An Update on S100A16 in Human Cancer
Abstract
1. Introduction
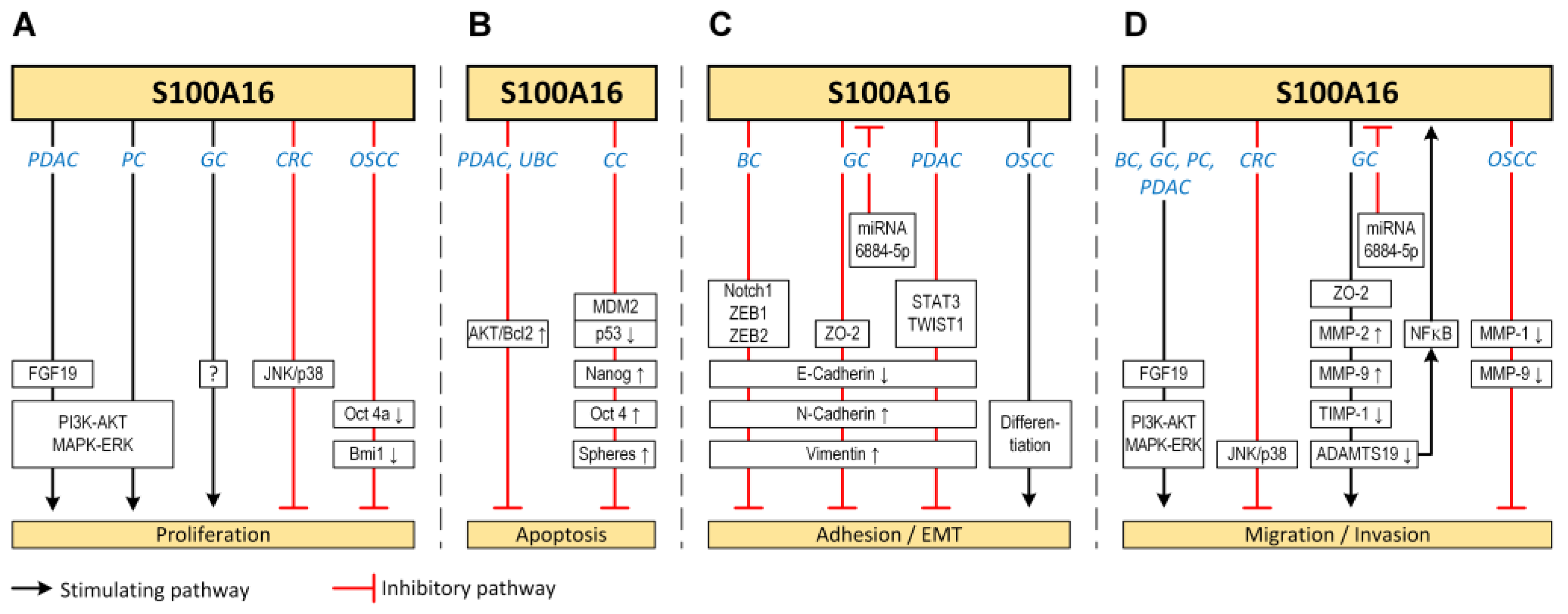
2. S100A16 in Human Cancer
2.1. Lung Cancer
2.2. Gastric Cancer
2.3. Colorectal Cancer
2.4. Pancreatic Cancer
2.5. Breast Cancer
2.6. Oral Squamous Cell Carcinoma
2.7. Prostate Cancer
2.8. Urinary Bladder Cancer
2.9. Renal Cell Carcinoma
2.10. Cervical Cancer
2.11. Ovarian Cancer
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAMTS19 | A disintegrin and metalloproteinase with thrombospondin motifs 19 |
| AKT | RAC-alpha serine/threonine-protein kinase |
| BC | breast cancer |
| Bcl2 | apoptosis regulator Bcl-2 (B-cell lymphoma 2) |
| Bmi1 | polycomb complex protein BMI-1 (B lymphoma Mo-MLV insertion region 1 homolog) |
| CC | cervical cancer |
| CRC | colorectal cancer |
| E-cadherin | epithelial cadherin |
| EMT | epithelial-mesenchymal transition |
| ERK | extracellular signal-regulated kinase |
| FANTOM | functional annotation of the mammalian genome |
| FGF19 | fibroblast growth factor 19 |
| FLG | filaggrin |
| GC | gastric cancer |
| GEPIA | Gene Expression Profiling Interactive Analysis |
| GO | gene ontology |
| GTEx | genotype-tissue expression |
| HPA | human protein atlas |
| IVL | involucrin |
| JNK | c-Jun N-terminal kinase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KRT13 | keratin 13 |
| LC | lung cancer |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| MAPK | mitogen-activated protein kinase |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 (mouse double minute 2 homolog) |
| miRNA | micro RNA |
| MMP | matrix metalloproteinase |
| Nanog | homeobox protein NANOG |
| N-cadherin | neural cadherin |
| NFκB | nuclear factor NF-kappa-B |
| Notch1 | neurogenic locus notch homolog protein 1 |
| pAKT | phosphorylated AKT |
| OC | ovarian cancer |
| Oct 4a | octamer-binding protein 4 |
| OSCC | oral squamous cell carcinoma |
| PC | prostate cancer |
| PDAC | pancreatic ductal adenocarcinoma |
| PI3K | phosphatidylinositol 3-kinase |
| p21 | cyclin-dependent kinase inhibitor 1 |
| p27 | cyclin-dependent kinase inhibitor 1B |
| p38 | p38 mitogen-activated protein kinase |
| p53 | cellular tumour antigen p53 |
| RCC | renal cell carcinoma |
| Slug | zinc finger protein SNAI2 (protein snail homolog 2/neural crest transcription factor Slug) |
| Snai1 | zinc finger protein SNAI1 (protein snail homolog 1/protein sna) |
| Stat3 | signal transducer and activator of transcription 3 |
| TCGA | The Cancer Genome Atlas |
| TGM1 | protein-glutamine gamma-glutamyltransferase K (transglutaminase-1) |
| TIMP-1 | metalloproteinase inhibitor 1 |
| TP53 | tumour protein p53 |
| TWIST1 | twist-related protein 1 |
| UBC | urinary bladder cancer |
| VEGF | vascular endothelial growth factor |
| VEGFR | vascular endothelial growth factor receptor |
| ZEB | zinc finger E-box-binding homeobox |
