Single-Cell RNA Sequencing Analysis of Gene Regulatory Network Changes in the Development of Lung Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
2.2. Data Integration, Unsupervised Dimensional Reduction and Clustering, and Cell Type Identification
2.3. Identification of Malignant Cells from Patients with Adenocarcinoma of the Lung
2.4. Functional Enrichment Analysis
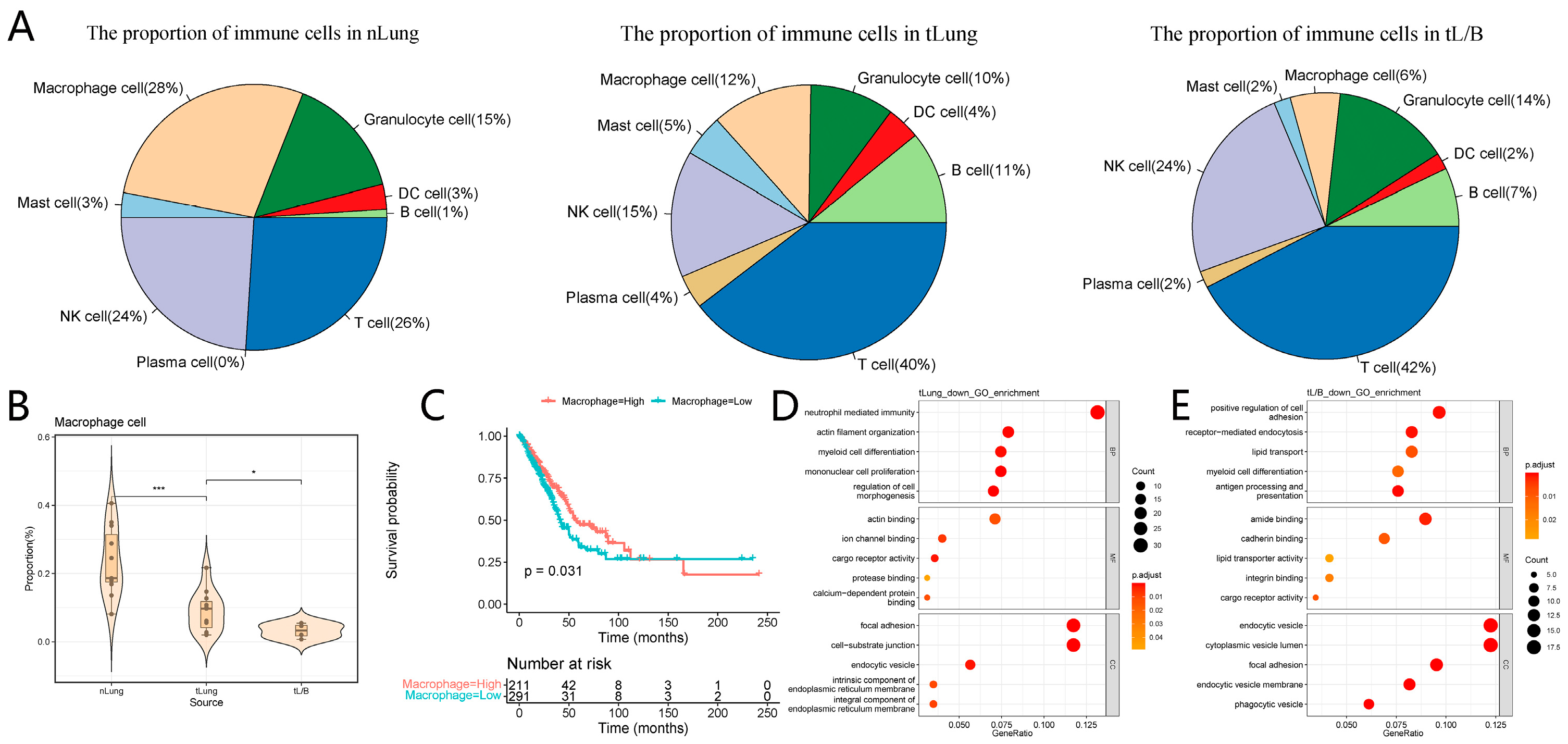
2.5. Survival Analysis of the Proportion of Macrophages in Patients with Lung Adenocarcinoma
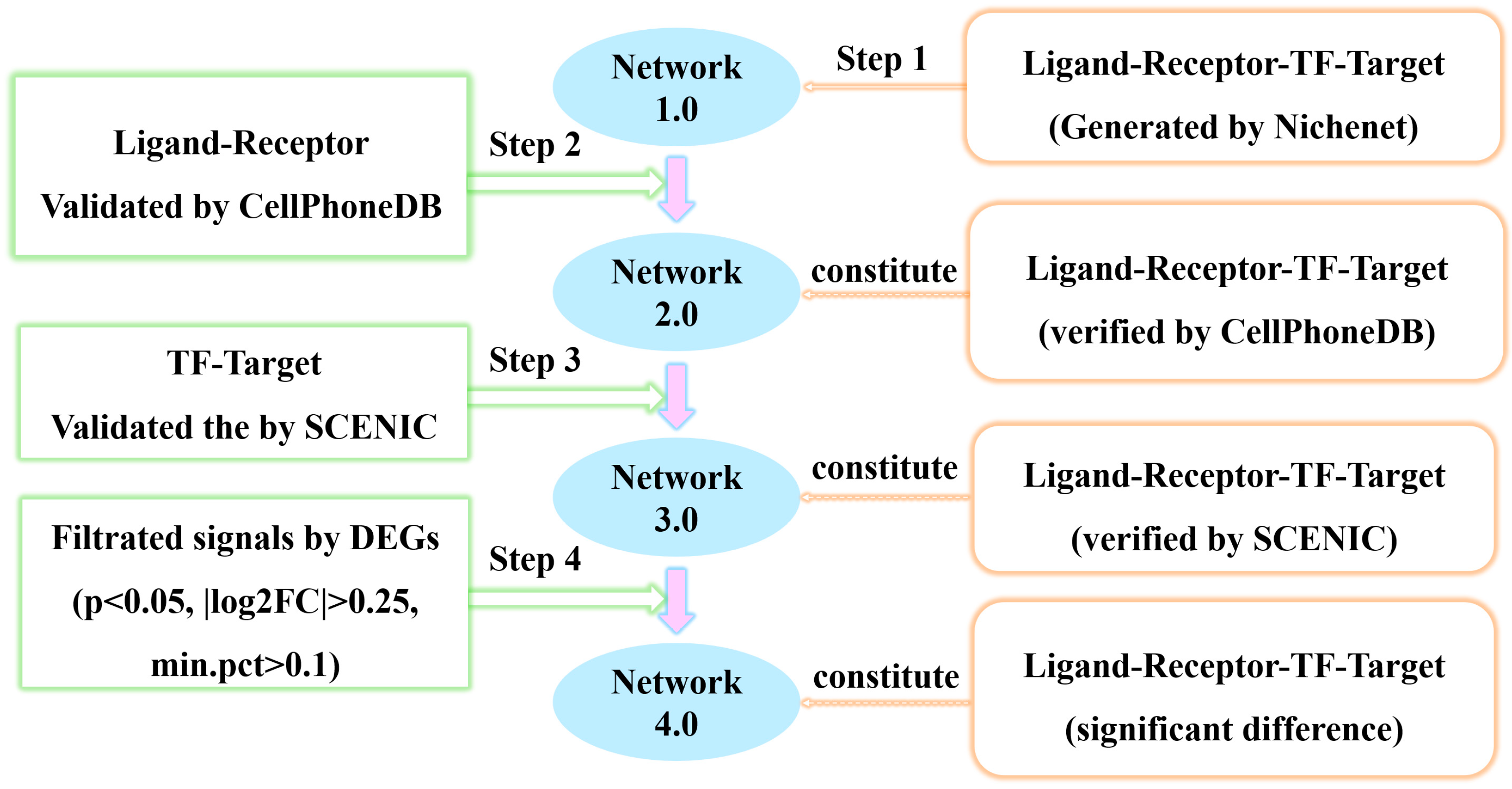
2.6. The Gene Regulatory Network of Tumor Cell-Macrophage Interaction Construction
2.7. Immune Cell Trajectory Is Constructed by Regulating the Signaling Molecules of the Network
3. Results
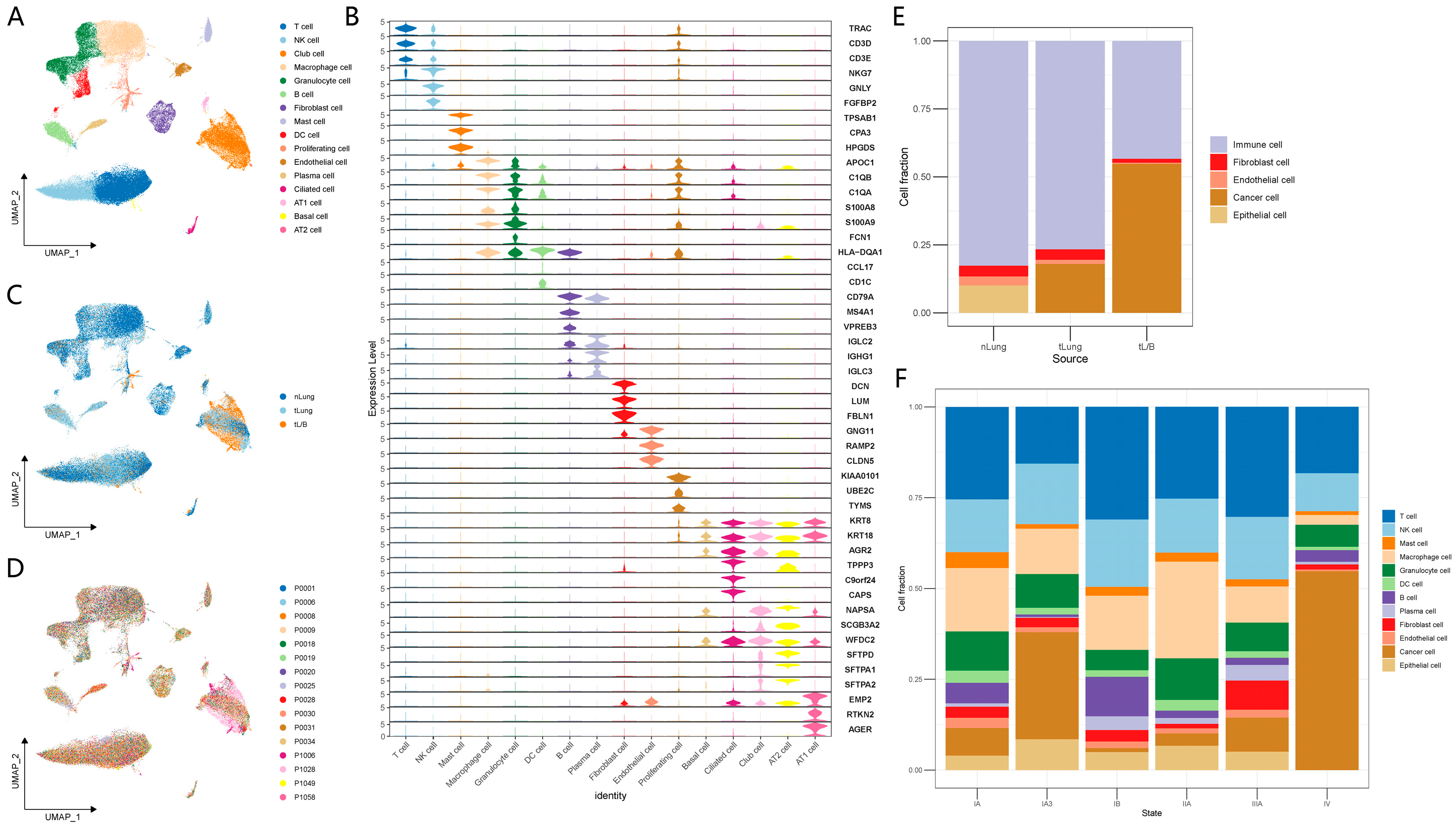
3.1. Single-Cell Expression Atlas and Cell Typing in Normal Lung, Early and Advanced LUAD Tissue Samples
3.2. The Role of Macrophages in the Immune Microenvironment
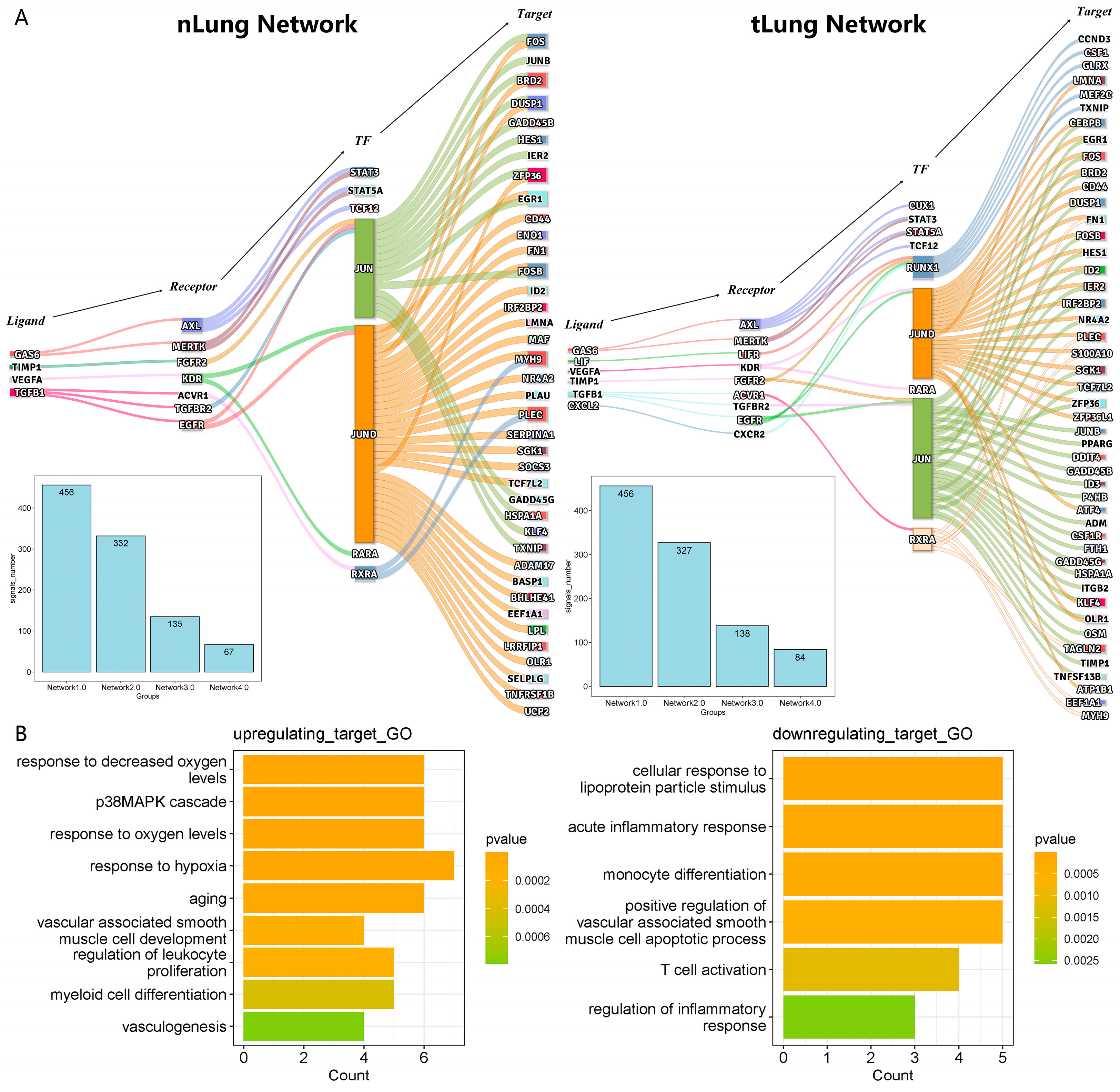
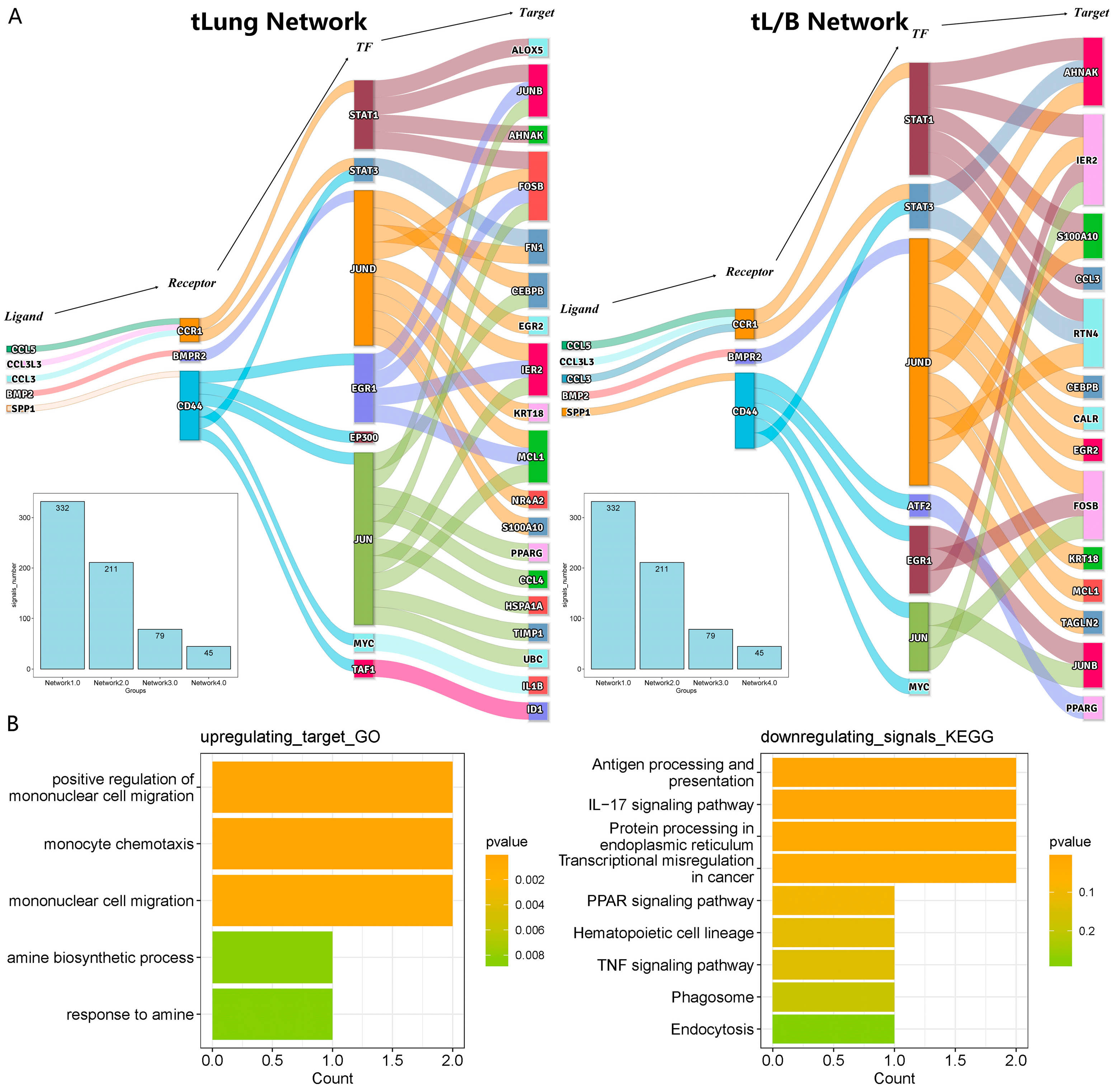
3.3. Construction of the Regulatory Network of Macrophages Regulated by Tumor Cells
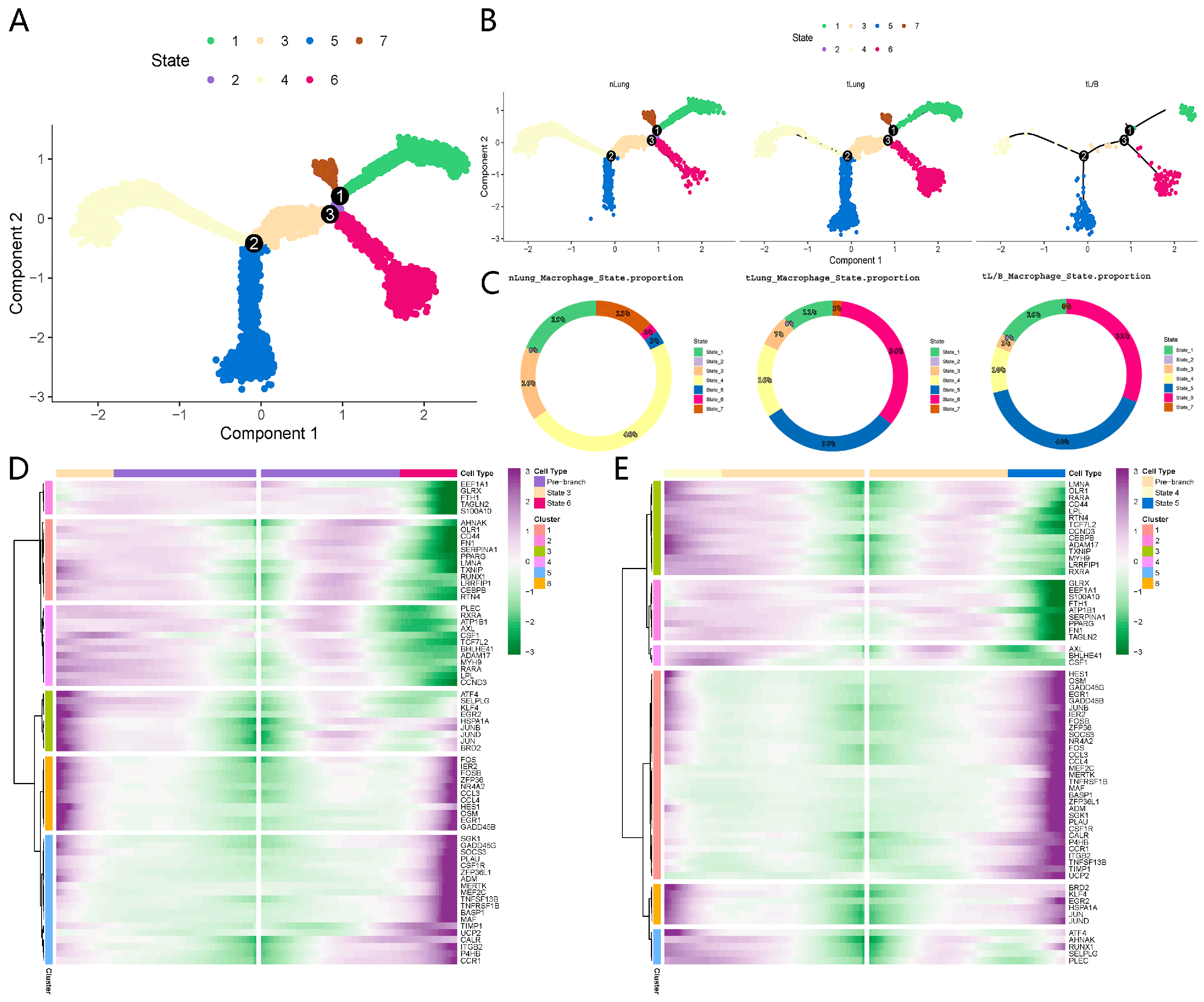
3.4. The Signal Changes in the Gene Regulatory Network Are Related to the Change in Macrophage State
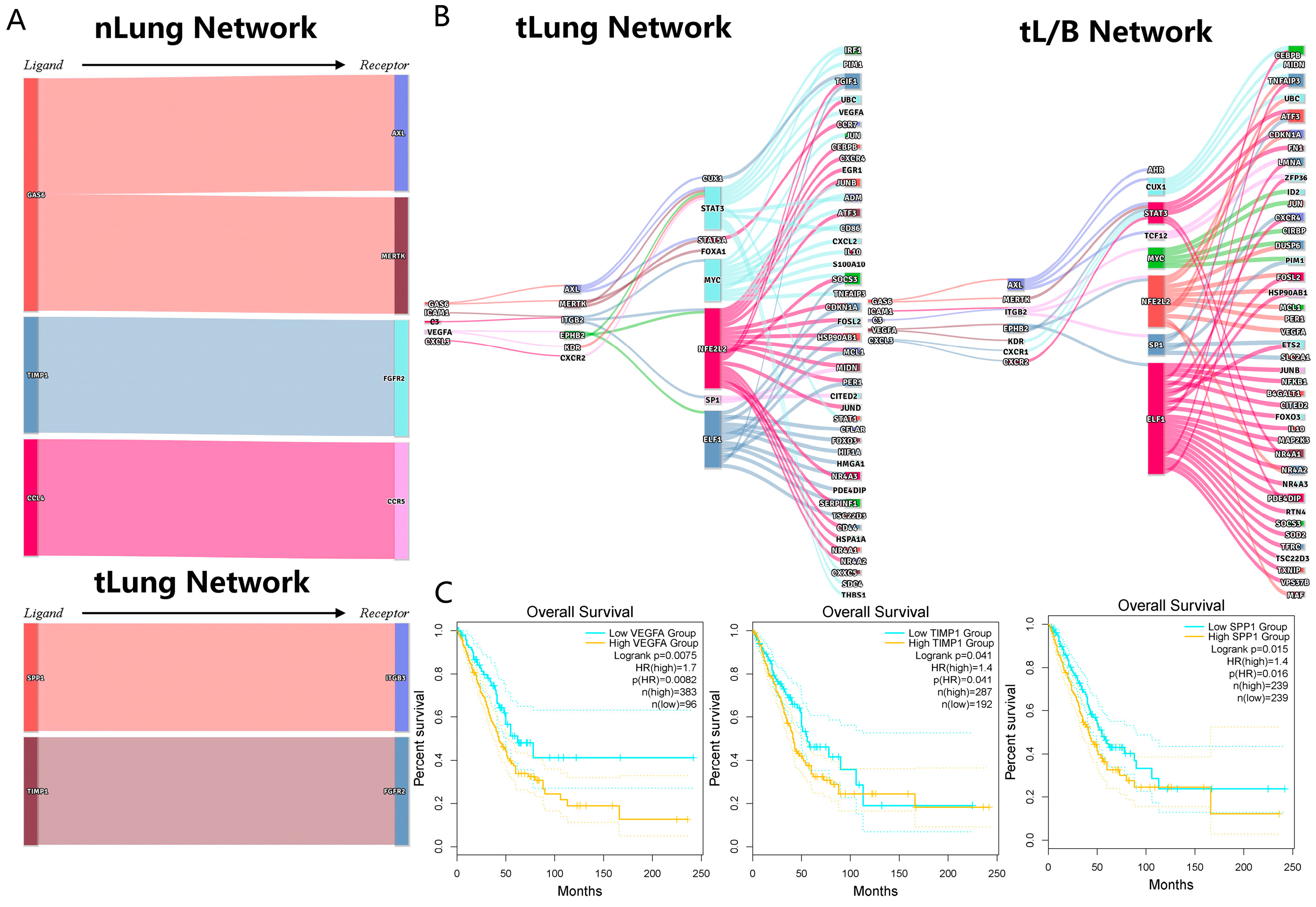
3.5. The Independent Validation Set of Lung Adenocarcinoma (nLung, tLung and tL/B) Was Used to Verify the Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seguin, L.; Durandy, M.; Feral, C.C. Lung Adenocarcinoma Tumor Origin: A Guide for Personalized Medicine. Cancers 2022, 14, 1759. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung Cancer Statistics. Adv. Exp. Med. Biol. 2016, 893, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.Y.; Huang, J.Y.; Chen, H.C.; Lin, C.H.; Lin, S.H.; Hung, W.H.; Cheng, Y.F. The comparison between adenocarcinoma and squamous cell carcinoma in lung cancer patients. J. Cancer Res. Clin. Oncol. 