Enhancing the Conformational Stability of the cl-Par-4 Tumor Suppressor via Site-Directed Mutagenesis
Abstract
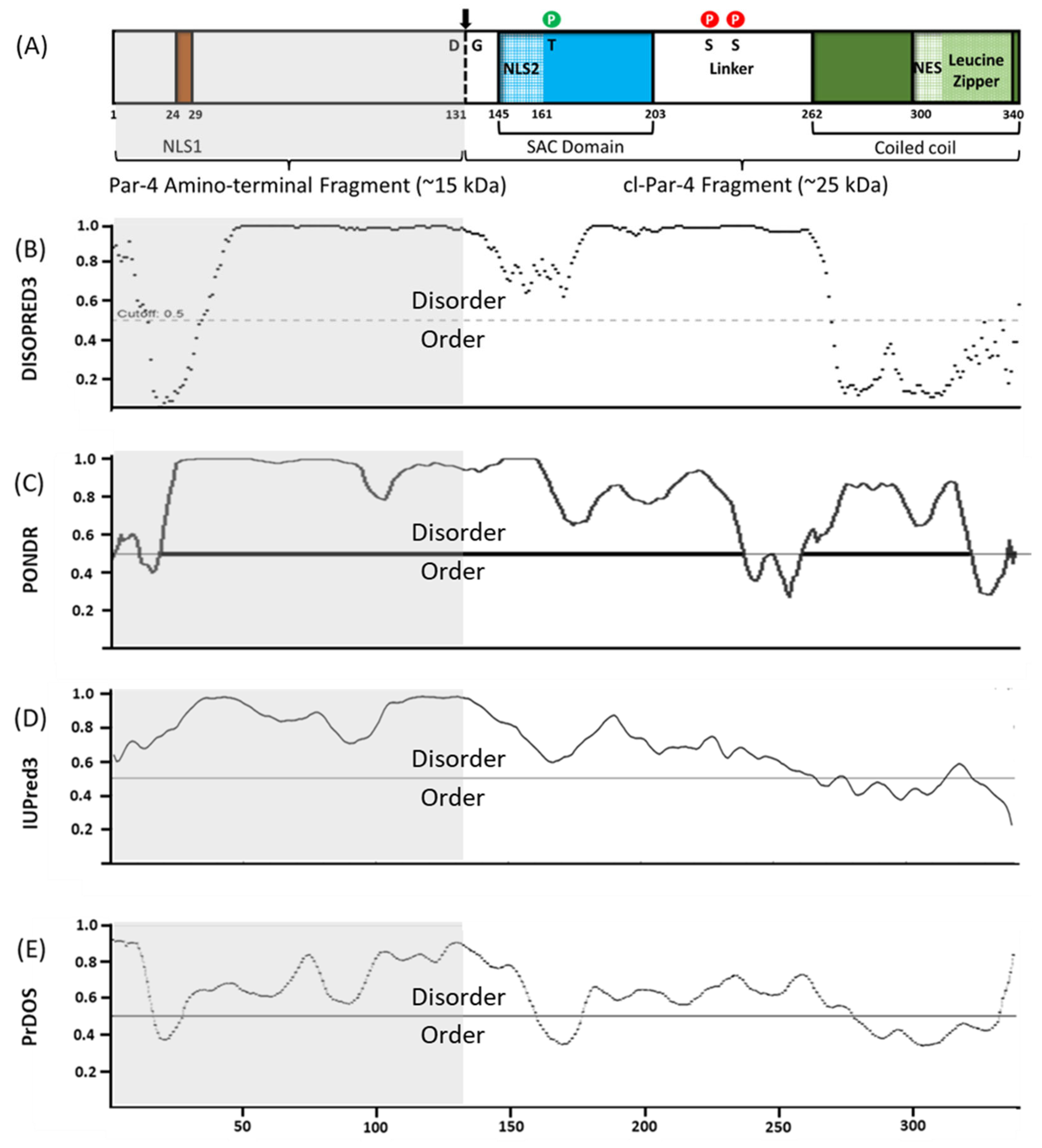
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of the D313K cl-Par-4 Mutant
2.2. Expression and Purification of Isotopically (15N) Labeled WT & D313K
2.3. Circular Dichroism Spectroscopy
2.4. Dynamic Light Scattering
2.5. Isotopically (15N) Labeled WT/D313K Sample Preparation for NMR
2.6. NMR Spectroscopy
3. Results
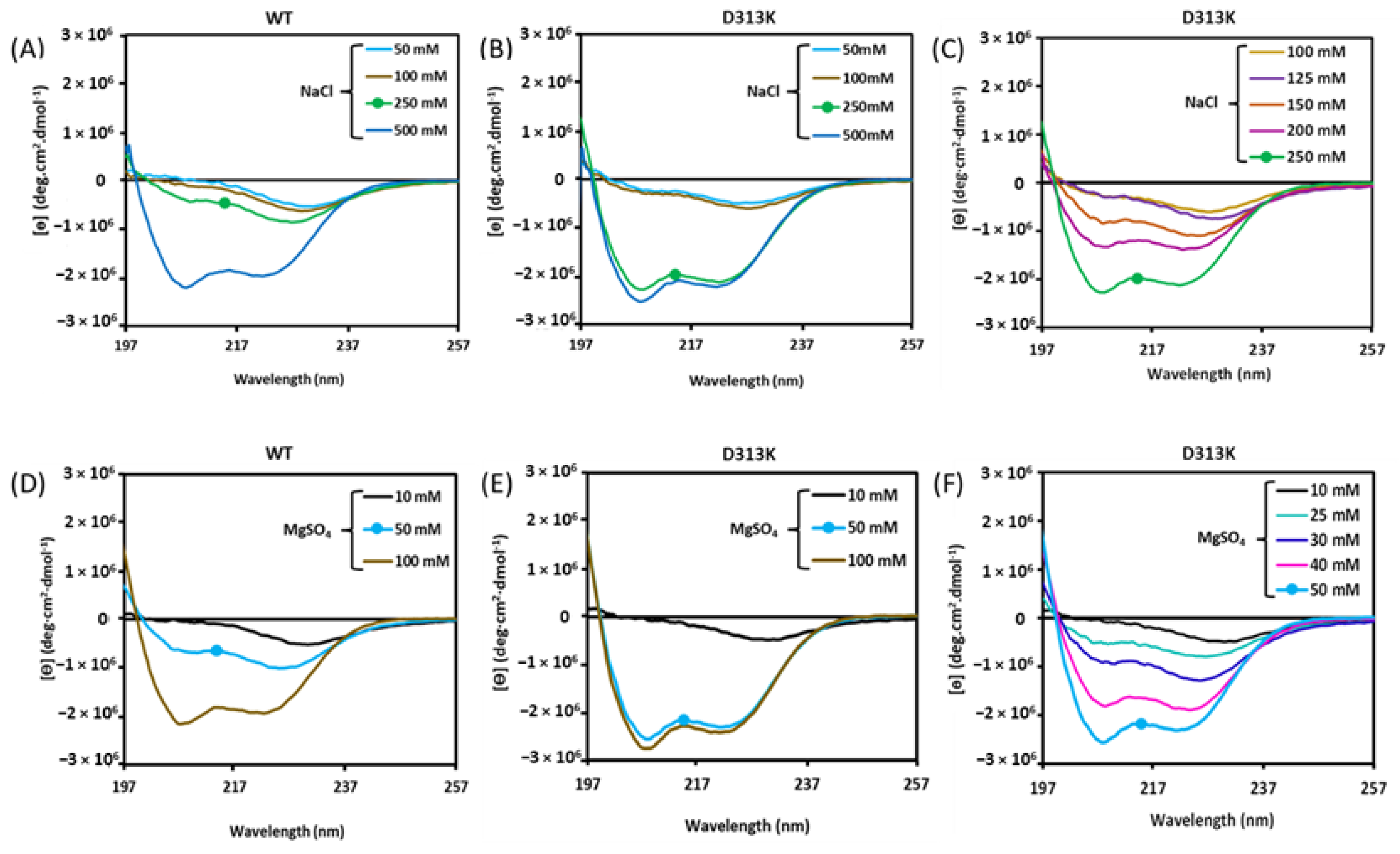
3.1. Effect of the D313K Mutation on CD Spectra
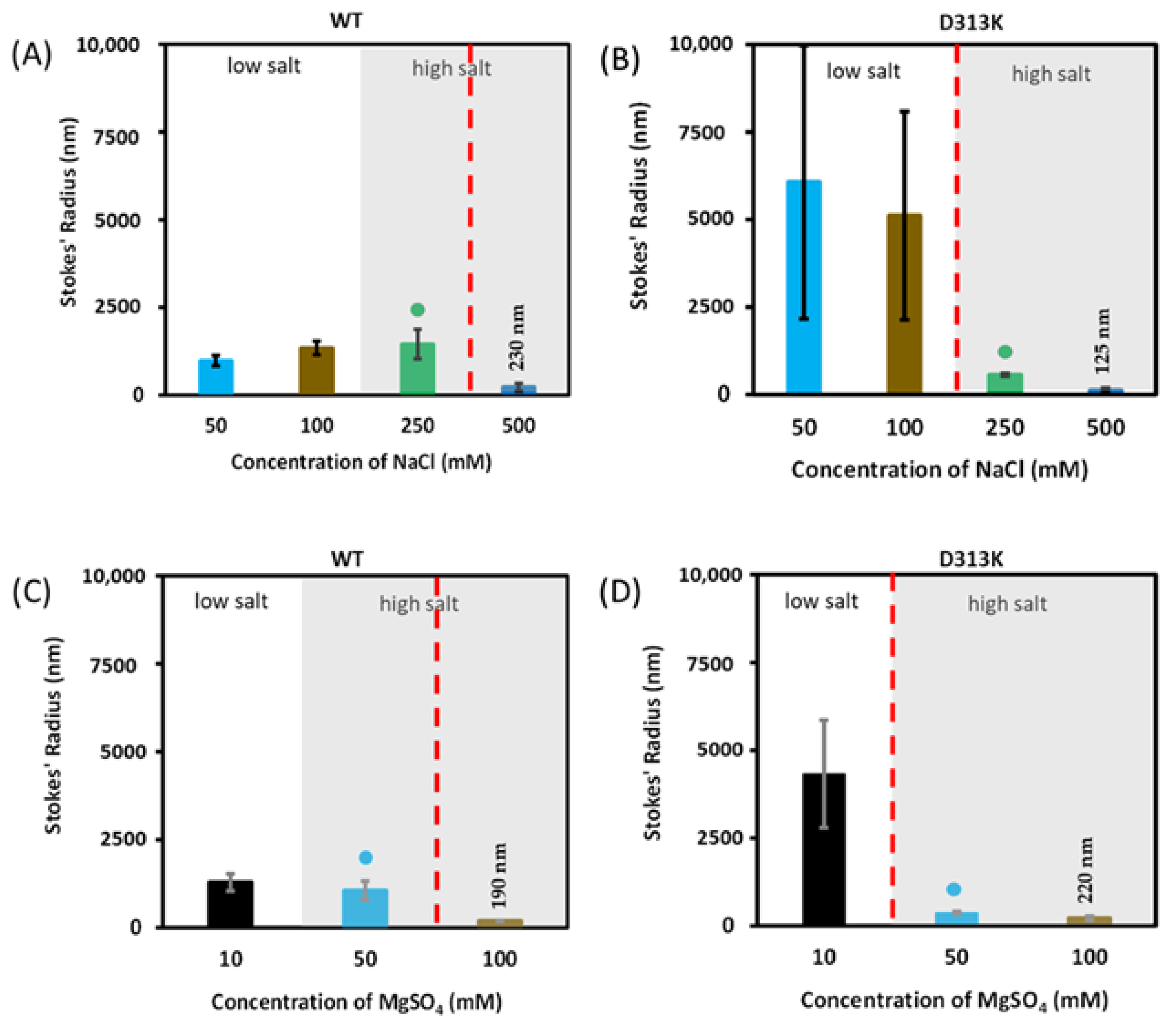
3.2. Effect of the D313K Mutation on Particle Size
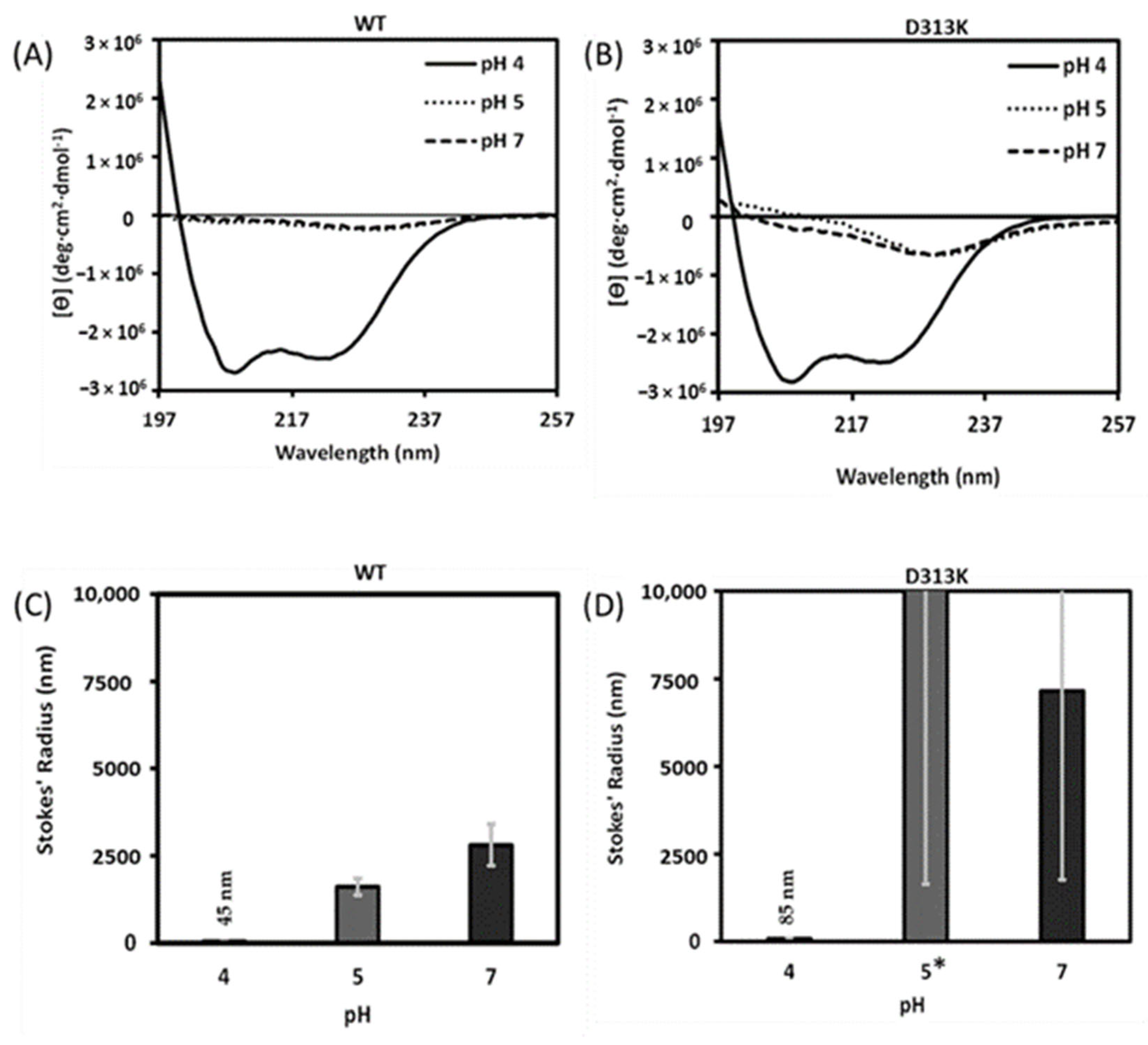
3.3. Effect of the D313K Mutation on CD Spectra and Particle Size at Low Salt as a Function of pH
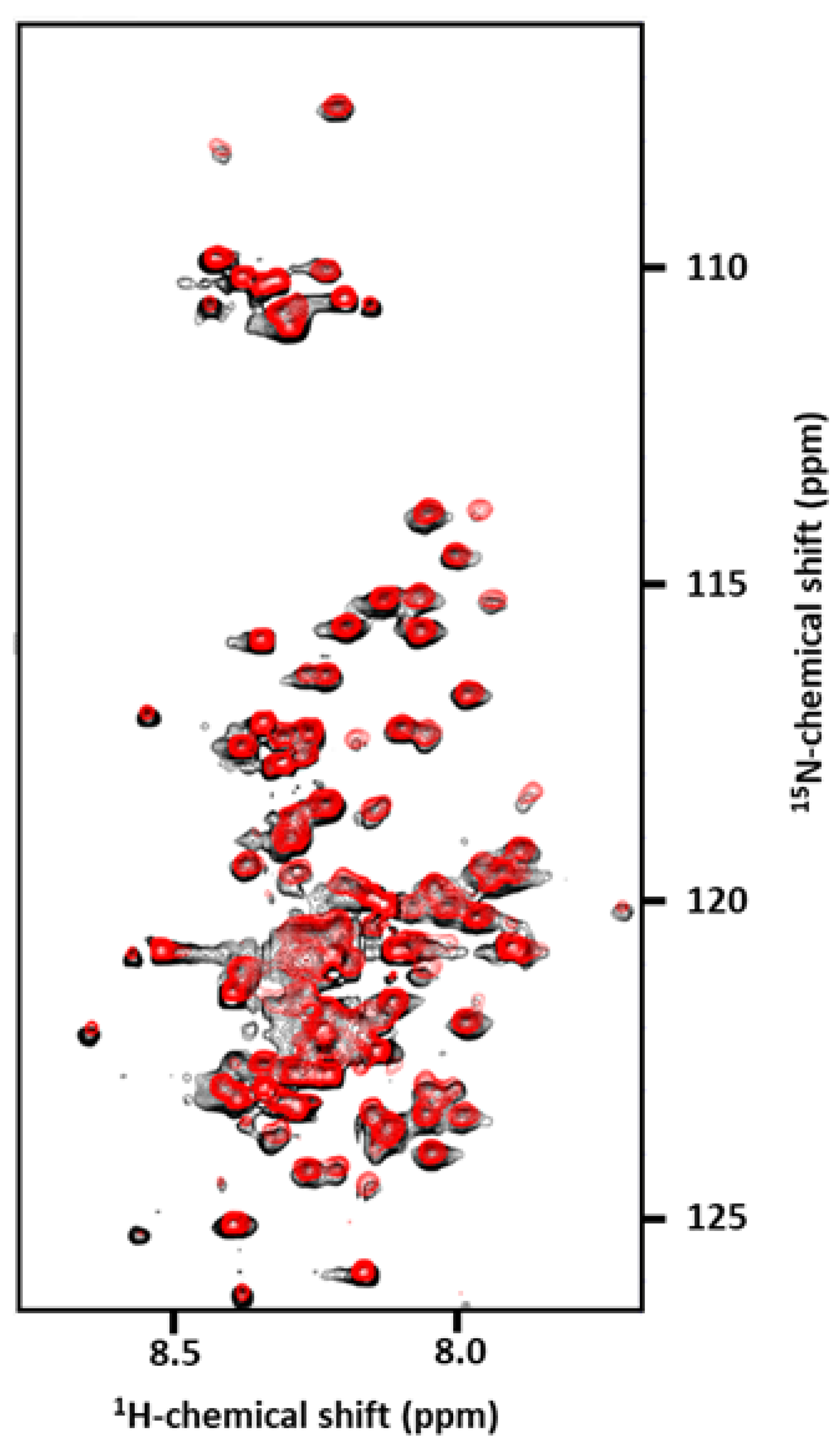
3.4. Effect of the D313K Mutation on NMR Spectra
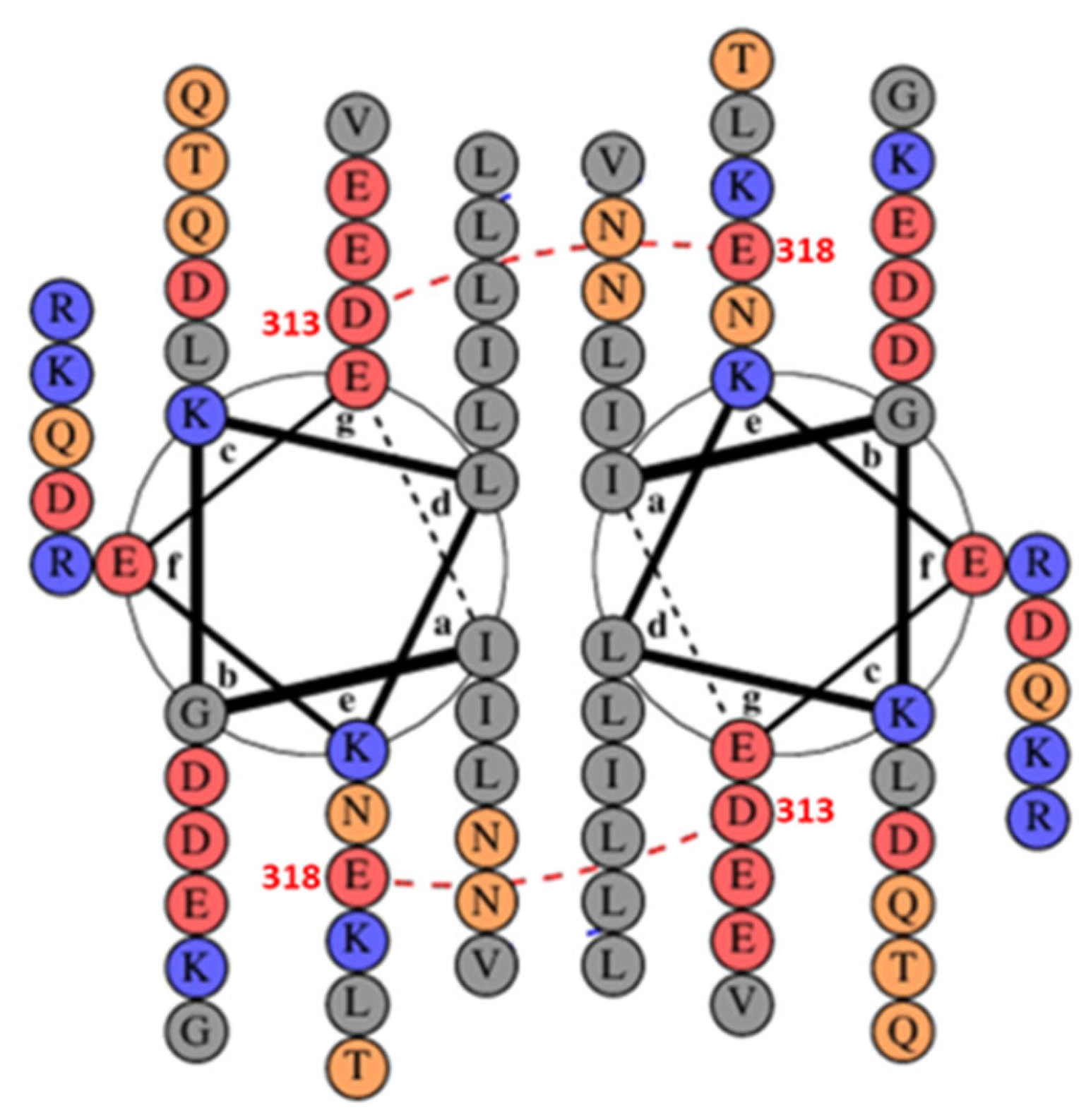
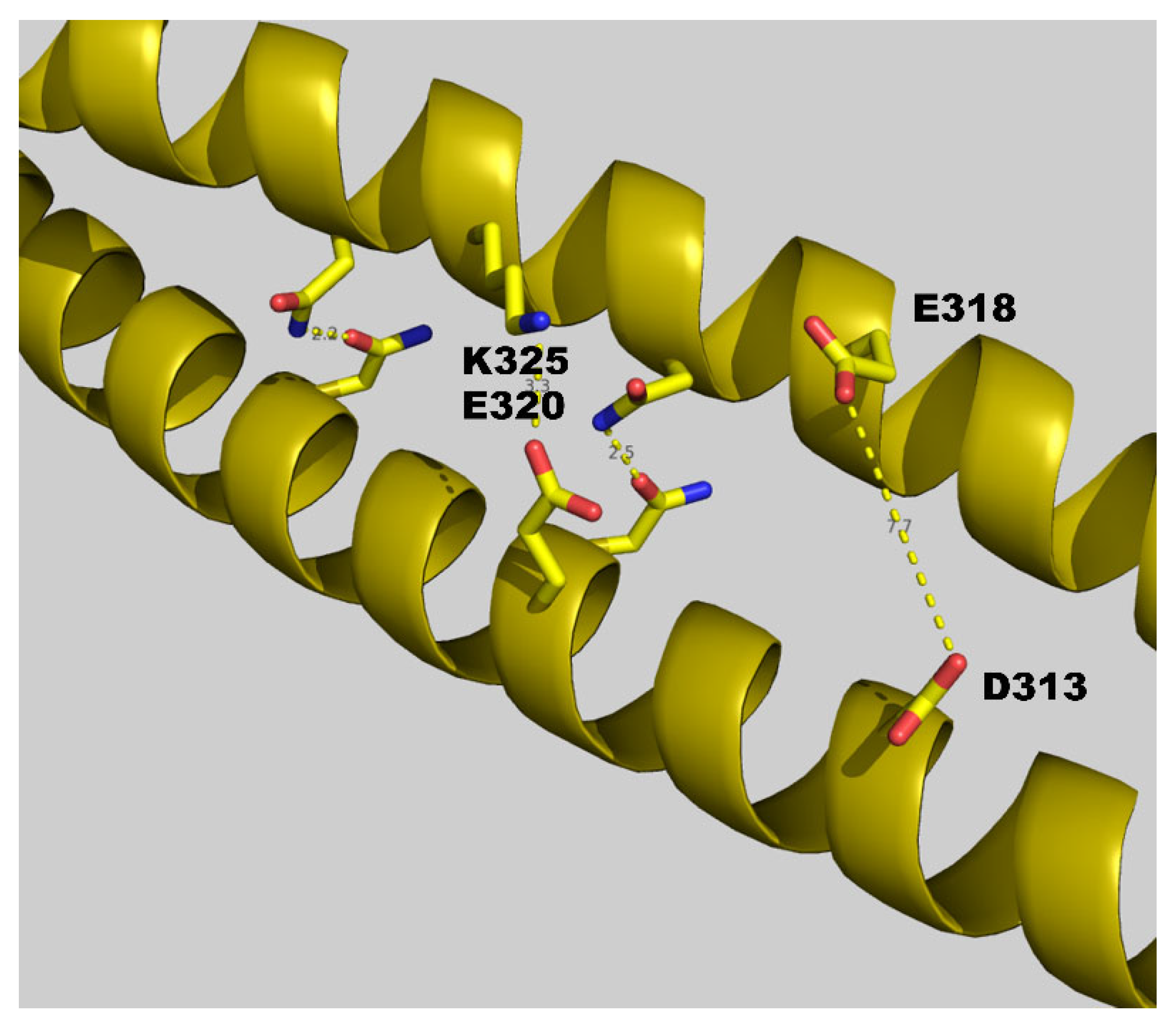
4. Discussion
Influence of Salts on D313K vs. WT
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liberles, D.A.; Teichmann, S.A.; Bahar, I.; Bastolla, U.; Bloom, J.; Bornberg-Bauer, E.; Colwell, L.J.; de Koning, A.P.J.; Dokholyan, N.V.; Echave, J.; et al. The interface of protein structure, protein biophysics, and molecular evolution. Protein Sci. 2012, 21, 769–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivedi, V.D. Protein structure–function exploration initiative in undergraduate biochemistry and independent research courses. Biochem. Mol. Biol. Educ. 2022, 50, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Sugase, K.; Dyson, H.J.; Wright, P.E. Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc. Natl. Acad. Sci. USA 2015, 112, 9614–9619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manukian, S.; Lindberg, G.E.; Punch, E.; Mudiyanselage, S.P.D.; Gage, M.J. pH-Dependent Compaction of the Intrinsically Disordered Poly-E Motif in Titin. Biology 2022, 11, 1302. [Google Scholar] [CrossRef]
