Exploring the Role of PI3P in Platelets: Insights from a Novel External PI3P Pool
Abstract
:1. Introduction
2. Materials and Methods
3. Results
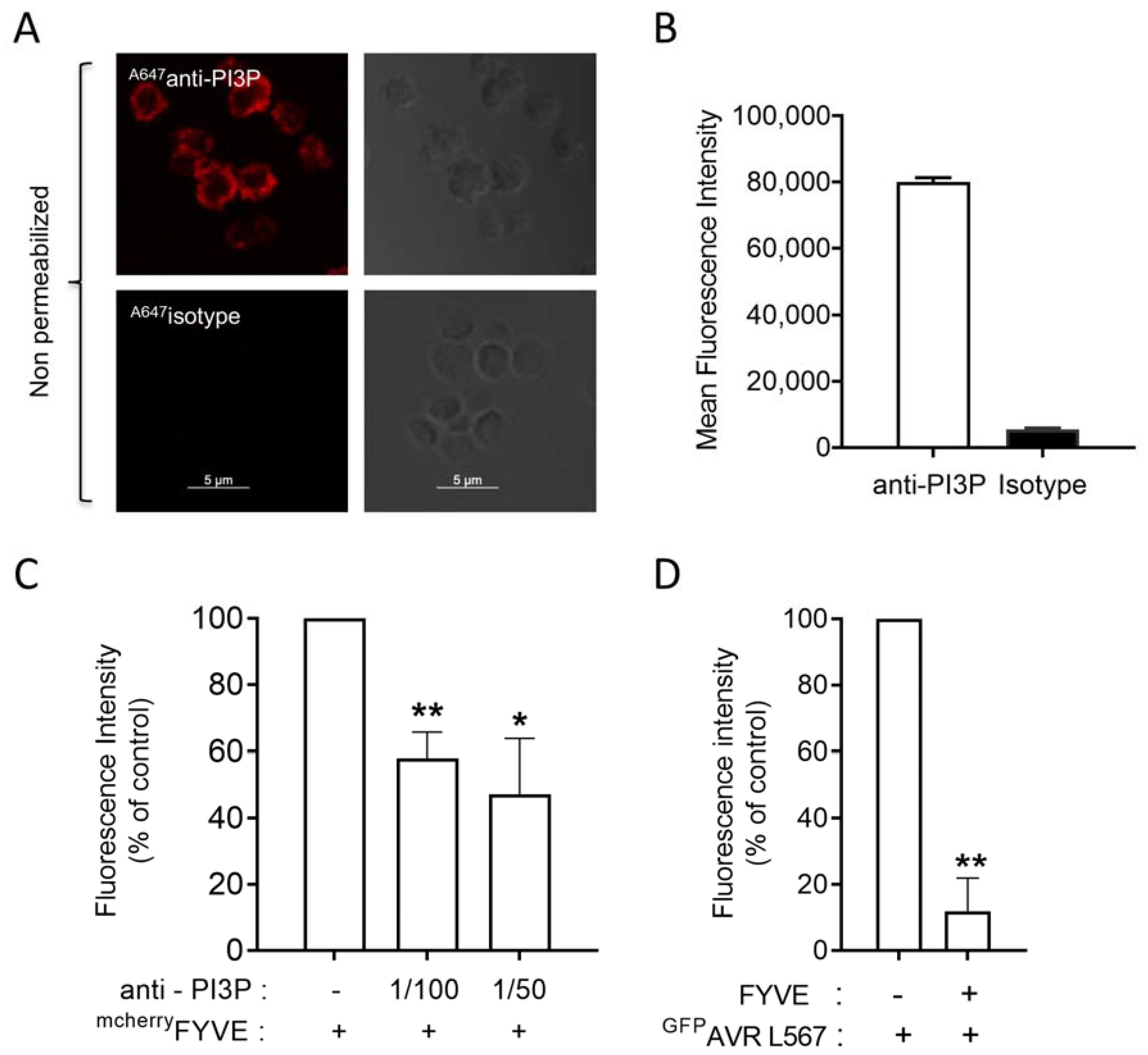
3.1. PI3P Is the Only PI Detected on the Surface of Resting Blood Platelets
3.2. The External PI3P Pool Can Be Metabolized by Exogenous MTM1 Phosphatase and ABH Phospholipase A1 and Involves Class III PI 3-Kinase and Class II PI 3-Kinase α
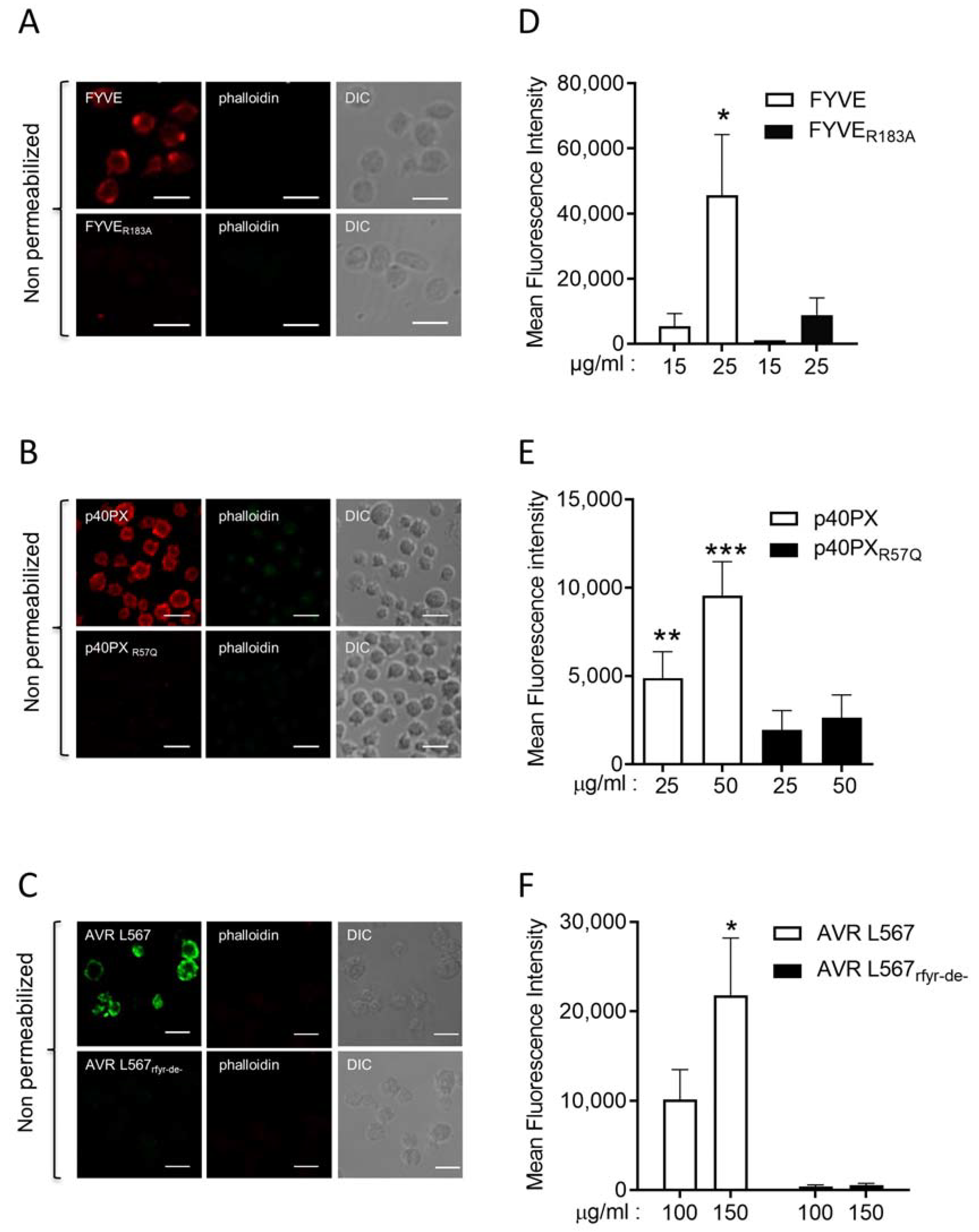
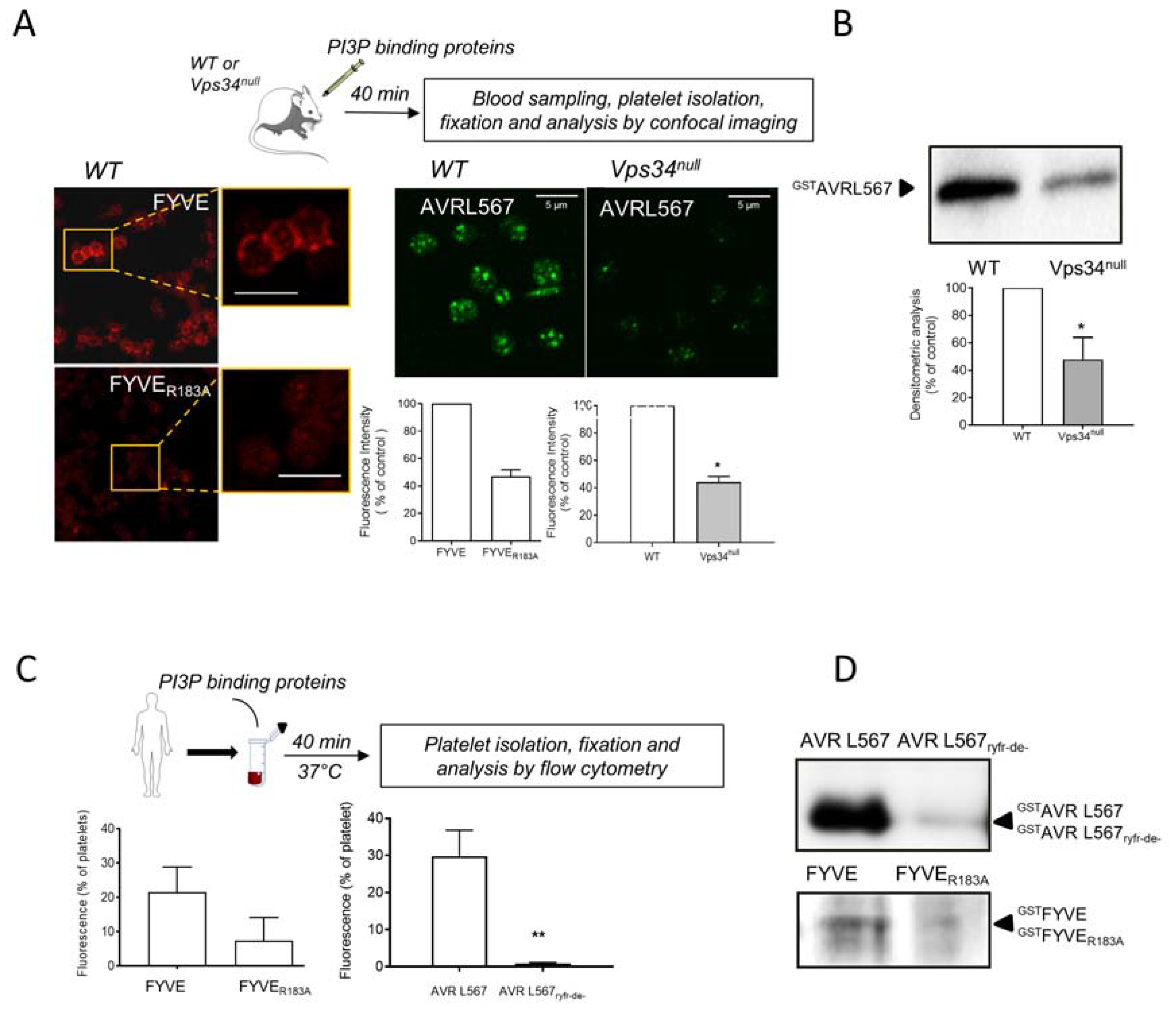
3.3. Interaction of PI3P Binding Domains with Platelets in Whole Blood In Vivo in Mouse and Ex Vivo in Human
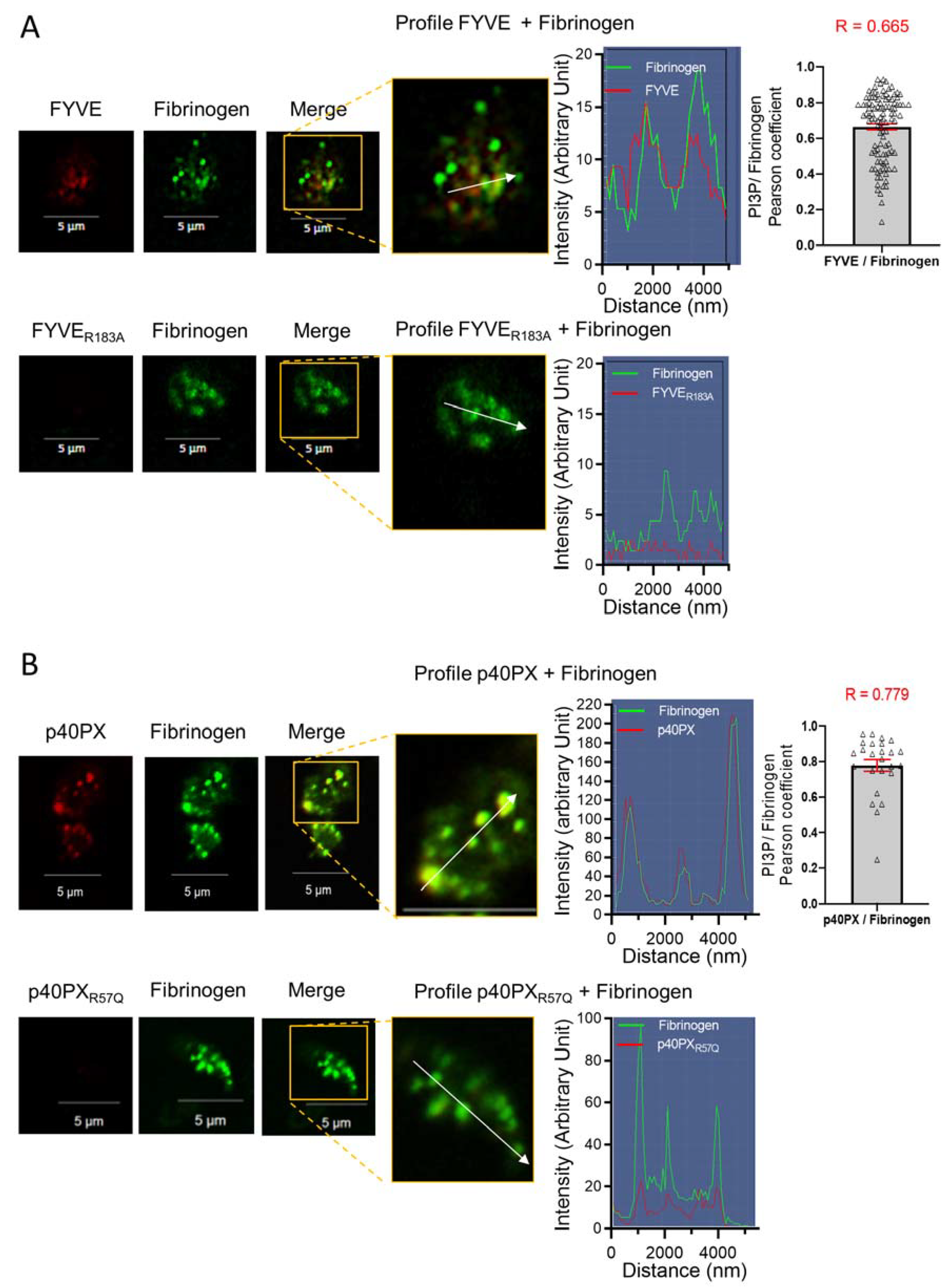
3.4. Endocytosis of the PI3P-Binding Domains to Platelet α-Granules
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Viaud, J.; Mansour, R.; Antkowiak, A.; Mujalli, A.; Valet, C.; Chicanne, G.; Xuereb, J.M.; Terrisse, A.D.; Séverin, S.; Gratacap, M.P.; et al. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 2016, 125, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Balla, T.; Szentpetery, Z.; Kim, Y.J. Phosphoinositide signaling: New tools and insights. Physiology 2009, 24, 231–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Craene, J.O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 634. [Google Scholar] [CrossRef] [Green Version]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef]