2020, 146, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Y.; Wang, C.; Yang, Y.; Ni, Y.; Wang, Z.; Song, T.; Yao, M.; Liu, Z.; Chao, N.; et al. Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal Transduct. Target. Ther. 2022, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, Q.; Song, T.; Wang, Z.; Song, L.; Yang, Y.; Shao, J.; Li, J.; Ni, Y.; Chao, N.; et al. The heterogeneous immune landscape between lung adenocarcinoma and squamous carcinoma revealed by single-cell RNA sequencing. Signal Transduct. Target. Ther. 2022, 7, 289. [Google Scholar] [CrossRef]
- Khullar, O.V.; Liu, Y.; Gillespie, T.; Higgins, K.A.; Ramalingam, S.; Lipscomb, J.; Fernandez, F.G. Survival After Sublobar Resection versus Lobectomy for Clinical Stage IA Lung Cancer: An Analysis from the National Cancer Data Base. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2015, 10, 1625–1633. [Google Scholar] [CrossRef]
- Hanna, N.; Johnson, D.; Temin, S.; Baker, S., Jr.; Brahmer, J.; Ellis, P.M.; Giaccone, G.; Hesketh, P.J.; Jaiyesimi, I.; Leighl, N.B.; et al. Systemic Therapy for Stage IV Non-Small-Cell Lung Cancer: American Society of Clinical Oncology Clinical Practice Guideline Update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 3484–3515. [Google Scholar] [CrossRef]
- Friemel, J.; Frick, L.; Weber, A. Intratumor heterogeneity in HCC. Aging 2015, 7, 350–351. [Google Scholar] [CrossRef]
- Navin, N.; Krasnitz, A.; Rodgers, L.; Cook, K.; Meth, J.; Kendall, J.; Riggs, M.; Eberling, Y.; Troge, J.; Grubor, V.; et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010, 20, 68–80. [Google Scholar] [CrossRef]