| ZO-2 | tight junction protein ZO-2 |
References
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys Acta Mol. Cell Res. 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W. S100A16, a ubiquitously expressed EF-hand protein which is up-regulated in tumors. Biochem. Biophys Res. Commun. 2004, 313, 237–244. [Google Scholar] [CrossRef]
- Marenholz, I.; Heizmann, C.W.; Fritz, G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys Res. Commun. 2004, 322, 1111–1122. [Google Scholar] [CrossRef]
- Babini, E.; Bertini, I.; Borsi, V.; Calderone, V.; Hu, X.; Luchinat, C.; Parigi, G. Structural characterization of human S100A16, a low-affinity calcium binder. J. Biol. Inorg. Chem. 2011, 16, 243–256. [Google Scholar] [CrossRef]
- Sturchler, E.; Cox, J.A.; Durussel, I.; Weibel, M.; Heizmann, C.W. S100A16, a novel calcium-binding protein of the EF-hand superfamily. J. Biol. Chem. 2006, 281, 38905–38917. [Google Scholar] [CrossRef]
- Consortium, P.-K.B. PDBe-KB: A community-driven resource for structural and functional annotations. Nucleic Acids Res. 2020, 48, D344–D353. [Google Scholar] [CrossRef]
- Simon, M.A.; Bartus, É.; Mag, B.; Boros, E.; Roszjár, L.; Gógl, G.; Travé, G.; Martinek, T.A.; Nyitray, L. Promiscuity mapping of the S100 protein family using a high-throughput holdup assay. Sci. Rep. 2022, 12, 5904. [Google Scholar] [CrossRef]
- Sapkota, D.; Costea, D.E.; Ibrahim, S.O.; Johannessen, A.C.; Bruland, O. S100A14 interacts with S100A16 and regulates its expression in human cancer cells. PLoS ONE 2013, 8, e76058. [Google Scholar] [CrossRef]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Sapkota, D.; Bruland, O.; Parajuli, H.; Osman, T.A.; Teh, M.T.; Johannessen, A.C.; Costea, D.E. S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer 2015, 15, 631. [Google Scholar] [CrossRef]
- Fornander, L.; Ghafouri, B.; Kihlström, E.; Åkerlind, B.; Schön, T.; Tagesson, C.; Lindahl, M. Innate immunity proteins and a new truncated form of SPLUNC1 in nasopharyngeal aspirates from infants with respiratory syncytial virus infection. Proteom. Clin. Appl. 2011, 5, 513–522. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Forrest, A.R.; Kawaji, H.; Rehli, M.; Baillie, J.K.; de Hoon, M.J.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A promoter-level mammalian expression atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef]
- Chen, D.; Luo, L.; Liang, C. Aberrant S100A16 expression might be an independent prognostic indicator of unfavorable survival in non-small cell lung adenocarcinoma. PLoS ONE 2018, 13, e0197402. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Z.; Yao, Y.; Li, W.Y.; Gu, J. Analysis of expression differences of immune genes in non-small cell lung cancer based on TCGA and ImmPort data sets and the application of a prognostic model. Ann. Transl. Med. 2020, 8, 550. [Google Scholar] [CrossRef]
