Spindle Position Checkpoint Kinase Kin4 Regulates Organelle Transport in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Plasmids
2.2. Growth Conditions
2.3. Setup for Construction of Mutant Collecion and Microscopy Analysis
2.4. Image Acquisition
2.5. Time-Lapse Imaging
2.6. Vacuolar Staining with FM4-64
2.7. Immunoblotting
3. Results
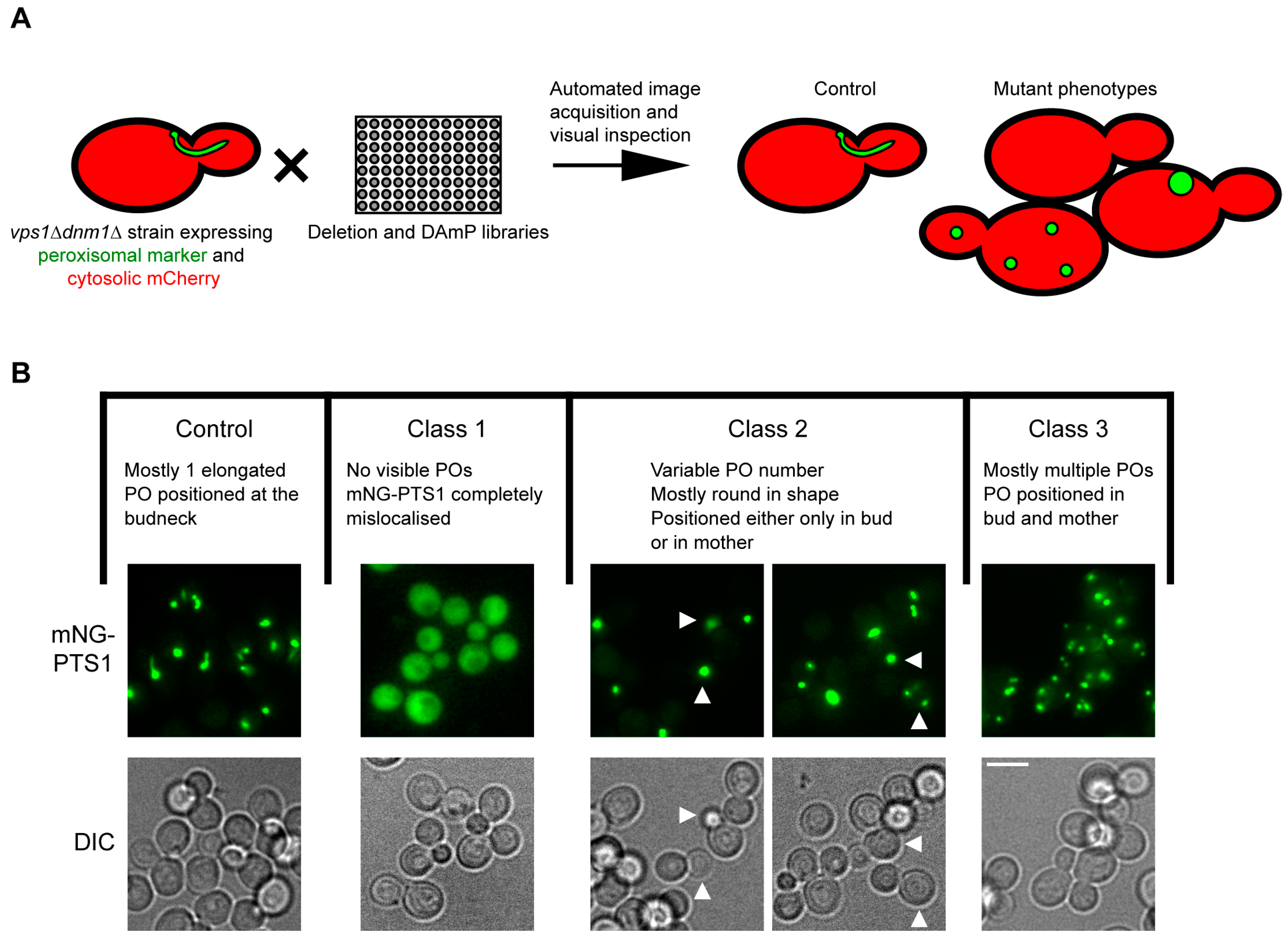