- Cohen, M.; Giladi, A.; Gorki, A.D.; Solodkin, D.G.; Zada, M.; Hladik, A.; Miklosi, A.; Salame, T.M.; Halpern, K.B.; David, E.; et al. Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 2018, 175, 1031–1044.e1018. [Google Scholar] [CrossRef]
- Boisset, J.C.; Vivié, J.; Grün, D.; Muraro, M.J.; Lyubimova, A.; van Oudenaarden, A. Mapping the physical network of cellular interactions. Nat. Methods 2018, 15, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Wang, W.; Laurenti, E.; Turinsky, A.L.; Wodak, S.J.; Bader, G.D.; Dick, J.E.; Zandstra, P.W. Intercellular network structure and regulatory motifs in the human hematopoietic system. Mol. Syst. Biol. 2014, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Paik, D.T.; Tian, L.; Lee, J.; Sayed, N.; Chen, I.Y.; Rhee, S.; Rhee, J.W.; Kim, Y.; Wirka, R.C.; Buikema, J.W.; et al. Large-Scale Single-Cell RNA-Seq Reveals Molecular Signatures of Heterogeneous Populations of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Circ. Res. 2018, 123, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.P.; Du, J.; Lagoudas, G.; Jiao, Y.; Sawyer, A.; Drummond, D.C.; Lauffenburger, D.A.; Raue, A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018, 25, 1458–1468.e1454. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Martin, J.C.; Chang, C.; Boschetti, G.; Ungaro, R.; Giri, M.; Grout, J.A.; Gettler, K.; Chuang, L.S.; Nayar, S.; Greenstein, A.J.; et al. Single-Cell Analysis of Crohn’s Disease Lesions Identifies a Pathogenic Cellular Module Associated with Resistance to Anti-TNF Therapy. Cell 2019, 178, 1493–1508.e1420. [Google Scholar] [CrossRef]
- AlMusawi, S.; Ahmed, M.; Nateri, A.S. Understanding cell-cell communication and signaling in the colorectal cancer microenvironment. Clin. Transl. Med. 2021, 11, e308. [Google Scholar] [CrossRef]