- Lindsay, R.J.; Mansbach, R.A.; Gnanakaran, S.; Shen, T. Effects of pH on an IDP conformational ensemble explored by molecular dynamics simulation. Biophys. Chem. 2021, 271, 106552. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Guo, Y.; Ning, S.; Duan, M. Phosphorylation regulates the binding of intrinsically disordered proteins via a flexible conformation selection mechanism. Commun. Chem. 2020, 3, 123. [Google Scholar] [CrossRef]
- Wicky, B.I.M.; Shammas, S.L.; Clarke, J. Affinity of IDPs to their targets is modulated by ion-specific changes in kinetics and residual structure. Proc. Natl. Acad. Sci. USA 2017, 114, 9882–9887. [Google Scholar] [CrossRef] [Green Version]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun. Signal. 2022, 20, 20. [Google Scholar] [CrossRef]
- Cheng, Y.; LeGall, T.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry 2006, 45, 10448–10460. [Google Scholar] [CrossRef]
- Du, Z.; Uversky, V.N. A Comprehensive Survey of the Roles of Highly Disordered Proteins in Type 2 Diabetes. Int. J. Mol. Sci. 2017, 18, 2010. [Google Scholar] [CrossRef] [Green Version]
- Mészáros, B.; Hajdu-Soltész, B.; Zeke, A.; Dosztányi, Z. Mutations of Intrinsically Disordered Protein Regions Can Drive Cancer but Lack Therapeutic Strategies. Biomolecules 2021, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, A.H.S.; Lopes, F.C.; John, E.B.O.; Carlini, C.R.; Ligabue-Braun, R. Modulation of Disordered Proteins with a Focus on Neurodegenerative Diseases and Other Pathologies. Int. J. Mol. Sci. 2019, 20, 1322. [Google Scholar] [CrossRef] [Green Version]
