The Hop2-Mnd1 Complex and Its Regulation of Homologous Recombination
Abstract
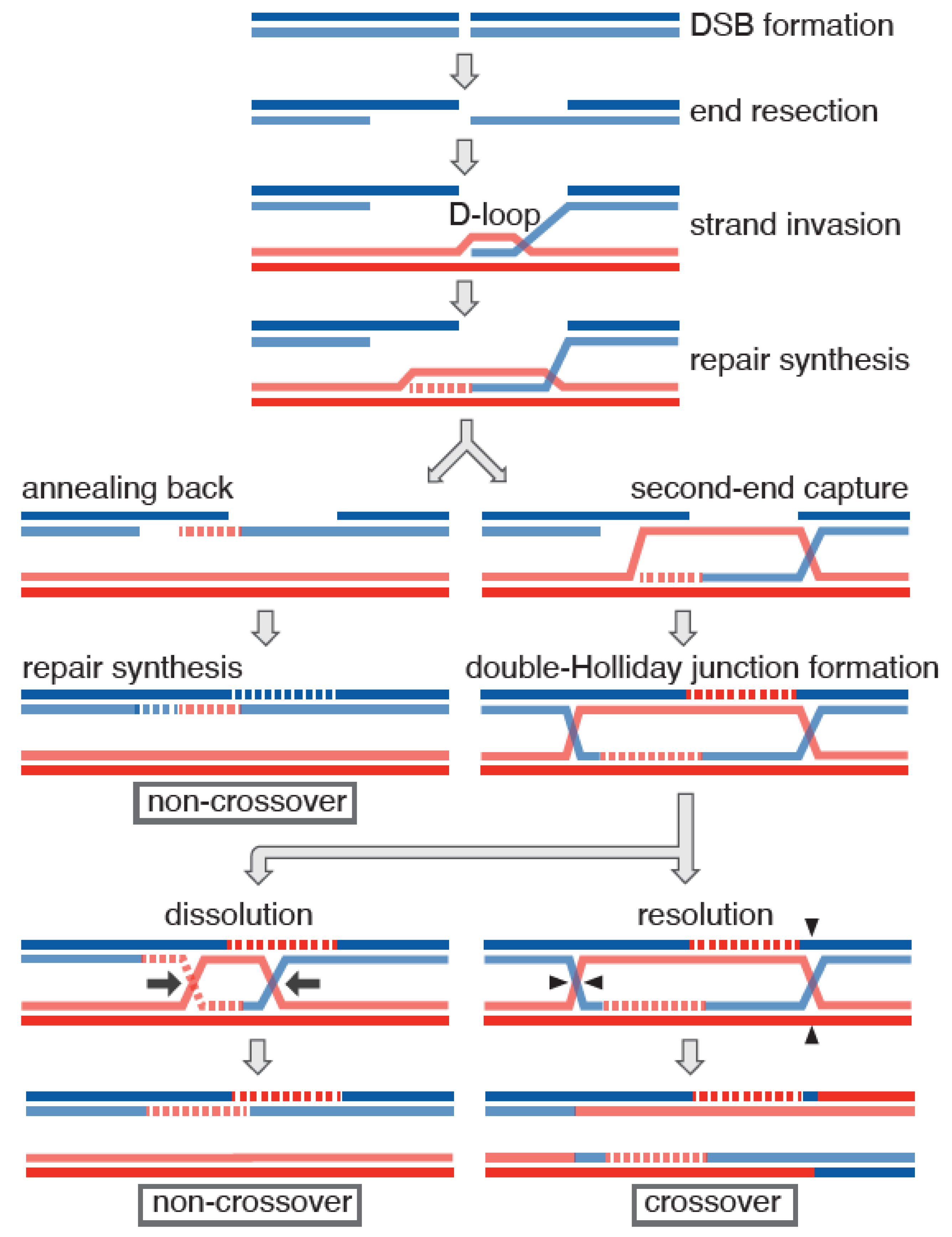
1. Introduction
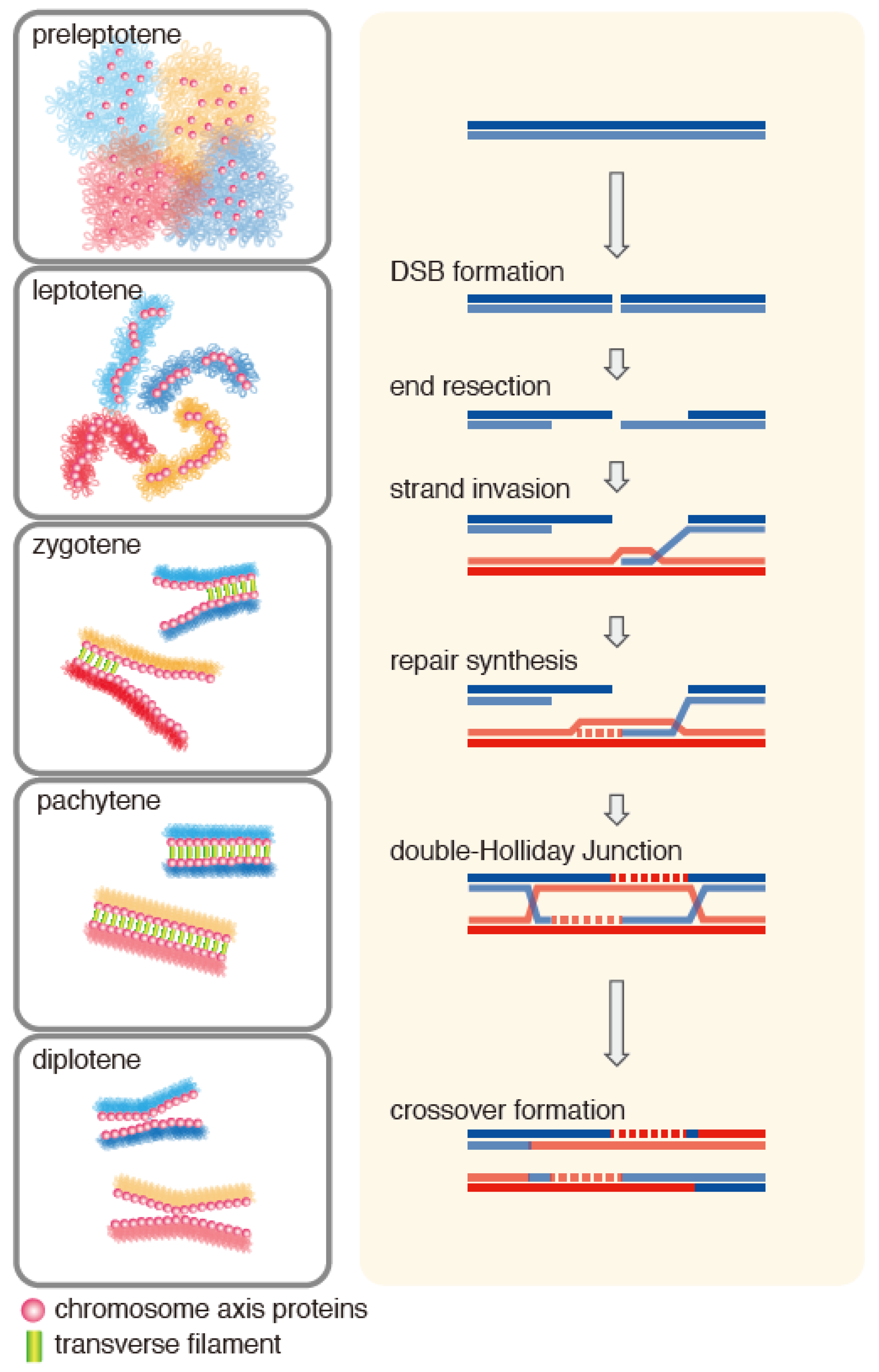
2. Meiotic Role of the Hop2-Mnd1 Complex
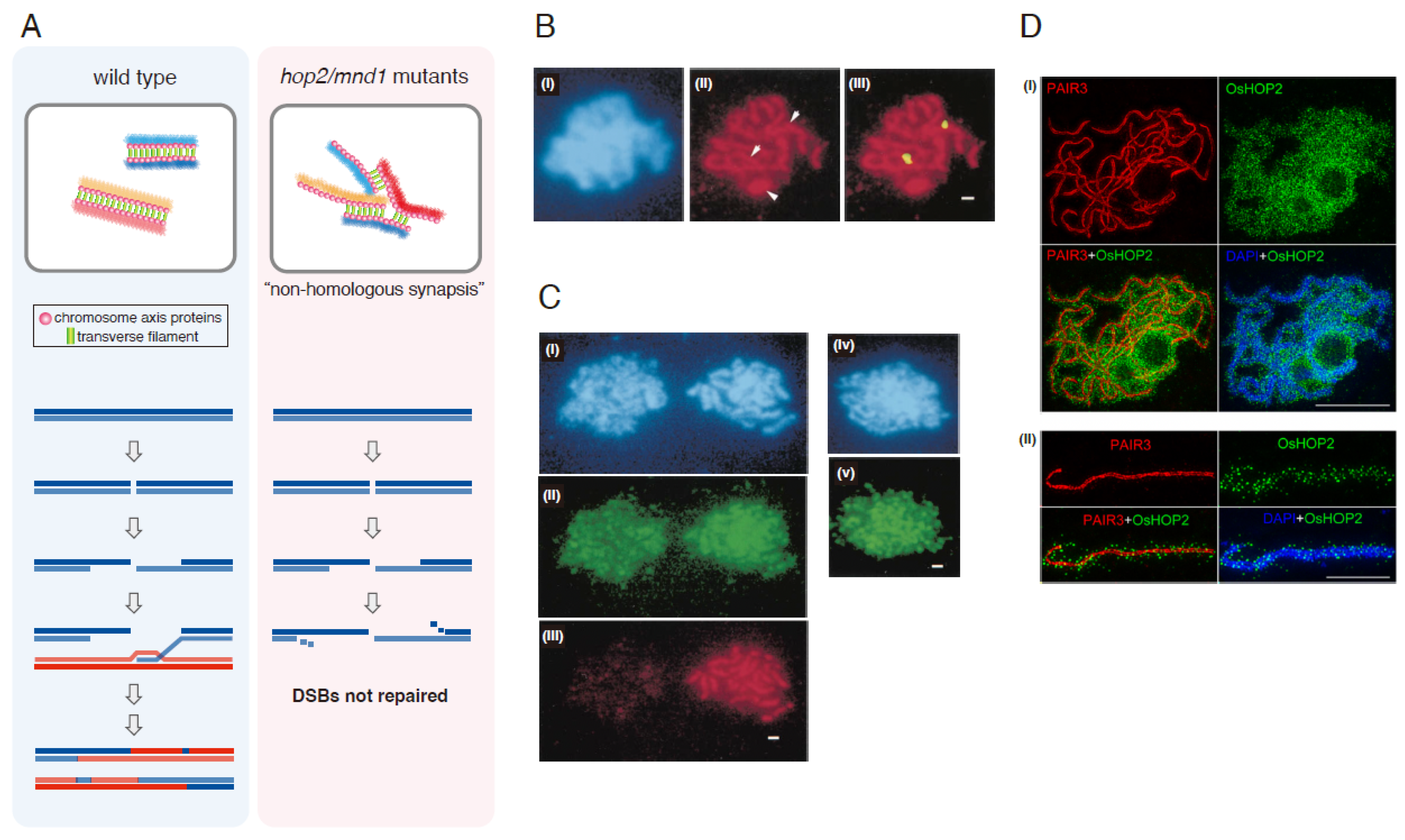
2.1. Mutant Phenotypes
2.2. Localization of Hop2-Mnd1
3. Structure and Biochemical Properties of the Hop2-Mnd1 Complex
3.1. Structure
3.2. Biochemical Activity
4. Interplay between the Hop2-Mnd1 Complex and Other Auxiliary Factors
5. Roles of the Hop2-Mnd1 Complex Outside Meiosis
6. Hop2-Mnd1 and Human Disease
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Wright, W.D.; Shah, S.S.; Heyer, W.D. Homologous Recombination and the Repair of DNA Double-Strand Breaks. J. Biol. Chem. 2018, 293, 10524–10535. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Rothstein, R.; Lisby, M. Mechanisms and Regulation of Mitotic Recombination in Saccharomyces Cerevisiae. Genetics 2014, 198, 795–835. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, J.S.; Harvey, C.S.; Wheeler, D.L.; Lichten, M. Repeated Strand Invasion and Extensive Branch Migration Are Hallmarks of Meiotic Recombination. Mol. Cell 2021, 81, 4258–4270.e4. [Google Scholar] [CrossRef] [PubMed]
- Marsolier-Kergoat, M.C.; Khan, M.M.; Schott, J.; Zhu, X.; Llorente, B. Mechanistic View and Genetic Control of DNA Recombination during Meiosis. Mol. Cell 2018, 70, 9–20.e6. [Google Scholar] [CrossRef]
- Grey, C.; de Massy, B. Chromosome Organization in Early Meiotic Prophase. Front. Cell Dev. Biol. 2021, 9, 688878. [Google Scholar] [CrossRef] [PubMed]
- Hunter, N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb. Perspect. Biol. 2015, 7, a016618. [Google Scholar] [CrossRef] [PubMed]
- Zickler, D.; Kleckner, N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016626. [Google Scholar] [CrossRef]
- Zickler, D.; Kleckner, N. A Few of Our Favorite Things: Pairing, the Bouquet, Crossover Interference and Evolution of Meiosis. Semin. Cell Dev. Biol. 2016, 54, 135–148. [Google Scholar] [CrossRef]
- Lam, I.; Keeney, S. Mechanism and Regulation of Meiotic Recombination Initiation. Cold Spring Harb. Perspect. Biol. 2014, 7, a016634. [Google Scholar] [CrossRef]
- Lake, C.M.; Hawley, R.S. Synaptonemal Complex. Curr. Biol. 2021, 31, R225–R227. [Google Scholar] [CrossRef]
- Brown, M.S.; Bishop, D.K. DNA Strand Exchange and RecA Homologs in Meiosis. Cold Spring Harb. Perspect. Biol. 2015, 7, a016659. [Google Scholar] [CrossRef]