- Hinshaw, D.C.; Shevde, L.A. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Chang, R.B.; Beatty, G.L. The interplay between innate and adaptive immunity in cancer shapes the productivity of cancer immunosurveillance. J. Leukoc. Biol. 2020, 108, 363–376. [Google Scholar] [CrossRef]
- Wei, K.; Ma, Z.; Yang, F.; Zhao, X.; Jiang, W.; Pan, C.; Li, Z.; Pan, X.; He, Z.; Xu, J.; et al. M2 macrophage-derived exosomes promote lung adenocarcinoma progression by delivering miR-942. Cancer Lett. 2022, 526, 205–216. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, J.; Xu, Q.; Foley, R.; Guo, J.; Zhang, X.; Tian, C.; Mu, M.; Xing, Y.; Liu, Y.; et al. Identification of Key Genes Driving Tumor Associated Macrophage Migration and Polarization Based on Immune Fingerprints of Lung Adenocarcinoma. Front. Cell Dev. Biol. 2021, 9, 751800. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, P.; Trinks, A.; Obermayer, B.; Pett, J.P.; Wiederspahn, J.; Uhlitz, F.; Liang, X.; Lehmann, A.; Jurmeister, P.; Elsner, A.; et al. Single-cell RNA sequencing reveals distinct tumor microenvironmental patterns in lung adenocarcinoma. Oncogene 2021, 40, 6748–6758. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Z.; Zhou, K.; Wang, C.; Jiang, L.; Zhang, L.; Yang, Y.; Luo, W.; Qiao, W.; Wang, G.; et al. Deciphering cell lineage specification of human lung adenocarcinoma with single-cell RNA sequencing. Nat. Commun. 2021, 12, 6500. [Google Scholar] [CrossRef] [PubMed]
- Sinjab, A.; Han, G.; Treekitkarnmongkol, W.; Hara, K.; Brennan, P.M.; Dang, M.; Hao, D.; Wang, R.; Dai, E.; Dejima, H.; et al. Resolving the Spatial and Cellular Architecture of Lung Adenocarcinoma by Multiregion Single-Cell Sequencing. Cancer Discov. 2021, 11, 2506–2523. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, Y.; Hu, Z.; Zhao, M.; Li, M.; Bi, G.; Zheng, Y.; Liang, J.; Lu, T.; Jiang, W.; et al. Landscape and dynamics of single tumor and immune cells in early and advanced-stage lung adenocarcinoma. Clin. Transl. Med. 2021, 11, e350. [Google Scholar] [CrossRef]