3.1. vps1Δdnm1Δ Cells Show a Weak Peroxisome Segregation
3.2. Identification of Genes Affecting Peroxisome Dynamics Using a High-Content Microscopy Setup
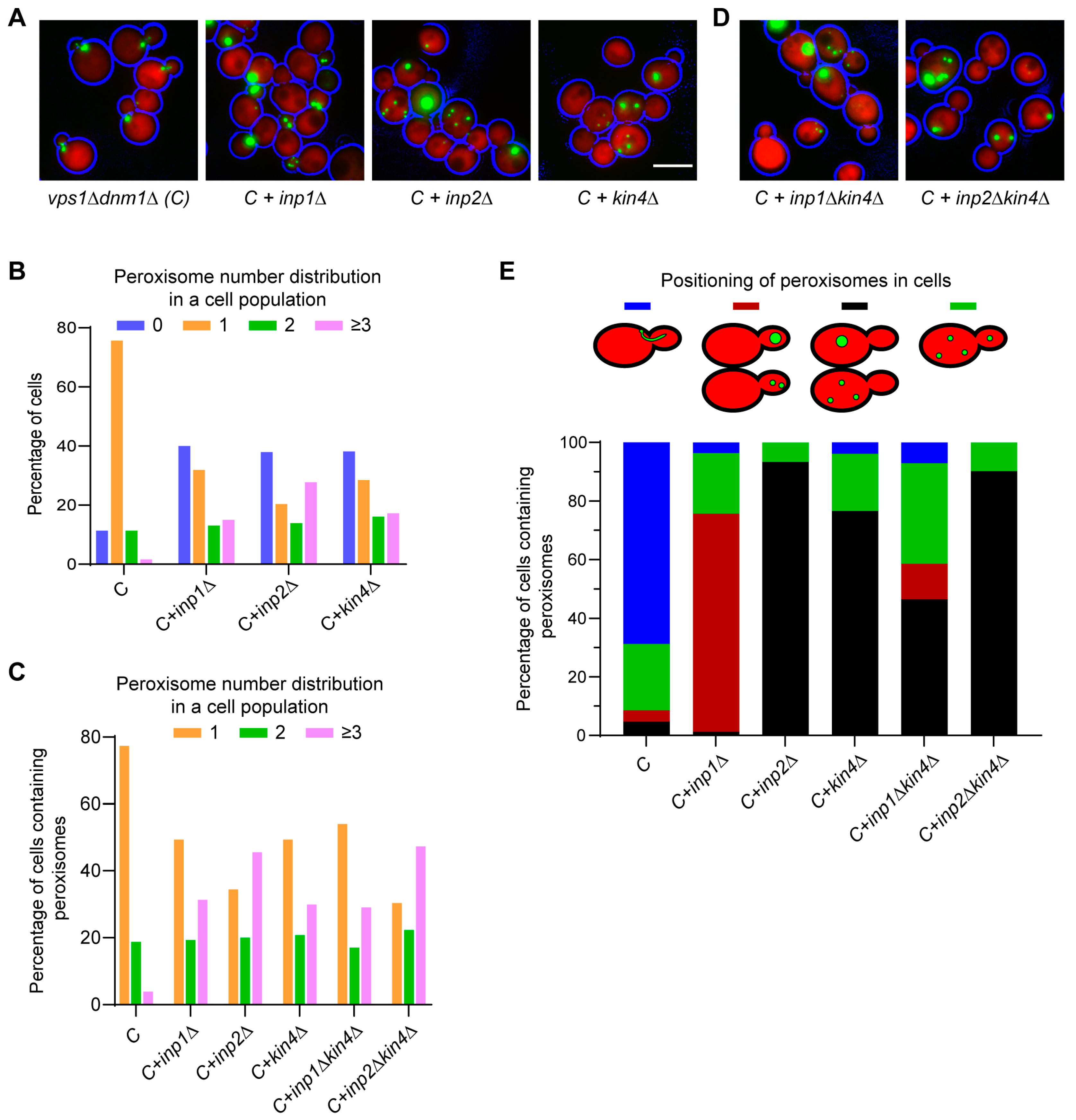
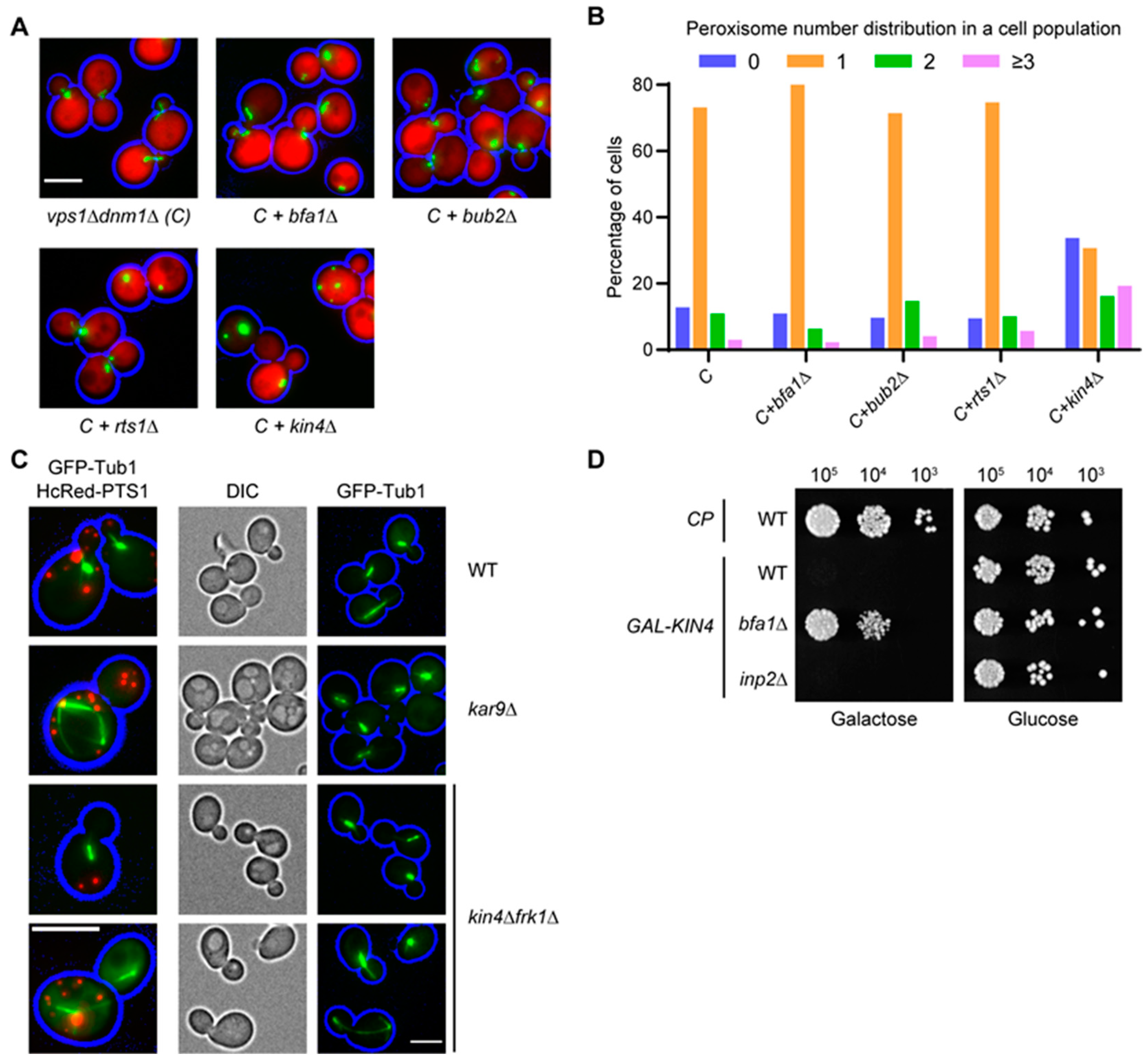
3.3. The SPOC Kinase Kin4 Is Required for Peroxisome Transport into the Bud
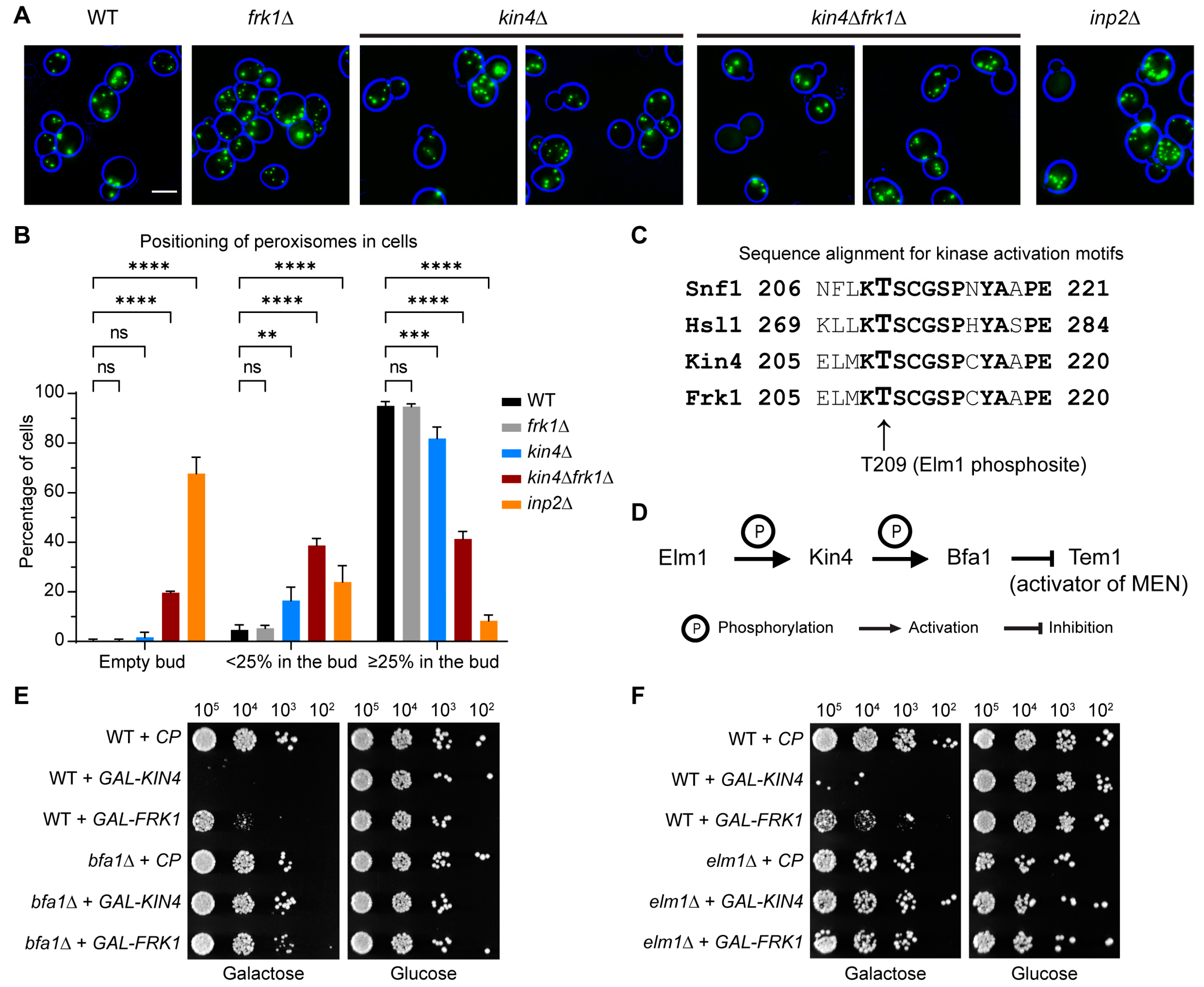
