Genome-Wide Analysis of microRNA Expression Profile in Roots and Leaves of Three Wheat Cultivars under Water and Drought Conditions
Abstract
1. Introduction
2. Methods and Materials
2.1. Wheat Cultivars, Growth and Drought Treatment
2.2. sRNA Isolation, Library Construction and Next-Generation Sequencing
2.3. Processing and Bioinformatics Analysis
2.4. Identification of miRNAs
2.5. Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
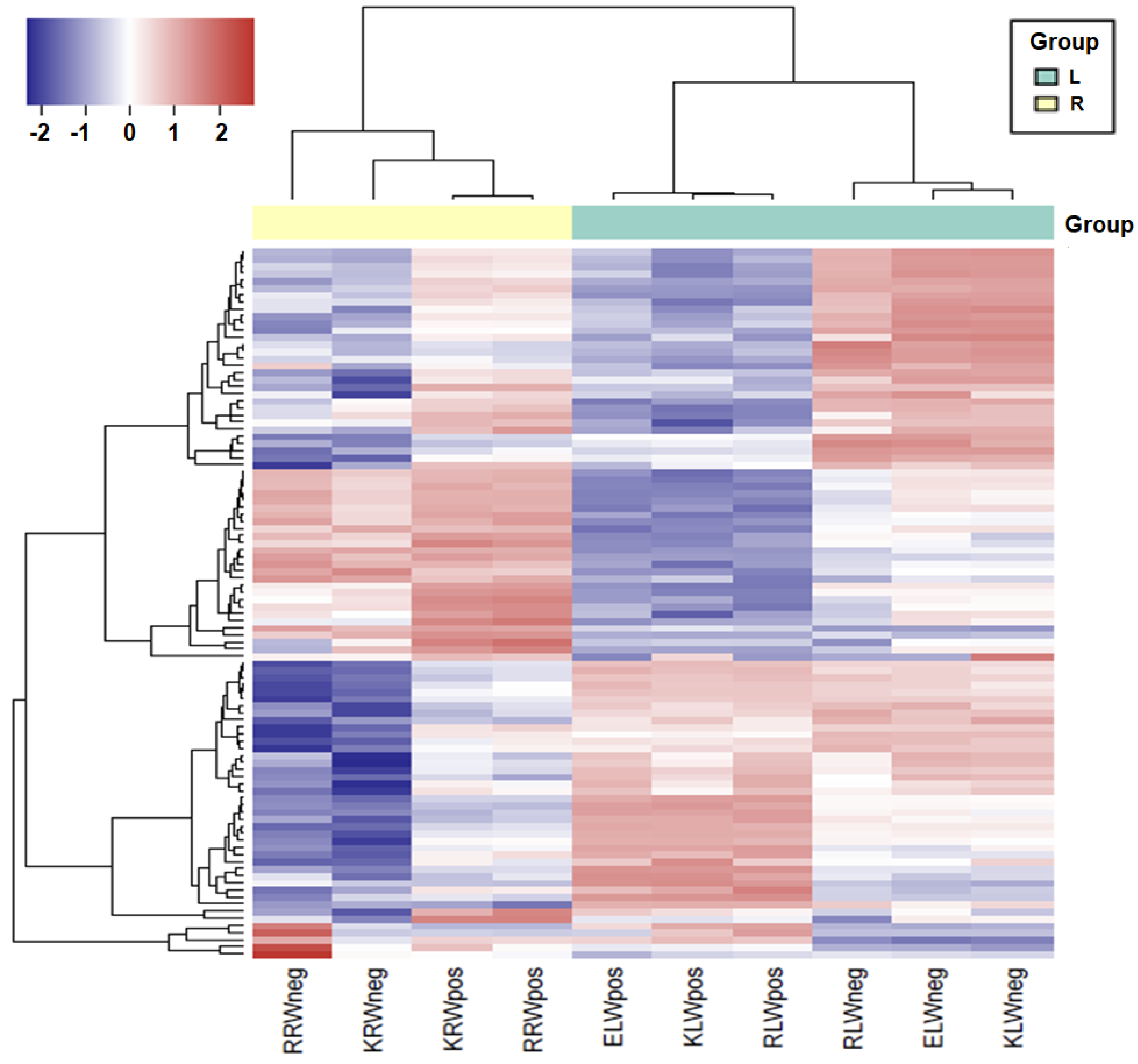
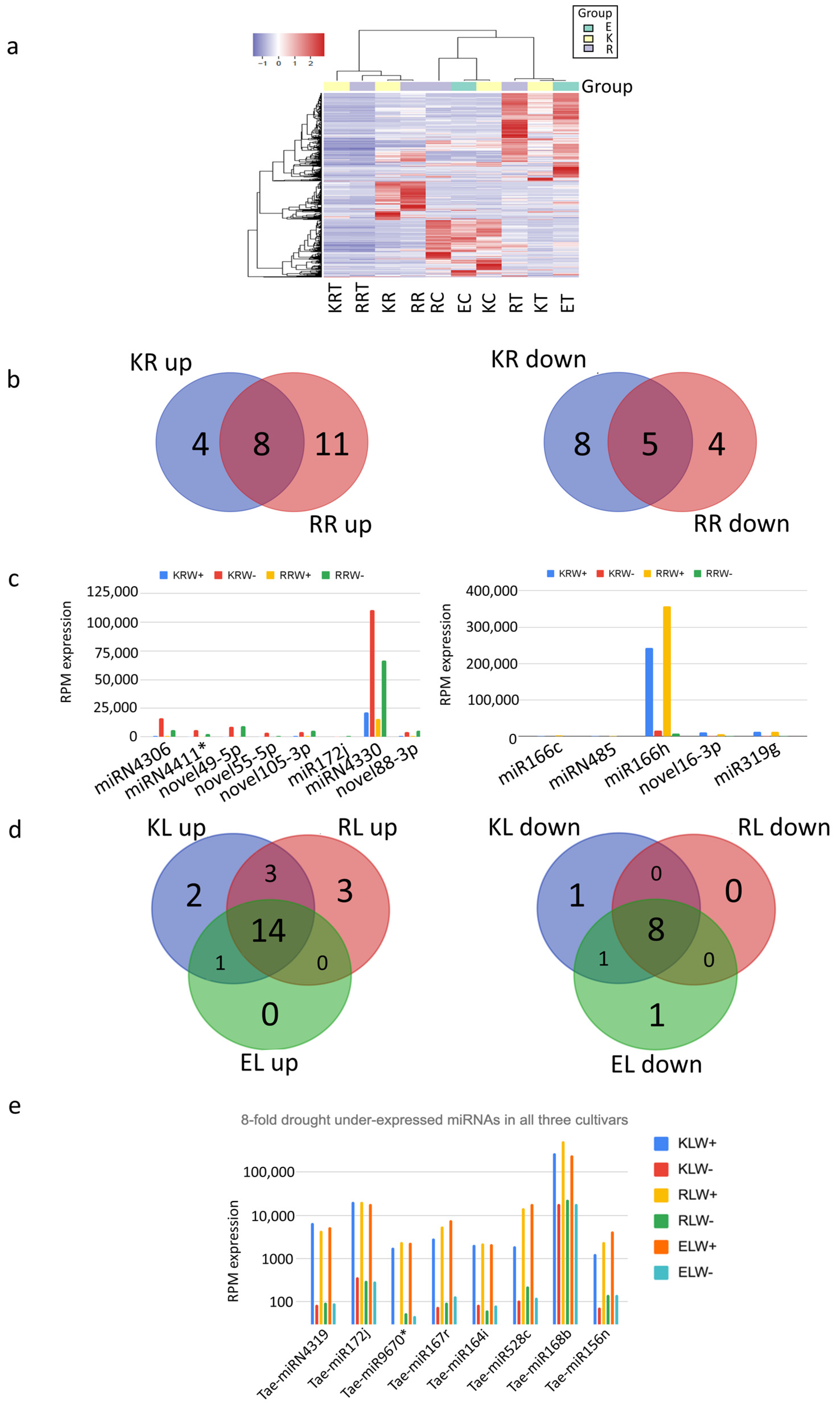
2.6. Hierarchical Cluster Analysis of Expression of sRNAs and miRNAs
2.7. Prediction and Verification of miRNAs’ Targets
2.8. Gene Ontology Analysis of miRNAs’ Targets
2.9. Network Analysis of miRNAs’ Target Genes
3. Results
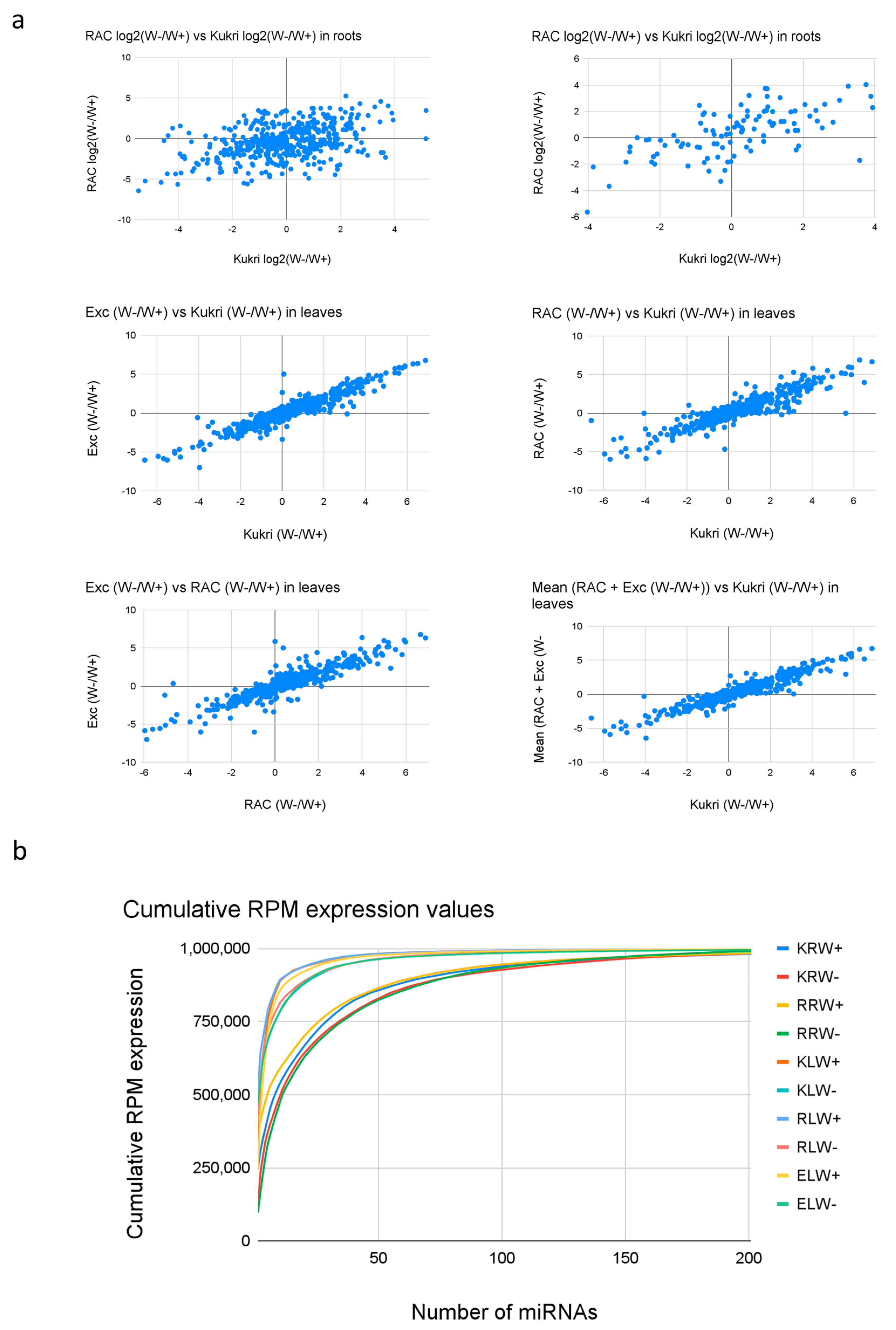
3.1. Profiles and Classification of sRNAs from the sRNA Libraries
3.2. Length Distribution of sRNA Reads from the sRNA Libraries
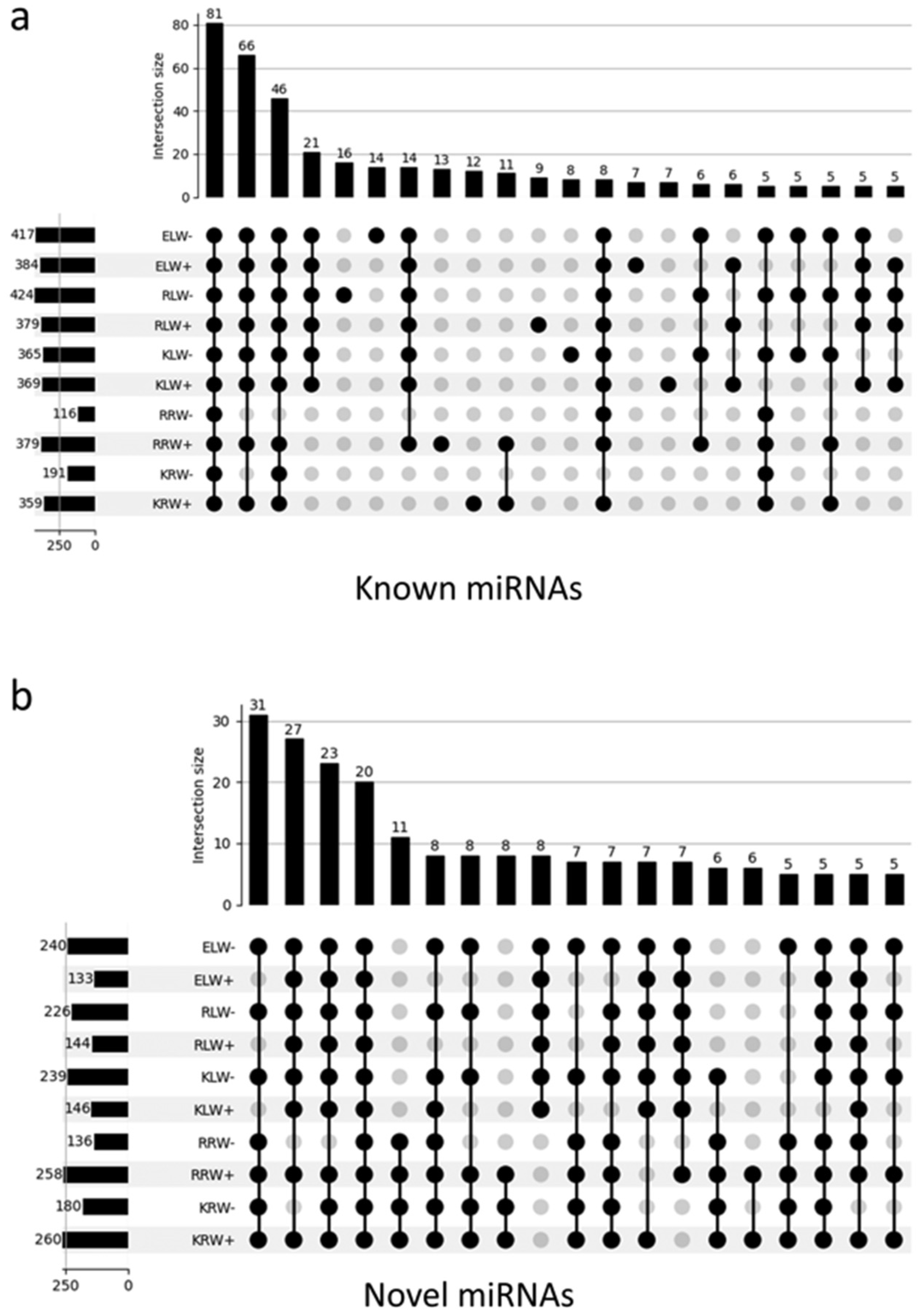
3.3. Identification of Known miRNAs and Novel miRNAs in the sRNA Libraries
3.4. Differential Expression of miRNAs between Tissues, Cultivars and Water and Drought Conditions
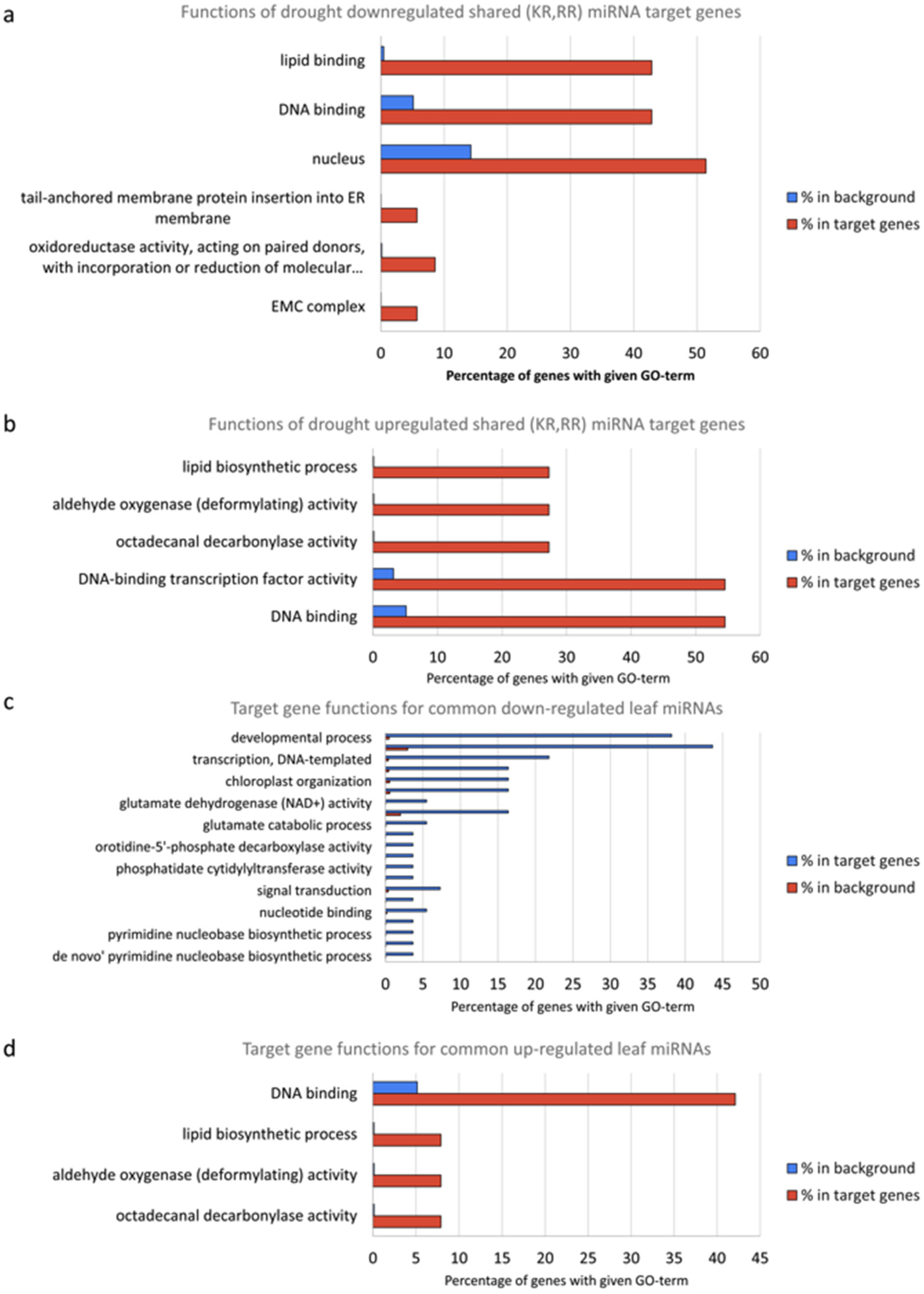
3.5. Functional Analysis of miRNAs
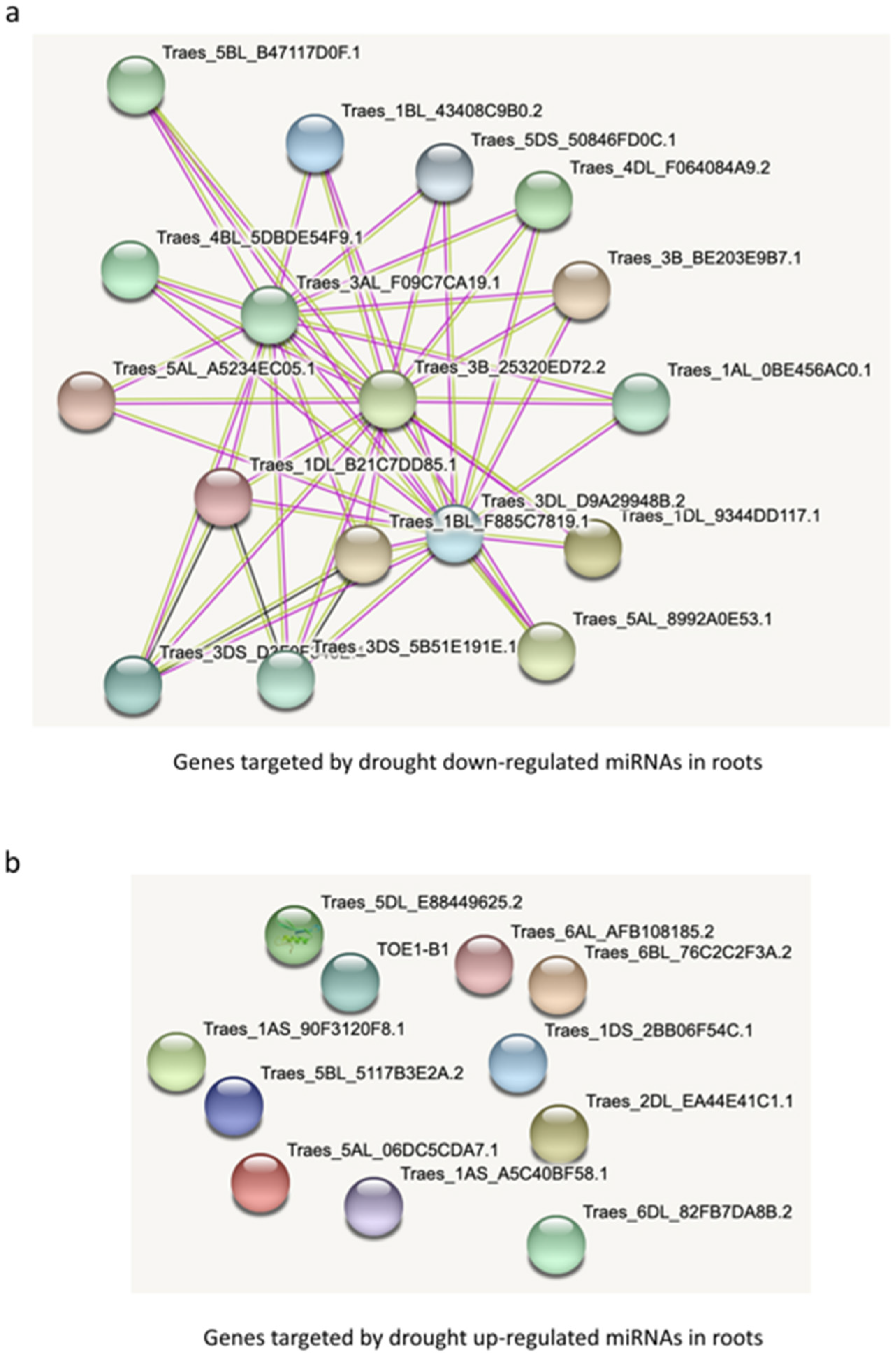
3.6. Network of Target Genes by Common Drought-Regulated miRNAs in Roots and Leaves
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Consultative Group for International Agricultural Research (CGIAR). Wheat in the World. In CGIAR Research Program on Wheat; CIMMYT: Texcoco, MÉXICO, 2019. [Google Scholar]