- Bell, J.C.; Kowalczykowski, S.C. RecA: Regulation and Mechanism of a Molecular Search Engine. Trends Biochem. Sci. 2016, 41, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, J.B.; Beláň, O.; Kwon, Y.; Terakawa, T.; Al-Zain, A.; Smith, M.J.; Crickard, J.B.; Qi, Z.; Zhao, W.; Rothstein, R.; et al. Defining the Influence of Rad51 and Dmc1 Lineage-Specific Amino Acids on Genetic Recombination. Genes Dev. 2019, 33, 1191–1207. [Google Scholar] [CrossRef] [PubMed]
- Kowalczykowski, S.C. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016410. [Google Scholar] [CrossRef]
- Crickard, J.B.; Greene, E.C. Biochemical Attributes of Mitotic and Meiotic Presynaptic Complexes. DNA Repair. 2018, 71, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Huselid, E.; Bunting, S.F. The Regulation of Homologous Recombination by Helicases. Genes 2020, 11, 498. [Google Scholar] [CrossRef]
- Tsubouchi, H.; Argunhan, B.; Iwasaki, H. Biochemical Properties of Fission Yeast Homologous Recombination Enzymes. Curr. Opin. Genet. Dev. 2021, 71, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.K.; Park, D.; Xu, L.; Kleckner, N. DMC1: A Meiosis-Specific Yeast Homolog of E. Coli RecA Required for Recombination, Synaptonemal Complex Formation, and Cell Cycle Progression. Cell 1992, 69, 439–456. [Google Scholar] [CrossRef]
- Bishop, D.K. RecA Homologs Dmc1 and Rad51 Interact to Form Multiple Nuclear Complexes Prior to Meiotic Chromosome Synapsis. Cell 1994, 79, 1081–1092. [Google Scholar] [CrossRef]
- Fukushima, K.; Tanaka, Y.; Nabeshima, K.; Yoneki, T.; Tougan, T.; Tanaka, S.; Nojima, H. Dmc1 of Schizosaccharomyces Pombe Plays a Role in Meiotic Recombination. Nucleic Acids Res. 2000, 28, 2709–2716. [Google Scholar] [CrossRef]
- Grishchuk, A.L.; Kohli, J. Five RecA-like Proteins of Schizosaccharomyces Pombe Are Involved in Meiotic Recombination. Genetics 2003, 165, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Cloud, V.; Chan, Y.-L.; Grubb, J.; Budke, B.; Bishop, D.K. Rad51 Is an Accessory Factor for Dmc1-Mediated Joint Molecule Formation During Meiosis. Science 2012, 337, 1222–1225. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, H.; Roeder, G.S. The Budding Yeast Mei5 and Sae3 Proteins Act Together with Dmc1 during Meiotic Recombination. Genetics 2004, 168, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Hayase, A.; Takagi, M.; Miyazaki, T.; Oshiumi, H.; Shinohara, M.; Shinohara, A. A Protein Complex Containing Mei5 and Sae3 Promotes the Assembly of the Meiosis-Specific RecA Homolog Dmc1. Cell 2004, 119, 927–940. [Google Scholar] [CrossRef]
