ||||
| Cinnamomum cassia extract [69] | TGF-β | ||||
| Dehydroandrogranpholide [243] | SATB2 | ||||
| Delphinidin [75] | ERK, MAPK | ||||
| Gamabufotalin [71] | PI3K/Akt, TGF-β | ||||
| Glaucocalyxin A [72] | TGF-β, Smad | ||||
| Magnoflorine [244] | miR-410-3p, HMGB1, NF-κB | ||||
| Nitidine Chloride [46] | Akt, GSK-3β, Snail | ||||
| Oridonin [58] | TGF-β, Smad, Snail | ||||
| Piperlongumine [82] | miR-30d-5p, SOCS3, JAK2/STAT3 | ||||
| Polyphillin I [245] | NF-κB, c-Myc | ||||
| Rosmarinic acid [246] | DJ-1, PI3K/Akt | ||||
| Salvia 13landestine extract [247] | Akt/PKB | ||||
| Sauchinone [248] | Sonic hedgehog | ||||
| Triptolide [249] | ↑ | miR-181a, PTEN |
 : Significant association; ↑: Inverse association;
: Significant association; ↑: Inverse association;  : Not studied/reported.
: Not studied/reported.Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hinton, K.; Kirk, A.; Paul, P.; Persad, S. Regulation of the Epithelial to Mesenchymal Transition in Osteosarcoma. Biomolecules 2023, 13, 398. https://doi.org/10.3390/biom13020398
Hinton K, Kirk A, Paul P, Persad S. Regulation of the Epithelial to Mesenchymal Transition in Osteosarcoma. Biomolecules. 2023; 13(2):398. https://doi.org/10.3390/biom13020398
Chicago/Turabian StyleHinton, Kristin, Andrew Kirk, Paulose Paul, and Sujata Persad. 2023. "Regulation of the Epithelial to Mesenchymal Transition in Osteosarcoma" Biomolecules 13, no. 2: 398. https://doi.org/10.3390/biom13020398
APA StyleHinton, K., Kirk, A., Paul, P., & Persad, S. (2023). Regulation of the Epithelial to Mesenchymal Transition in Osteosarcoma. Biomolecules, 13(2), 398. https://doi.org/10.3390/biom13020398