- Libich, D.S.; Schwalbe, M.; Kate, S.; Venugopal, H.; Claridge, J.K.; Edwards, P.J.; Dutta, K.; Pascal, S.M. Intrinsic disorder and coiled-coil formation in prostate apoptosis response factor 4. FEBS J. 2009, 276, 3710–3728. [Google Scholar] [CrossRef]
- Ward, J.J.; McGuffin, L.J.; Bryson, K.; Buxton, B.F.; Jones, D.T. The DISOPRED server for the prediction of protein disorder. Bioinformatics 2004, 20, 2138–2139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.T.; Cozzetto, D. DISOPRED3: Precise disordered region predictions with annotated protein-binding activity. Bioinformatics 2015, 31, 857–863. [Google Scholar] [CrossRef] [Green Version]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: An overview. Cell Res. 2009, 19, 929–949. [Google Scholar] [CrossRef] [Green Version]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef] [Green Version]
- Burikhanov, R.; Zhao, Y.; Goswami, A.; Qiu, S.; Schwarze, S.R.; Rangnekar, V.M. The tumor suppressor Par-4 activates an extrinsic pathway for apoptosis. Cell 2009, 138, 377–388. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Marcos, P.J.; Abu-Baker, S.; Joshi, J.; Galvez, A.; Castilla, E.A.; Cañamero, M.; Collado, M.; Saez, C.; Moreno-Bueno, G.; Palacios, J.; et al. Simultaneous inactivation of Par-4 and PTEN in vivo leads to synergistic NF-κB activation and invasive prostate carcinoma. Proc. Natl. Acad. Sci. USA 2009, 106, 12962–12967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, J.V.; Pan, T.C.; Ruth, J.; Feng, Y.; Zhou, A.; Pant, D.; Grimley, J.S.; Wandless, T.J.; DeMichele, A.; Chodosh, L.A. Par-4 Downregulation Promotes Breast Cancer Recurrence by Preventing Multinucleation following Targeted Therapy. Cancer Cell 2013, 24, 30–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelke, G.V.; Jagtap, J.C.; Kim, D.