- Krauss, M.; Haucke, V. Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 2007, 8, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Saarikangas, J.; Zhao, H.; Lappalainen, P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 2010, 90, 259–289. [Google Scholar] [CrossRef] [Green Version]
- Choy, C.H.; Han, B.K.; Botelho, R.J. Phosphoinositide Diversity, Distribution, and Effector Function: Stepping Out of the Box. Bioessays 2017, 39, 1700121. [Google Scholar] [CrossRef]
- Posor, Y.; Jang, W.; Haucke, V. Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 2022, 23, 797–816. [Google Scholar] [CrossRef]
- Hammond, G.R.; Balla, T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim. Biophys. Acta 2015, 1851, 746–758. [Google Scholar] [CrossRef] [Green Version]
- Schink, K.O.; Raiborg, C.; Stenmark, H. Phosphatidylinositol 3-phosphate, a lipid that regulates membrane dynamics, protein sorting and cell signalling. Bioessays 2013, 35, 900–912. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017, 284, 1267–1278. [Google Scholar] [CrossRef] [Green Version]
- Birkeland, H.C.; Stenmark, H. Protein targeting to endosomes and phagosomes via FYVE and PX domains. Curr. Top Microbiol. Immunol. 2004, 282, 89–115. [Google Scholar] [CrossRef]
- Lodhi, I.J.; Bridges, D.; Chiang, S.H.; Zhang, Y.; Cheng, A.; Geletka, L.M.; Weisman, L.S.; Saltiel, A.R. Insulin stimulates phosphatidylinositol 3-phosphate production via the activation of Rab5. Mol. Biol. Cell 2008, 19, 2718–2728. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, Y. Activation Mechanisms of the VPS34 Complexes. Cells 2021, 10, 3124. [Google Scholar] [CrossRef]