- Hackenberg, M.; Shi, B.-J.; Gustafson, P.; Langridge, P. A Transgenic Transcription Factor (TaDREB3) in Barley Affects the Expression of MicroRNAs and Other Small Non-Coding RNAs. PLoS ONE 2012, 7, e42030. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, M.; Shi, B.J.; Gustafson, P.; Langridge, P. Characterization of Phosphorus-Regulated MiR399 and MiR827 and Their Isomirs in Barley under Phosphorus-Sufficient and Phosphorus-Deficient Conditions. BMC Plant Biol. 2013, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, M.; Gustafson, P.; Langridge, P.; Shi, B.J. Differential Expression of MicroRNAs and Other Small RNAs in Barley between Water and Drought Conditions. Plant Biotechnol. J. 2015, 13, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Hackenberg, M.; Rueda, A.; Gustafson, P.; Langridge, P.; Shi, B.J. Generation of different sizes and classes of small RNAs in barley is locus, chromosome and/or cultivar-dependent. BMC Genom. 2016, 17, 735. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J. Deep Sequencing of Small RNAs in Plants: Applied Bioinformatics. Brief. Funct. Genom. 2012, 11, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene Silencing by MicroRNAs: Contributions of Translational Repression and MRNA Decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.; Lendeckel, W.; Tuschl, T. RNA Interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef]
- Moazed, D. Small RNAs in Transcriptional Gene Silencing and Genome Defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef]