- Tsubouchi, H.; Roeder, G.S. The Mnd1 Protein Forms a Complex with Hop2 To Promote Homologous Chromosome Pairing and Meiotic Double-Strand Break Repair. Mol. Cell Biol. 2002, 22, 3078–3088. [Google Scholar] [CrossRef] [PubMed]
- Argunhan, B.; Murayama, Y.; Iwasaki, H. The Differentiated and Conserved Roles of Swi5-Sfr1 in Homologous Recombination. FEBS Lett. 2017, 591, 2035–2047. [Google Scholar] [CrossRef]
- Akamatsu, Y.; Dziadkowiec, D.; Ikeguchi, M.; Shinagawa, H.; Iwasaki, H. Two Different Swi5-Containing Protein Complexes Are Involved in Mating-Type Switching and Recombination Repair in Fission Yeast. Proc. Natl. Acad. Sci. USA 2003, 100, 15770–15775. [Google Scholar] [CrossRef]
- Akamatsu, Y.; Tsutsui, Y.; Morishita, T.; Siddique, M.D.S.P.; Kurokawa, Y.; Ikeguchi, M.; Yamao, F.; Arcangioli, B.; Iwasaki, H. Fission Yeast Swi5/Sfr1 and Rhp55/Rhp57 Differentially Regulate Rhp51-Dependent Recombination Outcomes. EMBO J. 2007, 26, 1352–1362. [Google Scholar] [CrossRef]
- Haruta, N.; Kurokawa, Y.; Murayama, Y.; Akamatsu, Y.; Unzai, S.; Tsutsui, Y.; Iwasaki, H. The Swi5-Sfr1 Complex Stimulates Rhp51/Rad51—and Dmc1-Mediated DNA Strand Exchange in Vitro. Nat. Struct. Mol. Biol. 2006, 13, 823–830. [Google Scholar] [CrossRef]
- Akamatsu, Y.; Jasin, M. Role for the Mammalian Swi5-Sfr1 Complex in DNA Strand Break Repair through Homologous Recombination. PLoS Genet. 2010, 6, e1001160. [Google Scholar] [CrossRef]
- Leu, J.-Y.; Chua, P.R.; Roeder, G.S. The Meiosis-Specific Hop2 Protein of S. Cerevisiae Ensures Synapsis between Homologous Chromosomes. Cell 1998, 94, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Rabitsch, K.P.; Tóth, A.; Gálová, M.; Schleiffer, A.; Schaffner, G.; Aigner, E.; Rupp, C.; Penkner, A.M.; Moreno-Borchart, A.C.; Primig, M.; et al. A Screen for Genes Required for Meiosis and Spore Formation Based on Whole-Genome Expression. Curr. Biol. 2001, 11, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Gerton, J.L.; DeRisi, J.L. Mnd1p: An Evolutionarily Conserved Protein Required for Meiotic Recombination. Proc. Natl. Acad. Sci. USA 2002, 99, 6895–6900. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, C.; Berlinger, M.; Rupp, C.; Shinohara, A.; Klein, F. Mnd1 Is Required for Meiotic Interhomolog Repair. Curr. Biol. 2004, 14, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.M.; Camahort, R.; Rice, D.A.