- Kale, S.D.; Gu, B.; Capelluto, D.G.; Dou, D.; Feldman, E.; Rumore, A.; Arredondo, F.D.; Hanlon, R.; Fudal, I.; Rouxel, T.; et al. External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 2010, 142, 284–295. [Google Scholar] [CrossRef] [Green Version]
- Sun, F.; Kale, S.D.; Azurmendi, H.F.; Li, D.; Tyler, B.M.; Capelluto, D.G. Structural basis for interactions of the Phytophthora sojae RxLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Mol. Plant Microbe Interact. 2013, 26, 330–344. [Google Scholar] [CrossRef] [Green Version]
- Yoneda, A.; Kanemaru, K.; Matsubara, A.; Takai, E.; Shimozawa, M.; Satow, R.; Yamaguchi, H.; Nakamura, Y.; Fukami, K. Phosphatidylinositol 4,5-bisphosphate is localized in the plasma membrane outer leaflet and regulates cell adhesion and motility. Biochem. Biophys. Res. Commun. 2020, 527, 1050–1056. [Google Scholar] [CrossRef]
- Kim, O.H.; Kang, G.H.; Hur, J.; Lee, J.; Jung, Y.; Hong, I.S.; Lee, H.; Seo, S.Y.; Lee, D.H.; Lee, C.S.; et al. Externalized phosphatidylinositides on apoptotic cells are eat-me signals recognized by CD14. Cell Death Differ. 2022, 29, 1423–1432. [Google Scholar] [CrossRef]
- Min, S.H.; Abrams, C.S. Regulation of platelet plug formation by phosphoinositide metabolism. Blood 2013, 122, 1358–1365. [Google Scholar] [CrossRef] [Green Version]