- Zhao, B.T.; Liang, R.Q.; Ge, L.F.; Li, W.; Xiao, H.S.; Lin, H.X.; Ruan, K.C.; Jin, Y.X. Identification of Drought-Induced MicroRNAs in Rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-Responsive MicroRNAs in Populus. Plant J. 2008, 55, 131–151. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel Stress-Regulated MicroRNAs Other Small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-Based Analysis of Stress-Regulated MicroRNAs in Arabidopsis Thali-Ana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 Transcription Factor Is Regulated Transcriptionally and Posttranscriptionally to Promote Drought Resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Qin, Y.; Duan, H.; Yin, W.; Xia, X. Genome-Wide Characterization of New and Drought Stress Responsive MicroRNAs in Populus Euphratica. J. Exp. Bot. 2011, 62, 3765–3779. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Ni, Z.; Peng, H.; Sun, F.; Xin, M.; Sunkar, R.; Zhu, J.-K.; Sun, Q. Noncoding Small RNAs Responsive to Abiotic Stress in Wheat (Triticum aestivum L.). Funct. Integr. Genom. 2010, 10, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.; Lucas, S.J.; Budak, H. MiRNA Expression Patterns of Triticum Dicoccoides in Response to Shock Drought Stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Kantar, M.; Unver, T.; Budak, H. Regulation of Barley MiRNAs upon Dehydration Stress Correlated with Target Gene Expression. Funct. Integr. Genom. 2010, 10, 493–507. [Google Scholar] [CrossRef]
- Kulcheski, F.R.; Oliveira, L.F.; Molina, L.G.; Almerao, M.P.; Rodrigues, F.A.; Marcolino, J. Identification of Novel Soybean MicroRNAs Involved in Abiotic and Biotic Stresses. BMC Genom. 2011, 12, 307. [Google Scholar] [CrossRef]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of Drought-Responsive MicroRNAs in Medicago truncatula by Genome-Wide High-Throughput Sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef]