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Gerton, J.L. Mnd1/Hop2 Facilitates Dmc1-Dependent Interhomolog Crossover Formation in Meiosis of Budding Yeast. Mol. Cell Biol. 2006, 26, 2913–2923. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-K.; Leng, C.-H.; Olivares, H.; Lee, M.-H.; Chang, Y.-C.; Kung, W.-M.; Ti, S.-C.; Lo, Y.-H.; Wang, A.H.-J.; Chang, C.-S.; et al. Heterodimeric Complexes of Hop2 and Mnd1 Function with Dmc1 to Promote Meiotic Homolog Juxtaposition and Strand Assimilation. Proc. Natl. Acad. Sci. USA 2004, 101, 10572–10577. [Google Scholar] [CrossRef]
- Shi, W.; Tang, D.; Shen, Y.; Xue, Z.; Zhang, F.; Zhang, C.; Ren, L.; Liu, C.; Du, G.; Li, Y.; et al. OsHOP2 Regulates the Maturation of Crossovers by Promoting Homologous Pairing and Synapsis in Rice Meiosis. New Phytol. 2019, 222, 805–819. [Google Scholar] [CrossRef]
- Tsubouchi, H.; Roeder, G.S. The Importance of Genetic Recombination for Fidelity of Chromosome Pairing in Meiosis. Dev. Cell 2003, 5, 915–925. [Google Scholar] [CrossRef]
- Nabeshima, K.; Kakihara, Y.; Hiraoka, Y.; Nojima, H. A Novel Meiosis-Specific Protein of Fission Yeast, Meu13p, Promotes Homologous Pairing Independently of Homologous Recombination. EMBO J. 2001, 20, 3871–3881. [Google Scholar] [CrossRef]
- Saito, T.T.; Tougan, T.; Kasama, T.; Okuzaki, D.; Nojima, H. Mcp7, a Meiosis-Specific Coiled-Coil Protein of Fission Yeast, Associates with Meu13 and Is Required for Meiotic Recombination. Nucleic Acids Res. 2004, 32, 3325–3339. [Google Scholar] [CrossRef]
- Shimada, M.; Nabeshima, K.; Tougan, T.; Nojima, H. The Meiotic Recombination Checkpoint Is Regulated by Checkpoint Rad+genes in Fission Yeast. EMBO J. 2002, 21, 2807–2818. [Google Scholar] [CrossRef] [PubMed]
- Petukhova, G.; Romanienko, P.J.; Camerini-Otero, R.D. The Hop2 Protein Has a Direct Role in Promoting Interhomolog Interactions during Mouse Meiosis. Dev. Cell 2003, 5, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Beven, A.; Lawrenson, T.; Shaw, P.; Sablowski, R. AHP2 Is Required for Bivalent Formation and for Segregation of Homologous Chromosomes in Arabidopsis Meiosis. Plant J. 2003, 36, 1–11. [Google Scholar] [CrossRef]
- Stronghill, P.; Pathan, N.; Ha, H.; Supijono, E.; Hasenkampf, C. Ahp2 (Hop2) Function in Arabidopsis Thaliana (Ler) Is Required for Stabilization of Close Alignment and Synaptonemal Complex Formation except for the Two Short Arms That Contain Nucleolus Organizer Regions. Chromosoma 2010, 119, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Vignard, J.; Siwiec, T.; Chelysheva, L.; Vrielynck, N.; Gonord, F.; Armstrong, S.J.; Schlögelhofer, P.; Mercier, R. The Interplay of RecA-Related Proteins and the MND1-HOP2 Complex during Meiosis in Arabidopsis Thaliana. PLoS Genet. 2007, 3, 1894–1906. [Google Scholar] [CrossRef] [PubMed]