-K.; Shah, R.D.; Das, G.; Shivayogi, M.; Pujari, R.; Shastry, P. TNF-α and IFN-γ Together Up-Regulates Par-4 Expression and Induce Apoptosis in Human Neuroblastomas. Biomedicines 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cook, J.; Krishnan, S.; Ananth, S.; Sells, S.F.; Shi, Y.; Walther, M.M.; Linehan, W.M.; Sukhatme, V.P.; Weinstein, M.H.; Rangnekar, V.M. Decreased expression of the pro-apoptotic protein Par-4 in renal cell carcinoma. Oncogene 1999, 18, 1205–1208. [Google Scholar] [CrossRef] [Green Version]
- Guo, Q.; Fu, W.; Xie, J.; Luo, H.; Sells, S.F.; Geddes, J.W.; Bondada, V.; Rangnekar, V.M.; Mattson, M.P. Par-4 is a mediator of neuronal degeneration associated with the pathogenesis of Alzheimer disease. Nat. Med. 1998, 4, 957–962. [Google Scholar] [CrossRef]
- El-Guendy, N.; Rangnekar, V.M. Apoptosis by Par-4 in cancer and neurodegenerative diseases. Exp. Cell Res. 2003, 283, 51–66. [Google Scholar] [CrossRef]
- Xie, J.; Guo, Q. PAR-4 is involved in regulation of beta-secretase cleavage of the Alzheimer amyloid precursor protein. J. Biol. Chem. 2005, 280, 13824–13832. [Google Scholar] [CrossRef] [Green Version]
- El-Guendy, N.; Zhao, Y.; Gurumurthy, S.; Burikhanov, R.; Rangnekar, V.M. Identification of a Unique Core Domain of Par-4 Sufficient for Selective Apoptosis Induction in Cancer Cells. Mol. Cell. Biol. 2003, 23, 5516–5525. [Google Scholar] [CrossRef] [Green Version]
- Shrestha-Bhattarai, T.; Rangnekar, V.M. Cancer-selective apoptotic effects of extracellular and intracellular Par-4. Oncogene 2010, 29, 3873–3880. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, S.; Goswami, A.; Vasudevan, K.M.; Rangnekar, V.M. Phosphorylation of Par-4 by protein kinase A is critical for apoptosis. Mol. Cell Biol. 2005, 25, 1146–1161. [Google Scholar] [CrossRef] [Green Version]