- Ribes, A.; Oprescu, A.; Viaud, J.; Hnia, K.; Chicanne, G.; Xuereb, J.M.; Severin, S.; Gratacap, M.P.; Payrastre, B. Phosphoinositide 3-kinases in platelets, thrombosis and therapeutics. Biochem. J. 2020, 477, 4327–4342. [Google Scholar] [CrossRef] [PubMed]
- Mujalli, A.; Chicanne, G.; Bertrand-Michel, J.; Viars, F.; Stephens, L.; Hawkins, P.; Viaud, J.; Gaits-Iacovoni, F.; Severin, S.; Gratacap, M.P.; et al. Profiling of phosphoinositide molecular species in human and mouse platelets identifies new species increasing following stimulation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Ceccato, L.; Chicanne, G.; Nahoum, V.; Pons, V.; Payrastre, B.; Gaits-Iacovoni, F.; Viaud, J. PLIF: A rapid, accurate method to detect and quantitatively assess protein-lipid interactions. Sci. Signal. 2016, 9, rs2. [Google Scholar] [CrossRef] [PubMed]
- Valet, C.; Chicanne, G.; Severac, C.; Chaussade, C.; Whitehead, M.A.; Cabou, C.; Gratacap, M.P.; Gaits-Iacovoni, F.; Vanhaesebroeck, B.; Payrastre, B.; et al. Essential role of class II PI3K-C2α in platelet membrane morphology. Blood 2015, 126, 1128–1137. [Google Scholar] [CrossRef] [Green Version]
- Valet, C.; Levade, M.; Chicanne, G.; Bilanges, B.; Cabou, C.; Viaud, J.; Gratacap, M.P.; Gaits-Iacovoni, F.; Vanhaesebroeck, B.; Payrastre, B.; et al. A dual role for the class III PI3K, Vps34, in platelet production and thrombus growth. Blood 2017, 130, 2032–2042. [Google Scholar] [CrossRef] [Green Version]
- Taylor, G.S.; Dixon, J.E. An assay for phosphoinositide phosphatases utilizing fluorescent substrates. Anal. Biochem. 2001, 295, 122–126. [Google Scholar] [CrossRef]