- Panoli, A.P.; Ravi, M.; Sebastian, J.; Nishal, B.; Reddy, T.; Marimuthu, M.P.; Subbiah, V.; Vijaybhaskar, V.; Siddiqi, I. AtMND1 Is Required for Homologous Pairing during Meiosis in Arabidopsis. BMC Mol. Biol. 2006, 7, 24. [Google Scholar] [CrossRef]
- Kerzendorfer, C.; Vignard, J.; Pedrosa-Harand, A.; Siwiec, T.; Akimcheva, S.; Jolivet, S.; Sablowski, R.; Armstrong, S.; Schweizer, D.; Mercier, R.; et al. The Arabidopsis Thaliana MND1 Homologue Plays a Key Role in Meiotic Homologous Pairing, Synapsis and Recombination. J. Cell Sci. 2006, 119, 2486–2496. [Google Scholar] [CrossRef]
- Pathan, N.; Stronghill, P.; Hasenkampf, C. Transmission Electron Microscopy and Serial Reconstructions Reveal Novel Meiotic Phenotypes for the Ahp2 Mutant of Arabidopsis Thaliana. Genome 2013, 56, 139–145. [Google Scholar] [CrossRef]
- Farahani-Tafreshi, Y.; Wei, C.; Gan, P.; Daradur, J.; Riggs, C.D.; Hasenkampf, C.A. The Arabidopsis HOP2 Gene Has a Role in Preventing Illegitimate Connections between Nonhomologous Chromosome Regions. Chromosome Res. 2022, 30, 59–75. [Google Scholar] [CrossRef]
- Couteau, F.; Belzile, F.; Horlow, C.; Grandjean, O.; Vezon, D.; Doutriaux, M.P. Random Chromosome Segregation without Meiotic Arrest in Both Male and Female Meiocytes of a Dmc1 Mutant of Arabidopsis. Plant Cell 1999, 11, 1623–1634. [Google Scholar] [CrossRef]
- Uanschou, C.; Ronceret, A.; von Harder, M.; de Muyt, A.; Vezon, D.; Pereira, L.; Chelysheva, L.; Kobayashi, W.; Kurumizaka, H.; Schlögelhofer, P.; et al. Sufficient Amounts of Functional HOP2/MND1 Complex Promote Interhomolog DNA Repair but Are Dispensable for Intersister DNA Repair during Meiosis in Arabidopsis. Plant Cell 2013, 25, 4924–4940. [Google Scholar] [CrossRef]
- Mochizuki, K.; Novatchkova, M.; Loidl, J. DNA Double-Strand Breaks, but Not Crossovers, Are Required for the Reorganization of Meiotic Nuclei in Tetrahymena. J. Cell Sci. 2008, 121, 2148–2158. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.A.; Shin, H.C.; Kalantzi, A.S.; Toseland, C.P.; Kim, H.M.; Gruber, S.; Peraro, M.D.; Oh, B.H. Crystal Structure of Hop2-Mnd1 and Mechanistic Insights into Its Role in Meiotic Recombination. Nucleic Acids Res. 2015, 43, 3841–3856. [Google Scholar] [CrossRef] [PubMed]