- Chaudhry, P.; Singh, M.; Parent, S.; Asselin, E. Prostate apoptosis response 4 (Par-4), a novel substrate of caspase-3 during apoptosis activation. Mol. Cell. Biol. 2012, 32, 826–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camandola, S.; Mattson, M.P. Pro-apoptotic action of PAR-4 involves inhibition of NF-kappaB activity and suppression of BCL-2 expression. J. Neurosci. Res. 2000, 61, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Ahmed, M.; Sells, S.F.; Mohiuddin, M.; Weinstein, M.H.; Rangnekar, V.M. Mutually exclusive expression patterns of Bcl-2 and Par-4 in human prostate tumors consistent with down-regulation of Bcl-2 by Par-4. Oncogene 1999, 18, 623–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheema, S.K.; Mishra, S.K.; Rangnekar, V.M.; Tari, A.M.; Kumar, R.; Lopez-Berestein, G. Par-4 transcriptionally regulates Bcl-2 through a WT1-binding site on the bcl-2 promoter. J. Biol. Chem. 2003, 278, 19995–20005. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Sahasrabuddhe, A.A.; Szankasi, P.; Chung, F.; Basrur, V.; Rangnekar, V.M.; Pagano, M.; Lim, M.S.; Elenitoba-Johnson, K.S. Fbxo45-mediated degradation of the tumor-suppressor Par-4 regulates cancer cell survival. Cell Death Differ. 2014, 21, 1535–1545. [Google Scholar] [CrossRef] [Green Version]
- Cheratta, A.R.; Thayyullathil, F.; Pallichankandy, S.; Subburayan, K.; Alakkal, A.; Galadari, S. Prostate apoptosis response-4 and tumor suppression: It’s not just about apoptosis anymore. Cell Death Dis. 2021, 12, 47. [Google Scholar] [CrossRef]