- Arenas-Huertero, C.; Pérez, B.; Rabanal, F.; Blanco-Melo, D.; Rosa, C.; Estrada-Navarrete, G. Conserved and Novel MiRNAs in the Legume Phaseolus Vulgaris in Response to Stress. Plant Mol. Biol. 2009, 70, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Frazier, T.P.; Sun, G.L.; Burklew, C.E.; Zhang, B.H. Salt and Drought Stresses Induce the Aberrant Expression of MicroRNA Genes in Tobacco. Mol. Biotechnol. 2011, 14, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-Expression of MicroRNA169 Confers Enhanced Drought Tolerance to Tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Bakhshi, B.; Fard, E.M.; Gharechahi, J.; Safarzadeh, M.; Nikpay, N.; Fotovat, R.; Azimi, M.R.; Salekdeh, G.H. The Contrasting MicroRNA Content of a Drought Tolerant and a Drought Susceptible Wheat Cultivar. J. Plant Physiol. 2017, 216, 35–43. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, C.; Yuan, J.; Li, H.; Han, X.; Chen, R.; Guan, W.; Zhong, N. Sputum Exosomal MicroRNAs Profiling Reveals Critical Pathways Modulated By Pseudomonas Aeruginosa Colonization In Bronchiectasis. Int. J. Chron. Obstruct. Pulmon. Dis. 2019, 14, 2563–2573. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Gómez-Martín, C.; Giannoukakos, S.; Medina, J.M.; Scheepbouwer, C.; García-Moreno, A.; Carmona-Saez, P.; Fromm, B.; Pegtel, M.; Keller, A.; et al. SRNAbench and SRNAtoolbox 2022 Update: Accurate MiRNA and SncRNA Profiling for Model and Non-Model Organisms. Nucleic Acids Res. 2022, 50, W710–W717. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Lebrón, R.; Rueda, A.; Gómez-Martín, C.; Giannoukakos, S.; Jaspez, D.; Medina, J.M.; Zubkovic, A.; Jurak, I.; Fromm, B.; et al. SRNAbench and SRNAtoolbox 2019: Intuitive Fast Small RNA Profiling and Differential Expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef]
- Rueda, A.; Barturen, G.; Lebrón, R.; Gómez-Martín, C.; Alganza, Á.; Oliver, J.L.; Hackenberg, M. SRNAtoolbox: An Integrated Collection of Small RNA Research Tools. Nucleic Acids Res. 2015, 43, W467–W473. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Wang, Y.; Zhao, Y.; Tao, Y.; Cheng, C.; Yang, J.; Lu, X.; Hao, C.; Wang, T.; et al. PmiREN: A Comprehensive Encyclopedia of Plant MiRNAs. Nucleic Acids Res. 2020, 48, D1114–D1121. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-Directed Phasing during Trans-Acting SiRNA Biogenesis in Plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; He, Y.; Billiau, K.; Van de Peer, Y. TAPIR, a Web Server for the Prediction of Plant MicroRNA Targets, Including Target Mimics. Bioinformatics 2010, 26, 1566–1568. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- The International Wheat Genome Sequencing Consortium. Shifting the Limits in Wheat Research and Breeding Using a Fully Annotated Reference Genome. Science 2018, 361, 7191. [Google Scholar] [CrossRef] [PubMed]