- Pezza, R.J.; Petukhova, G.; Ghirlando, R.; Camerini-Otero, R.D. Molecular Activities of Meiosis-Specific Proteins Hop2, Mnd1, and the Hop2-Mnd1 Complex. J. Biol. Chem. 2006, 281, 18426–18434. [Google Scholar] [CrossRef] [PubMed]
- Moktan, H.; Guiraldelli, M.F.; Eyster, C.A.; Zhao, W.; Lee, C.Y.; Mather, T.; Camerini-Otero, R.D.; Sung, P.; Zhou, D.H.; Pezza, R.J. Solution Structure and DNA-Binding Properties of the Winged Helix Domain of the Meiotic Recombination HOP2 Protein. J. Biol. Chem. 2014, 289, 14682–14691. [Google Scholar] [CrossRef]
- Zhao, W.; Saro, D.; Hammel, M.; Kwon, Y.; Xu, Y.; Rambo, R.P.; Williams, G.J.; Chi, P.; Lu, L.; Pezza, R.J.; et al. Mechanistic Insights into the Role of Hop2-Mnd1 in Meiotic Homologous DNA Pairing. Nucleic Acids Res. 2014, 42, 906–917. [Google Scholar] [CrossRef]
- Moktan, H.; Zhou, D.H. Wing 1 of Protein HOP2 Is as Important as Helix 3 in DNA Binding by MD Simulation. J. Biomol. Struct. Dyn. 2018, 36, 1853–1866. [Google Scholar] [CrossRef] [PubMed]
- Rampler, E.; Stranzl, T.; Orban-Nemeth, Z.; Hollenstein, D.M.; Hudecz, O.; Schloegelhofer, P.; Mechtler, K. Comprehensive Cross-Linking Mass Spectrometry Reveals Parallel Orientation and Flexible Conformations of Plant HOP2-MND1. J. Proteome Res. 2015, 14, 5048–5062. [Google Scholar] [CrossRef]
- Zhao, W.; Sung, P. Significance of Ligand Interactions Involving Hop2-Mnd1 and the RAD51 and DMC1 Recombinases in Homologous DNA Repair and XX Ovarian Dysgenesis. Nucleic Acids Res. 2015, 43, 4055–4066. [Google Scholar] [CrossRef]
- Petukhova, G.; Pezza, R.J.; Vanevski, F.; Ploquin, M.; Masson, J.Y.; Camerini-Otero, R.D. The Hop2 and Mnd1 Proteins Act in Concert with Rad51 and Dmc1 in Meiotic Recombination. Nat. Struct. Mol. Biol. 2005, 12, 449–453. [Google Scholar] [CrossRef]
- Enomoto, R.; Kinebuchi, T.; Sato, M.; Yagi, H.; Kurumizaka, H.; Yokoyama, S. Stimulation of DNA Strand Exchange by the Human TBPIP/Hop2-Mnd1 Complex. J. Biol. Chem. 2006, 281, 5575–5581. [Google Scholar] [CrossRef] [PubMed]
- Pezza, R.J.; Voloshin, O.N.; Vanevski, F.; Camerini-Otero, R.D. Hop2/Mnd1 Acts on Two Critical Steps in Dmc1-Promoted Homologous Pairing. Genes Dev. 2007, 21, 1758–1766. [Google Scholar] [CrossRef] [PubMed]
- Chi, P.; San Filippo, J.; Sehorn, M.G.; Petukhova, G.; Sung, P. Bipartite Stimulatory Action of the Hop2-Mnd1 Complex on the Rad51 Recombinase. Genes Dev. 2007, 21, 1747–1757. [Google Scholar] [CrossRef]