- Clark, A.M.; Ponniah, K.; Warden, M.S.; Raitt, E.M.; Yawn, A.C.; Pascal, S.M. pH-Induced Folding of the Caspase-Cleaved Par-4 Tumor Suppressor: Evidence of Structure Outside of the Coiled Coil Domain. Biomolecules 2018, 8, 162. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.M.; Ponniah, K.; Warden, M.S.; Raitt, E.M.; Smith, B.G.; Pascal, S.M. Tetramer formation by the caspase-activated fragment of the Par-4 tumor suppressor. FEBS J. 2019, 286, 4060–4073. [Google Scholar] [CrossRef]
- Raut, K.K.; Ponniah, K.; Pascal, S.M. Structural Analysis of the cl-Par-4 Tumor Suppressor as a Function of Ionic Environment. Biomolecules 2021, 11, 386. [Google Scholar] [CrossRef]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of intrinsically disordered regions and proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Dutta, K.; Engler, F.A.; Cotton, L.; Alexandrov, A.; Bedi, G.S.; Colquhoun, J.; Pascal, S.M. Stabilization of a pH-sensitive apoptosis-linked coiled coil through single point mutations. Protein Sci. 2003, 12, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Dutta, K.; Alexandrov, A.; Huang, H.; Pascal, S.M. pH-induced folding of an apoptotic coiled coil. Protein Sci. 2001, 10, 2531–2540. [Google Scholar] [CrossRef]
- Edelheit, O.; Hanukoglu, A.; Hanukoglu, I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, A.; Dutta, K.; Pascal, S.M. MBP fusion protein with a viral protease cleavage site: One-step cleavage/purification of insoluble proteins. Biotechniques 2001, 30, 1194–1198. [Google Scholar] [CrossRef] [Green Version]
- Skinner, S.P.; Fogh, R.H.; Boucher, W.; Ragan, T.J.; Mureddu, L.G.; Vuister, G.W. CcpNmr AnalysisAssign: A flexible platform for integrated NMR analysis. J. Biomol. NMR 2016, 66, 111–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wuo, M.G.; Mahon, A.B.; Arora, P.S. An Effective Strategy for Stabilizing Minimal Coiled Coil Mimetics. J. Am. Chem. Soc. 2015, 137, 11618–11621. [Google Scholar] [CrossRef] [PubMed]