- Kawa, D.; Testerink, C. Regulation of mRNA decay in plant responses to salt and osmotic stress. Cell Mol Life Sci 2017, 74, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Loss-Morais, G.; Waterhouse, P.M.; Margis, R. Description of Plant TRNA-Derived RNA Fragments (TRFs) Associated with Argo-Naute and Identification of Their Putative Targets. Biol. Direct 2013, 8. [Google Scholar] [CrossRef]
- Park, E.J.; Kim, T.H. Fine-Tuning of Gene Expression by TRNA-Derived Fragments during Abiotic Stress Signal Transduction. Int. J. Mol. Sci. 2018, 19, 518. [Google Scholar] [CrossRef]
- Lex, A.; Gehlenborg, N.; Strobelt, H.; Vuillemot, R.; Pfister, H. UpSet: Visualization of Intersecting Sets. IEEE Trans. Vis. Comput. Graph. 2014, 20, 1983–1992. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Gómez-Martín, C.; Giannoukakos, S.; Medina, J.M.; Marchal, J.A.; Hackenberg, M. mirnaQC: A webserver for comparative quality contrl of miRNA-seq data. Nucleic Acids Res. 2020, 48. [Google Scholar] [CrossRef]
- Zhou, H.; Hussain, S.S.; Hackenberg, M.; Bazanova, N.; Eini, O.; Li, J.; Gustafson, P.; Shi, B. Identification and Characterisation of a Previously Unknown Drought Tolerance-Associated MicroRNA in Barley. Plant J. 2018, 95, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Hussain, S.S.; Shi, B.J. One Vector-Based Method to Verify Predicted Plant MiRNAs, Target Sequences, and Function Modes. Biotechnol. Bioeng. 2021, 118, 3105–3116. [Google Scholar] [CrossRef] [PubMed]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and Function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Latchman, D.S. Transcription Factors: An Overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and Functional Diversification of MIRNA Genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Du, Q.; Wang, H. The Role of HD-ZIP III Transcription Factors and MiR165/166 in Vascular Development and Secondary Cell Wall Formation. Plant Signal. Behav. 