- Burd, C.G.; Emr, S.D. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 1998, 2, 157–162. [Google Scholar] [CrossRef]
- Levade, M.; David, E.; Garcia, C.; Laurent, P.A.; Cadot, S.; Michallet, A.S.; Bordet, J.C.; Tam, C.; Sié, P.; Ysebaert, L.; et al. Ibrutinib treatment affects collagen and von Willebrand factor-dependent platelet functions. Blood 2014, 124, 3991–3995. [Google Scholar] [CrossRef]
- Séverin, S.; Gratacap, M.P.; Lenain, N.; Alvarez, L.; Hollande, E.; Penninger, J.M.; Gachet, C.; Plantavid, M.; Payrastre, B. Deficiency of Src homology 2 domain-containing inositol 5-phosphatase 1 affects platelet responses and thrombus growth. J. Clin. Investig. 2007, 117, 944–952. [Google Scholar] [CrossRef]
- Patki, V.; Lawe, D.C.; Corvera, S.; Virbasius, J.V.; Chawla, A. A functional PtdIns(3)P-binding motif. Nature 1998, 394, 433–434. [Google Scholar] [CrossRef]
- Morris, J.B.; Hinchliffe, K.A.; Ciruela, A.; Letcher, A.J.; Irvine, R.F. Thrombin stimulation of platelets causes an increase in phosphatidylinositol 5-phosphate revealed by mass assay. FEBS Lett. 2000, 475, 57–60. [Google Scholar] [CrossRef]
- Chap, H.J.; Zwaal, R.F.; van Deenen, L.L. Action of highly purified phospholipases on blood platelets. Evidence for an asymmetric distribution of phospholipids in the surface membrane. Biochim. Biophys. Acta 1977, 467, 146–164. [Google Scholar] [CrossRef]