- Thayyullathil, F.; Pallichankandy, S.; Rahman, A.; Kizhakkayil, J.; Chathoth, S.; Patel, M.; Galadari, S. Caspase-3 mediated release of SAC domain containing fragment from Par-4 is necessary for the sphingosine-induced apoptosis in Jurkat cells. J. Mol. Signal. 2013, 8, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ko, J.; Park, H.; Heo, L.; Seok, C. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012, 40, W294–W297. [Google Scholar] [CrossRef]
- Tiruttani Subhramanyam, U.K.; Kubicek, J.; Eidhoff, U.B.; Labahn, J. Structural basis for the regulatory interactions of proapoptotic Par-4. Cell Death Differ. 2017, 24, 1540–1547. [Google Scholar] [CrossRef] [Green Version]
- Felicori, L.; Jameson, K.H.; Roblin, P.; Fogg, M.J.; Garcia-Garcia, T.; Ventroux, M.; Cherrier, M.V.; Bazin, A.; Noirot, P.; Wilkinson, A.J.; et al. Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners. Nucleic Acids Res. 2016, 44, 449–463. [Google Scholar] [CrossRef] [Green Version]
- Arakawa, T.; Tokunaga, M.; Kita, Y.; Niikura, T.; Baker, R.W.; Reimer, J.M.; Leschziner, A.E. Structure Analysis of Proteins and Peptides by Difference Circular Dichroism Spectroscopy. Protein J. 2021, 40, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Micsonai, A.; Wien, F.; Kernya, L.; Lee, Y.-H.; Goto, Y.; Réfrégiers, M.; Kardos, J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA 2015, 112, E3095–E3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [Green Version]
- March, D.; Bianco, V.; Franzese, G. Protein Unfolding and Aggregation near a Hydrophobic Interface. Polymers 2021, 13, 156. [Google Scholar] [CrossRef]