2015, 10, 1078955. [Google Scholar] [CrossRef]
- Seeholzer, S.; Tsuchimatsu, T.; Jordan, T.; Bieri, S.; Pajonk, S. Diversity at the Mla Powdery Mildew Resistance Locus from Cultivated Barley Reveals Sites of Positive Selection. Mol. Plant Microbe Interact. 2010, 23, 497–509. [Google Scholar] [CrossRef]
- Oh, I.H.; Reddy, E. The Myb Gene Family in Cell Growth, Differentiation and Apoptosis. Oncogene 1999, 18, 3017–3033. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, X. MiR156 Regulates Anthocyanin Biosynthesis through SPL Targets and Other MicroRNAs in Poplar. Hortic. Res. 2020, 7, 118. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. MiRNA-Based Drought Regulation in Wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, X.; Sun, B.; Hao, L.; Liu, C.; Zhang, D.; Tang, H.; Li, C.; Li, Y.; Shi, Y.; et al. Ge-Nome-Wide Identification and Comparative Analysis of Drought-Related MicroRNAs in Two Maize Inbred Lines with Con-Trasting Drought Tolerance by Deep Sequencing. PLoS ONE 2019, 14, 0219176. [Google Scholar]
- Zhang, F.; Luo, X.; Zhou, Y.; Xie, J. Genome-Wide Identification of Conserved MicroRNA and Their Response to Drought Stress in Dongxiang Wild Rice (Oryza rufipogon Griff.). Biotechnol. Lett. 2016, 38, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Sharif, R.; Raza, A.; Chen, P.; Li, Y.; El-Ballat, E.M.; Rauf, A.; Hano, C.; El-Esawi, M.A. HD-ZIP Gene Family: Potential Roles in Im-Proving Plant Growth and Regulating Stress-Responsive Mechanisms in Plants. Genes 2021, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, S.; Hao, W.; Song, G.; Li, Y.; Li, W.; Gao, J.; Zheng, Y.; Li, G. Lineage-Specific Evolved MicroRNAs Reg-Ulating NB-LRR Defense Genes in Triticeae. Int. J. Mol. Sci. 2019, 20, 3128. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Martín, C.; Zhou, H.; Medina, J.M.; Aparicio-Puerta, E.; Shi, B.; Hackenberg, M. Genome-Wide Analysis of microRNA Expression Profile in Roots and Leaves of Three Wheat Cultivars under Water and Drought Conditions. Biomolecules 2023, 13, 440. https://doi.org/10.3390/biom13030440
Gómez-Martín C, Zhou H, Medina JM, Aparicio-Puerta E, Shi B, Hackenberg M. Genome-Wide Analysis of microRNA Expression Profile in Roots and Leaves of Three Wheat Cultivars under Water and Drought Conditions. Biomolecules. 2023; 13(3):440. https://doi.org/10.3390/biom13030440
Chicago/Turabian StyleGómez-Martín, Cristina, Hui Zhou, José María Medina, Ernesto Aparicio-Puerta, Bujun Shi, and Michael Hackenberg. 2023. "Genome-Wide Analysis of microRNA Expression Profile in Roots and Leaves of Three Wheat Cultivars under Water and Drought Conditions" Biomolecules 13, no. 3: 440. https://doi.org/10.3390/biom13030440
APA StyleGómez-Martín, C., Zhou, H., Medina, J. M., Aparicio-Puerta, E., Shi, B., & Hackenberg, M. (2023). Genome-Wide Analysis of microRNA Expression Profile in Roots and Leaves of Three Wheat Cultivars under Water and Drought Conditions. Biomolecules, 13(3), 440. https://doi.org/10.3390/biom13030440