- Verkleij, A.J.; Zwaal, R.F.; Roelofsen, B.; Comfurius, P.; Kastelijn, D.; van Deenen, L.L. The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta 1973, 323, 178–193. [Google Scholar] [CrossRef]
- Blondeau, F.; Laporte, J.; Bodin, S.; Superti-Furga, G.; Payrastre, B.; Mandel, J.L. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 2000, 9, 2223–2229. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, S.; Kim, H.; Chan, R.B.; Agarwal, S.; Williamson, R.; Cho, W.; Paolo, G.D.; Satchell, K.J. Autophagy and endosomal trafficking inhibition by Vibrio cholerae MARTX toxin phosphatidylinositol-3-phosphate-specific phospholipase A1 activity. Nat. Commun. 2015, 6, 8745. [Google Scholar] [CrossRef]
- Taylor, G.S.; Maehama, T.; Dixon, J.E. Myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl. Acad. Sci. USA 2000, 97, 8910–8915. [Google Scholar] [CrossRef] [Green Version]
- Handagama, P.; Scarborough, R.M.; Shuman, M.A.; Bainton, D.F. Endocytosis of fibrinogen into megakaryocyte and platelet alpha-granules is mediated by alpha IIb beta 3 (glycoprotein IIb-IIIa). Blood 1993, 82, 135–138. [Google Scholar] [CrossRef] [Green Version]
- Ambrosio, A.L.; Di Pietro, S.M. Mechanism of platelet α-granule biogenesis: Study of cargo transport and the VPS33B-VPS16B complex in a model system. Blood Adv. 2019, 3, 2617–2626. [Google Scholar] [CrossRef]
- Caux, M.; Mansour, R.; Xuereb, J.M.; Chicanne, G.; Viaud, J.; Vauclard, A.; Boal, F.; Payrastre, B.; Tronchère, H.; Severin, S. PIKfyve-Dependent Phosphoinositide Dynamics in Megakaryocyte/Platelet Granule Integrity and Platelet Functions. Arter. Thromb. Vasc. Biol. 2022, 42, 987–1004. [Google Scholar] [CrossRef]