- Kim, N.; Kim, H.K.; Lee, K.; Hong, Y.; Cho, J.H.; Choi, J.W.; Lee, J.I.; Suh, Y.L.; Ku, B.M.; Eum, H.H.; et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020, 11, 2285. [Google Scholar] [CrossRef]
- Laughney, A.M.; Hu, J.; Campbell, N.R.; Bakhoum, S.F.; Setty, M.; Lavallée, V.P.; Xie, Y.; Masilionis, I.; Carr, A.J.; Kottapalli, S.; et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 2020, 26, 259–269. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Y.; Hao, Y.; Li, X.; Qi, Y.; Xin, M.; Xiao, Q.; Wang, P. Characterizing the Metabolic and Immune Landscape of Non-small Cell Lung Cancer Reveals Prognostic Biomarkers Through Omics Data Integration. Front. Cell Dev. Biol. 2021, 9, 702112. [Google Scholar] [CrossRef]
- Wu, F.; Fan, J.; He, Y.; Xiong, A.; Yu, J.; Li, Y.; Zhang, Y.; Zhao, W.; Zhou, F.; Li, W.; et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 2021, 12, 2540. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lan, Y.; Xu, J.; Quan, F.; Zhao, E.; Deng, C.; Luo, T.; Xu, L.; Liao, G.; Yan, M.; et al. CellMarker: A manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019, 47, D721–D728. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Bowers, R.R.; Delaney, J.R. Single-cell analysis of copy-number alterations in serous ovarian cancer reveals substantial heterogeneity in both low- and high-grade tumors. Cell Cycle (Georget. Tex.) 2020, 19, 3154–3166. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef]