- Routledge, K.E.; Tartaglia, G.G.; Platt, G.W.; Vendruscolo, M.; Radford, S.E. Competition between intramolecular and intermolecular interactions in an amyloid-forming protein. J. Mol. Biol. 2009, 389, 776–786. [Google Scholar] [CrossRef] [Green Version]
- Fersht, A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding; Macmillan: New York, NY, USA, 1999. [Google Scholar]
- Razvi, A.; Scholtz, J.M. Lessons in stability from thermophilic proteins. Protein Sci. 2006, 15, 1569–1578. [Google Scholar] [CrossRef] [Green Version]
- Johnstone, R.W.; See, R.H.; Sells, S.F.; Wang, J.; Muthukkumar, S.; Englert, C.; Haber, D.A.; Licht, J.D.; Sugrue, S.P.; Roberts, T.; et al. A novel repressor, par-4, modulates transcription and growth suppression functions of the Wilms’ tumor suppressor WT1. Mol. Cell. Biol. 1996, 16, 6945–6956. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, S.; Raut, K.K.; Clark, A.M.; Baudin, A.; Djemri, L.; Libich, D.S.; Ponniah, K.; Pascal, S.M. Enhancing the Conformational Stability of the cl-Par-4 Tumor Suppressor via Site-Directed Mutagenesis. Biomolecules 2023, 13, 667. https://doi.org/10.3390/biom13040667
Pandey S, Raut KK, Clark AM, Baudin A, Djemri L, Libich DS, Ponniah K, Pascal SM. Enhancing the Conformational Stability of the cl-Par-4 Tumor Suppressor via Site-Directed Mutagenesis. Biomolecules. 2023; 13(4):667. https://doi.org/10.3390/biom13040667
Chicago/Turabian StylePandey, Samjhana, Krishna K. Raut, Andrea M. Clark, Antoine Baudin, Lamya Djemri, David S. Libich, Komala Ponniah, and Steven M. Pascal. 2023. "Enhancing the Conformational Stability of the cl-Par-4 Tumor Suppressor via Site-Directed Mutagenesis" Biomolecules 13, no. 4: 667. https://doi.org/10.3390/biom13040667