- Saito, K.; Kobayashi, M.; Nagashio, R.; Ryuge, S.; Katono, K.; Nakashima, H.; Tsuchiya, B.; Jiang, S.X.; Saegusa, M.; Satoh, Y.; et al. S100A16 is a prognostic marker for lung adenocarcinomas. Asian Pac. J. Cancer Prev. 2015, 16, 7039–7044. [Google Scholar] [CrossRef]
- You, X.; Li, M.; Cai, H.; Zhang, W.; Hong, Y.; Gao, W.; Liu, Y.; Liang, X.; Wu, T.; Chen, F.; et al. Calcium binding protein S100A16 expedites proliferation, invasion and epithelial-mesenchymal transition process in gastric cancer. Front. Cell Dev. Biol. 2021, 9, 736929. [Google Scholar] [CrossRef]
- Lv, H.; Hou, H.; Lei, H.; Nie, C.; Chen, B.; Bie, L.; Han, L.; Chen, X. MicroRNA-6884-5p regulates the proliferation, invasion, and EMT of gastric cancer cells by directly targeting S100A16. Oncol. Res. 2020, 28, 225–236. [Google Scholar] [CrossRef]
- Ou, S.; Liao, Y.; Shi, J.; Tang, J.; Ye, Y.; Wu, F.; Wang, W.; Fei, J.; Xie, F.; Bai, L. S100A16 suppresses the proliferation, migration and invasion of colorectal cancer cells in part via the JNK/p38 MAPK pathway. Mol. Med. Rep. 2021, 23, 164. [Google Scholar] [CrossRef]
- Sun, X.; Wang, T.; Zhang, C.; Ning, K.; Guan, Z.-R.; Chen, S.-X.; Hong, T.-T.; Hua, D. S100A16 is a prognostic marker for colorectal cancer. J. Surg. Oncol. 2018, 117, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Chen, X.; Dong, F.; Zhang, Z.; Zhou, Z.; Ma, Z.; Huang, S.; Chen, B.; Zhang, C.; Hou, B. Prognostic values and immune suppression of the S100A family in pancreatic cancer. J. Cell. Mol. Med. 2021, 25, 3006–3018. [Google Scholar] [CrossRef]
- Tu, G.; Gao, W.; Li, Y.; Dian, Y.; Xue, B.; Niu, L.; Yu, X.; Zhu, H. Expressional and prognostic value of S100A16 in pancreatic cancer via integrated bioinformatics analyses. Front. Cell Dev. Biol. 2021, 9, 645641. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Zhang, C.; Xu, P.; Liu, Y.; Mo, X.; Sun, Q.; Abdelatty, A.; Hu, C.; Xu, H.; Zhou, G.; et al. S100A16 promotes metastasis and progression of pancreatic cancer through FGF19-mediated AKT and ERK1/2 pathways. Cell Biol. Toxicol. 2021, 37, 555–571. [Google Scholar] [CrossRef]
- Chen, T.; Xia, D.M.; Qian, C.; Liu, S.R. Integrated analysis identifies S100A16 as a potential prognostic marker for pancreatic cancer. Am. J. Transl. Res. 2021, 13, 5720–5730. [Google Scholar]
- Zhou, W.; Pan, H.; Xia, T.; Xue, J.; Cheng, L.; Fan, P.; Zhang, Y.; Zhu, W.; Xue, Y.; Liu, X.; et al. Up-regulation of S100A16 expression promotes epithelial-mesenchymal transition via Notch1 pathway in breast cancer. J. Biomed. Sci. 2014, 21, 97. [Google Scholar] [CrossRef]