- Ploquin, M.; Petukhova, G.; Morneau, D.; Déry, U.; Bransi, A.; Stasiak, A.; Camerini-Otero, R.D.; Masson, J.Y. Stimulation of Fission Yeast and Mouse Hop2-Mnd1 of the Dmc1 and Rad51 Recombinases. Nucleic Acids Res. 2007, 35, 2719–2733. [Google Scholar] [CrossRef] [PubMed]
- Pezza, R.J.; Camerini-Otero, R.D.; Bianco, P.R. Hop2-Mnd1 Condenses DNA to Stimulate the Synapsis Phase of DNA Strand Exchange. Biophys. J. 2010, 99, 3763–3772. [Google Scholar] [CrossRef]
- Bugreev, D.; Huang, F.; Mazina, O.M.; Pezza, R.J.; Voloshin, O.N.; Daniel Camerini-Otero, R.; Mazin, A. HOP2-MND1 Modulates RAD51 Binding to Nucleotides and DNA. Nat. Commun. 2014, 5, 4198. [Google Scholar] [CrossRef] [PubMed]
- Pezza, R.J.; Voloshin, O.N.; Volodin, A.A.; Boateng, K.A.; Bellani, M.A.; Mazin, A.; Camerini-Otero, R.D. The Dual Role of HOP2 in Mammalian Meiotic Homologous Recombination. Nucleic Acids Res. 2014, 42, 2346–2357. [Google Scholar] [CrossRef]
- Enomoto, R.; Kinebuchi, T.; Sato, M.; Yagi, H.; Shibata, T.; Kurumizaka, H.; Yokoyama, S. Positive Role of the Mammalian TBPIP/HOP2 Protein in DMC1-Mediated Homologous Pairing. J. Biol. Chem. 2004, 279, 35263–35272. [Google Scholar] [CrossRef]
- Chan, Y.L.; Brown, M.S.; Qin, D.; Handa, N.; Bishop, D.K. The Third Exon of the Budding Yeast Meiotic Recombination Gene HOP2 Is Required for Calcium-Dependent and Recombinase Dmc1-Specific Stimulation of Homologous Strand Assimilation. J. Biol. Chem. 2014, 289, 18076–18086. [Google Scholar] [CrossRef]
- Crickard, J.B.; Kwon, Y.; Sung, P.; Greene, E.C. Dynamic Interactions of the Homologous Pairing 2 (Hop2)–Meiotic Nuclear Divisions 1 (Mnd1) Protein Complex with Meiotic Presynaptic Filaments in Budding Yeast. J. Biol. Chem. 2019, 294, 490–501. [Google Scholar] [CrossRef]
- Tsubouchi, H.; Argunhan, B.; Ito, K.; Takahashi, M.; Iwasaki, H. Two Auxiliary Factors Promote Dmc1-Driven DNA Strand Exchange via Stepwise Mechanisms. Proc. Natl. Acad. Sci. USA 2020, 117, 12062–12070. [Google Scholar] [CrossRef]
- Kelso, A.A.; Say, A.F.; Sharma, D.; Ledford, L.A.L.; Turchick, A.; Saski, C.A.; King, A.; Attaway, C.C.; Temesvari, L.A.; Sehorn, M.G.; et al. Entamoeba Histolytica Dmc1 Catalyzes Homologous DNA Pairing and Strand Exchange That Is Stimulated by Calcium and Hop2-Mnd1. PLoS ONE 2015, 10, e0139399. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.L.; Zhang, A.; Weissman, B.P.; Bishop, D.K. RPA Resolves Conflicting Activities of Accessory Proteins during Reconstitution of Dmc1-Mediated Meiotic Recombination. Nucleic Acids Res. 2019, 47, 747–761. [Google Scholar] [CrossRef]