- George, B.; Seals, S.; Aban, I. Survival analysis and regression models. J. Nucl. Cardiol. Off. Publ. Am. Soc. Nucl. Cardiol. 2014, 21, 686–694. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Efremova, M.; Vento-Tormo, M.; Teichmann, S.A.; Vento-Tormo, R. CellPhoneDB: Inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020, 15, 1484–1506. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Mishra, B.; Athar, M.; Mukhtar, S. Inference of Gene Regulatory Network from Single-Cell Transcriptomic Data Using pySCENIC. Methods Mol. Biol. 2021, 2328, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, D.; Ding, Q.; Zhu, X.; Zhu, R.; Chen, Y. Progress in Single-cell RNA Sequencing of Lung Adenocarcinoma. Zhongguo Fei Ai Za Zhi = Chin. J. Lung Cancer 2021, 24, 434–440. [Google Scholar] [CrossRef]
- Kurtenbach, S.; Cruz, A.M.; Rodriguez, D.A.; Durante, M.A.; Harbour, J.W. Uphyloplot2: Visualizing phylogenetic trees from single-cell RNA-seq data. BMC Genom. 2021, 22, 419. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, G.; Huang, L.; Zhang, C.; Sheng, Y.; Wu, B.; Han, L.; Wu, C.; Dong, B.; Qi, Y. Single-cell transcriptome analysis demonstrates inter-patient and intra-tumor heterogeneity in primary and metastatic lung adenocarcinoma. Aging 2020, 12, 21559–21581. [Google Scholar] [CrossRef]
- Sheu, K.M.; Hoffmann, A. Functional Hallmarks of Healthy Macrophage Responses: Their Regulatory Basis and Disease Relevance. Annu. Rev. Immunol. 2022, 40, 295–321. [Google Scholar] [CrossRef]
- Zhao, R.; Cao, J.; Yang, X.; Zhang, Q.; Iqbal, M.Z.; Lu, J.; Kong, X. Inorganic material based macrophage regulation for cancer therapy: Basic concepts and recent advances. Biomater. Sci. 2021, 9, 4568–4590. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, G.; Wang, L.; Zhang, J.; Li, W. Derivation of a Novel CIHI in Patients with Lung Adenocarcinoma for Estimating Tumor Microenvironment and Clinical Prognosis. Dis. Mrk. 2021, 2021, 4495489. [Google Scholar] [CrossRef]