- Bertović, I.; Kurelić, R.; Milošević, I.; Bender, M.; Krauss, M.; Haucke, V.; Jurak Begonja, A. Vps34 derived phosphatidylinositol 3-monophosphate modulates megakaryocyte maturation and proplatelet production through late endosomes/lysosomes. J. Thromb. Haemost. 2020, 18, 1756–1772. [Google Scholar] [CrossRef]
- Kale, S.D.; Tyler, B.M. Entry of oomycete and fungal effectors into plant and animal host cells. Cell. Microbiol. 2011, 13, 1839–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mujalli, A.; Viaud, J.; Severin, S.; Gratacap, M.-P.; Chicanne, G.; Hnia, K.; Payrastre, B.; Terrisse, A.-D. Exploring the Role of PI3P in Platelets: Insights from a Novel External PI3P Pool. Biomolecules 2023, 13, 583. https://doi.org/10.3390/biom13040583
Mujalli A, Viaud J, Severin S, Gratacap M-P, Chicanne G, Hnia K, Payrastre B, Terrisse A-D. Exploring the Role of PI3P in Platelets: Insights from a Novel External PI3P Pool. Biomolecules. 2023; 13(4):583. https://doi.org/10.3390/biom13040583
Chicago/Turabian StyleMujalli, Abdulrahman, Julien Viaud, Sonia Severin, Marie-Pierre Gratacap, Gaëtan Chicanne, Karim Hnia, Bernard Payrastre, and Anne-Dominique Terrisse. 2023. "Exploring the Role of PI3P in Platelets: Insights from a Novel External PI3P Pool" Biomolecules 13, no. 4: 583. https://doi.org/10.3390/biom13040583