- Tanaka, M.; Ichikawa-Tomikawa, N.; Shishito, N.; Nishiura, K.; Miura, T.; Hozumi, A.; Chiba, H.; Yoshida, S.; Ohtake, T.; Sugino, T. Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer 2015, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Xue, Y.; Liang, C.; Zhang, R.; Zhang, Z.; Li, H.; Su, D.; Liang, X.; Zhang, Y.; Huang, Q.; et al. S100A16 promotes cell proliferation and metastasis via AKT and ERK cell signaling pathways in human prostate cancer. Tumour Biol. 2016, 37, 12241–12250. [Google Scholar] [CrossRef]
- Wang, R.; Wu, Y.; Yu, J.; Yang, G.; Yi, H.; Xu, B. Plasma messenger RNAs identified through bioinformatics analysis are novel, non-invasive prostate cancer biomarkers. Onco Targets Ther. 2020, 13, 541–548. [Google Scholar] [CrossRef]
- Yao, R.; Lopez-Beltran, A.; Maclennan, G.T.; Montironi, R.; Eble, J.N.; Cheng, L. Expression of S100 protein family members in the pathogenesis of bladder tumors. Anticancer Res. 2007, 27, 3051–3058. [Google Scholar]
- Guo, Y.; Zheng, Z.; Mao, S.; Yang, F.; Wang, R.; Wang, H.; Liu, J.; Li, C.; Wang, Q.; Zhang, W.; et al. Metabolic-associated signature and hub genes associated with immune microenvironment and prognosis in bladder cancer. Mol. Carcinog. 2023, 62, 185–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, R.; Tang, J.; Gao, J.; Fang, Z.; Zhang, M.; Shen, X.; Lu, L.; Chen, Y. Calbindin S100A16 promotes renal cell carcinoma progression and angiogenesis via the VEGF/VEGFR2 signaling pathway. Contrast Media Mol. Imaging 2022, 2022, 5602011. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Song, H.M.; Zhou, Q. Comprehensive analysis of the expression and prognosis for S100 in human ovarian cancer: A STROBE study. Medicine 2020, 99, e22777. [Google Scholar] [CrossRef] [PubMed]
- Tomiyama, N.; Ikeda, R.; Nishizawa, Y.; Masuda, S.; Tajitsu, Y.; Takeda, Y. S100A16 up-regulates Oct4 and Nanog expression in cancer stem-like cells of Yumoto human cervical carcinoma cells. Oncol. Lett. 2018, 15, 9929–9933. [Google Scholar] [CrossRef]
- Li, T.; Ren, T.; Huang, C.; Li, Y.; Yang, P.; Che, G.; Luo, L.; Chen, Y.; Peng, S.; Lin, Y.; et al. S100A16 induces epithelial-mesenchymal transition in human PDAC cells and is a new therapeutic target for pancreatic cancer treatment that synergizes with gemcitabine. Biochem. Pharmacol. 2021, 189, 114396. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, X.; Li, A.; Yang, S.; Qiao, R.; Zhang, J. S100A16 regulated by Snail promotes the chemoresistance of nonmuscle invasive bladder cancer through the AKT/Bcl-2 pathway. Cancer Manag. Res. 2019, 11, 2449–2456. [Google Scholar] [CrossRef]
- Kobayashi, M.; Nagashio, R.; Saito, K.; Aguilar-Bonavides, C.; Ryuge, S.; Katono, K.; Igawa, S.; Tsuchiya, B.; Jiang, S.-X.; Ichinoe, M.; et al. Prognostic significance of S100A16 subcellular localization in lung adenocarcinoma. Hum. Pathol. 2018, 74, 148–155. [Google Scholar] [CrossRef]