- Dong, Y.; Arif, A.A.; Guo, J.; Ha, Z.; Lee-Sayer, S.S.M.; Poon, G.F.T.; Dosanjh, M.; Roskelley, C.D.; Huan, T.; Johnson, P. CD44 Loss Disrupts Lung Lipid Surfactant Homeostasis and Exacerbates Oxidized Lipid-Induced Lung Inflammation. Front. Immunol. 2020, 11, 29. [Google Scholar] [CrossRef]
- Viswanadhapalli, S.; Dileep, K.V.; Zhang, K.Y.J.; Nair, H.B.; Vadlamudi, R.K. Targeting LIF/LIFR signaling in cancer. Genes Dis. 2022, 9, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Hargreaves, D.C. The alternative macrophage relay: STAT6 passes the baton to EGR2. Genes Dev. 2020, 34, 1407–1409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, Z.; Wu, D.; Chen, L.; Xie, L. Single-Cell RNA-Seq Analysis Reveals Microenvironmental Infiltration of Plasma Cells and Hepatocytic Prognostic Markers in HCC With Cirrhosis. Front. Oncol. 2020, 10, 596318. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Simińska, D.; Gąssowska-Dobrowolska, M.; Listos, J.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation through HIF-1 and NF-κB Activation: A Review of the Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 701. [Google Scholar] [CrossRef]
- Justo, B.L.; Jasiulionis, M.G. Characteristics of TIMP1, CD63, and β1-Integrin and the Functional Impact of Their Interaction in Cancer. Int. J. Mol. Sci. 2021, 22, 9319. [Google Scholar] [CrossRef]
- Tang, H.; Chen, J.; Han, X.; Feng, Y.; Wang, F. Upregulation of SPP1 Is a Marker for Poor Lung Cancer Prognosis and Contributes to Cancer Progression and Cisplatin Resistance. Front. Cell Dev. Biol. 2021, 9, 646390. [Google Scholar] [CrossRef]
- Lopez-Yrigoyen, M.; Cassetta, L.; Pollard, J.W. Macrophage targeting in cancer. Ann. N. Y. Acad. Sci. 2021, 1499, 18–41. [Google Scholar] [CrossRef]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Cha, Y.J.; Koo, J.S. Role of Tumor-Associated Myeloid Cells in Breast Cancer. Cells 2020, 9, 1785. [Google Scholar] [CrossRef]