- Domenichini, S.; Raynaud, C.; Ni, D.A.; Henry, Y.; Bergounioux, C. Atmnd1-Δ1 Is Sensitive to Gamma-Irradiation and Defective in Meiotic DNA Repair. DNA Repair. 2006, 5, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Ko, L.; Cardona, G.R.; Henrion-Caude, A.; Chin, W.W. Identification and Characterization of a Tissue-Specific Coactivator, GT198, That Interacts with the DNA-Binding Domains of Nuclear Receptors. Mol. Cell Biol. 2002, 22, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.W.; Dilley, R.L.; Lampson, M.A.; Greenberg, R.A. Interchromosomal Homology Searches Drive Directional ALT Telomere Movement and Synapsis. Cell 2014, 159, 108–121. [Google Scholar] [CrossRef]
- Zangen, D.; Kaufman, Y.; Zeligson, S.; Perlberg, S.; Fridman, H.; Kanaan, M.; Abdulhadi-Atwan, M.; Abu Libdeh, A.; Gussow, A.; Kisslov, I.; et al. XX Ovarian Dysgenesis Is Caused by a PSMC3IP/HOP2 Mutation That Abolishes Coactivation of Estrogen-Driven Transcription. Am. J. Hum. Genet. 2011, 89, 572–579. [Google Scholar] [CrossRef]
- Gantchev, J.; Martínez Villarreal, A.; Gunn, S.; Zetka, M.; Ødum, N.; Litvinov, I. The Ectopic Expression of MeiCT Genes Promotes Meiomitosis and May Facilitate Carcinogenesis. Cell Cycle 2020, 19, 837–854. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Bakker, J.L.; DiCioccio, R.A.; Gille, J.J.P.; Zhao, H.; Odunsi, K.; Sucheston, L.; Jaafar, L.; Mivechi, N.F.; Waisfisz, Q.; et al. Inactivating Mutations in GT198 in Familial and Early-Onset Breast and Ovarian Cancers. Genes Cancer 2013, 4, 15–25. [Google Scholar] [CrossRef]
- Peng, M.; Yang, Z.; Zhang, H.; Jaafar, L.; Wang, G.; Liu, M.; Flores-Rozas, H.; Xu, J.; Mivechi, N.F.; Ko, L. GT198 Splice Variants Display Dominant-Negative Activities and Are Induced by Inactivating Mutations. Genes Cancer 2013, 4, 26–38. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsubouchi, H. The Hop2-Mnd1 Complex and Its Regulation of Homologous Recombination. Biomolecules 2023, 13, 662. https://doi.org/10.3390/biom13040662
Tsubouchi H. The Hop2-Mnd1 Complex and Its Regulation of Homologous Recombination. Biomolecules. 2023; 13(4):662. https://doi.org/10.3390/biom13040662
Chicago/Turabian StyleTsubouchi, Hideo. 2023. "The Hop2-Mnd1 Complex and Its Regulation of Homologous Recombination" Biomolecules 13, no. 4: 662. https://doi.org/10.3390/biom13040662
APA StyleTsubouchi, H. (2023). The Hop2-Mnd1 Complex and Its Regulation of Homologous Recombination. Biomolecules, 13(4), 662. https://doi.org/10.3390/biom13040662