- Katono, K.; Sato, Y.; Kobayashi, M.; Nagashio, R.; Ryuge, S.; Igawa, S.; Ichinoe, M.; Murakumo, Y.; Saegusa, M.; Masuda, N. S100A16, a promising candidate as a prognostic marker for platinum-based adjuvant chemotherapy in resected lung adenocarcinoma. Onco Targets Ther. 2017, 10, 5273–5279. [Google Scholar] [CrossRef]
- Xu, Z.H.; Miao, Z.W.; Jiang, Q.Z.; Gan, D.X.; Wei, X.G.; Xue, X.Z.; Li, J.Q.; Zheng, F.; Qin, X.X.; Fang, W.G.; et al. Brain microvascular endothelial cell exosome-mediated S100A16 up-regulation confers small-cell lung cancer cell survival in brain. FASEB J. 2019, 33, 1742–1757. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, X.; Zhao, Y.; Huang, J.; Li, T.; Chen, H.; Zhou, J.; Huang, Z.; Yang, Z. ADAMTS19 suppresses cell migration and invasion by targeting S100A16 via the NF-κB pathway in human gastric cancer. Biomolecules 2021, 11, 561. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Ma, X.; Xin, W.; Fan, X. S100A16 regulates HeLa cell through the phosphatidylinositol 3 kinase (PI3K)/AKT signaling pathway. Med. Sci. Monit. 2020, 26, e919757. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Li, L.D.; Li, J.; Lu, X. Prognostic values of S100 family members in ovarian cancer patients. BMC Cancer 2018, 18, 1256. [Google Scholar] [CrossRef] [PubMed]



| Cancer Type | Specimens | mRNA/Protein | Expression | Ref. |
| LC | Specimens * | mRNA and protein | Up | [15] |
| Specimens ** | mRNA | Up | [16] | |
| Specimens * | Protein | Up | [17] | |
| GC | GC specimens/cell lines | mRNA and protein | Up | [18,19] |
| CRC | CRC specimens | mRNA and protein | Down | [20,21] |
| PC | PC specimens/cell-lines | mRNA and protein | Up | [22,23,24,25] |
| BC | BC specimens/cell-lines | mRNA and protein | Up | [26,27] |
| OSCC | OSCC specimens | mRNA and protein | Down | [11] |
| PC | PC specimens/cell-lines | mRNA and protein | Up | [28] |
| PC specimens | mRNA | Down | [29] | |
| UBC | UBC specimens | mRNA | Up | [30,31] |
| RCC | RCC specimens/cell-lines | mRNA | Up | [32] |
| CC | CC specimens | mRNA | Up | [23] |
| OC | OC specimens | mRNA | Up | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basnet, S.; Vallenari, E.M.; Maharjan, U.; Sharma, S.; Schreurs, O.; Sapkota, D. An Update on S100A16 in Human Cancer. Biomolecules 2023, 13, 1070. https://doi.org/10.3390/biom13071070
Basnet S, Vallenari EM, Maharjan U, Sharma S, Schreurs O, Sapkota D. An Update on S100A16 in Human Cancer. Biomolecules. 2023; 13(7):1070. https://doi.org/10.3390/biom13071070
Chicago/Turabian StyleBasnet, Suyog, Evan Michael Vallenari, Urusha Maharjan, Sunita Sharma, Olaf Schreurs, and Dipak Sapkota. 2023. "An Update on S100A16 in Human Cancer" Biomolecules 13, no. 7: 1070. https://doi.org/10.3390/biom13071070
APA StyleBasnet, S., Vallenari, E. M., Maharjan, U., Sharma, S., Schreurs, O., & Sapkota, D. (2023). An Update on S100A16 in Human Cancer. Biomolecules, 13(7), 1070. https://doi.org/10.3390/biom13071070