- He, D.; Wang, D.; Lu, P.; Yang, N.; Xue, Z.; Zhu, X.; Zhang, P.; Fan, G. Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 2021, 40, 355–368. [Google Scholar] [CrossRef]
- Almet, A.A.; Cang, Z.; Jin, S.; Nie, Q. The landscape of cell-cell communication through single-cell transcriptomics. Curr. Opin. Syst. Biol. 2021, 26, 12–23. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, F.; Wang, J.; Hu, L.; Jiang, F.; Chen, J.; Chen, J.; Wang, L. lncRNA LOC100132354 promotes angiogenesis through VEGFA/VEGFR2 signaling pathway in lung adenocarcinoma. Cancer Manag. Res. 2018, 10, 4257–4266. [Google Scholar] [CrossRef]
- Zhang, Y.; Du, W.; Chen, Z.; Xiang, C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell Res. 2017, 359, 449–457. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, D.; Zhang, S.; Liu, Z.; Xu, L.; Chen, L.; Xie, L. Single-Cell RNA Sequencing Analysis of Gene Regulatory Network Changes in the Development of Lung Adenocarcinoma. Biomolecules 2023, 13, 671. https://doi.org/10.3390/biom13040671
Yu D, Zhang S, Liu Z, Xu L, Chen L, Xie L. Single-Cell RNA Sequencing Analysis of Gene Regulatory Network Changes in the Development of Lung Adenocarcinoma. Biomolecules. 2023; 13(4):671. https://doi.org/10.3390/biom13040671
Chicago/Turabian StyleYu, Dongshuo, Siwen Zhang, Zhenhao Liu, Linfeng Xu, Lanming Chen, and Lu Xie. 2023. "Single-Cell RNA Sequencing Analysis of Gene Regulatory Network Changes in the Development of Lung Adenocarcinoma" Biomolecules 13, no. 4: 671. https://doi.org/10.3390/biom13040671
APA StyleYu, D., Zhang, S., Liu, Z., Xu, L., Chen, L., & Xie, L. (2023). Single-Cell RNA Sequencing Analysis of Gene Regulatory Network Changes in the Development of Lung Adenocarcinoma. Biomolecules, 13(4), 671. https://doi.org/10.3390/biom13040671