3.4. Frk1 Is a Functional Paralog of Kin4
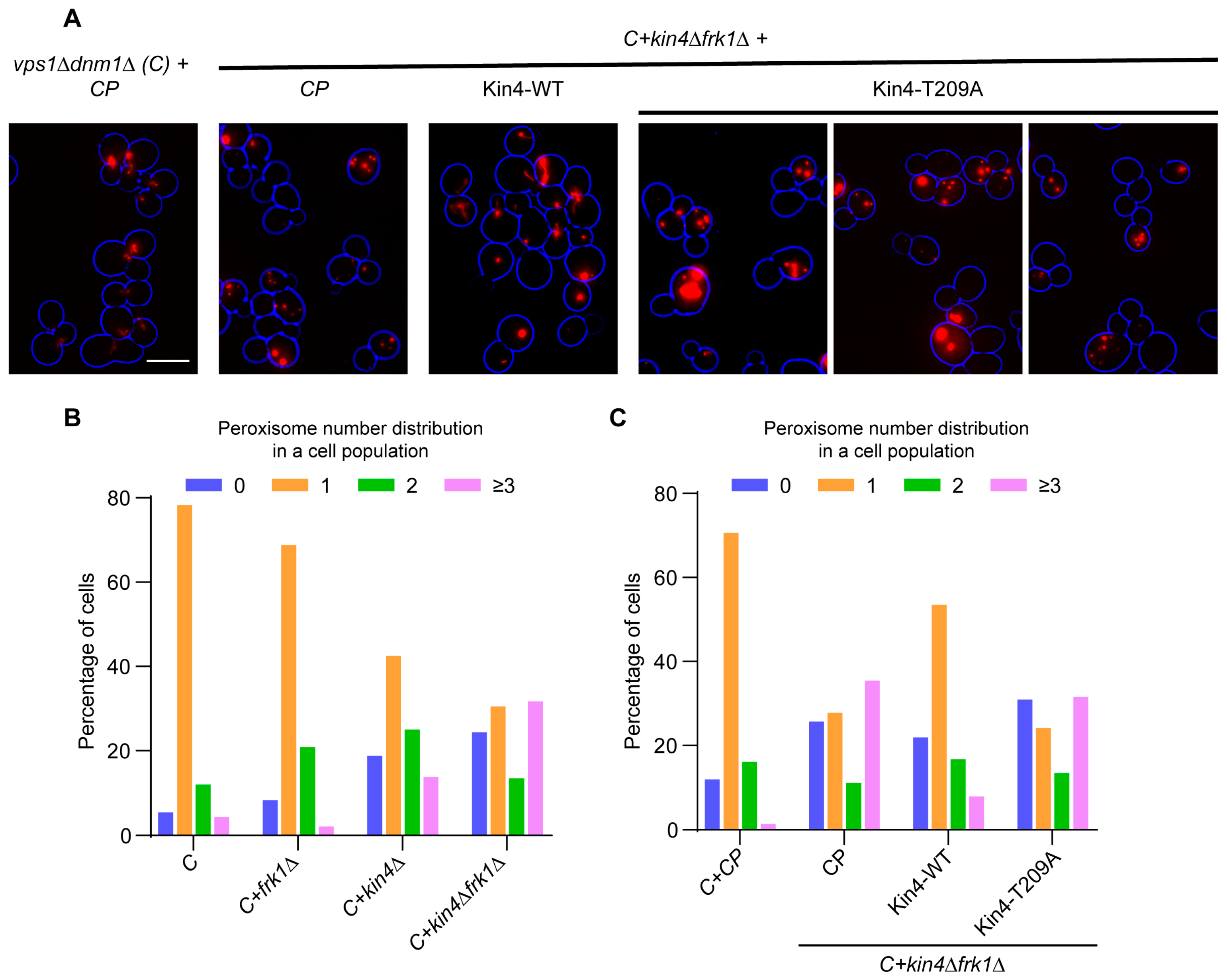
3.5. The Kinase Activity of Kin4 Is Required for Its Function in Peroxisome Segregation
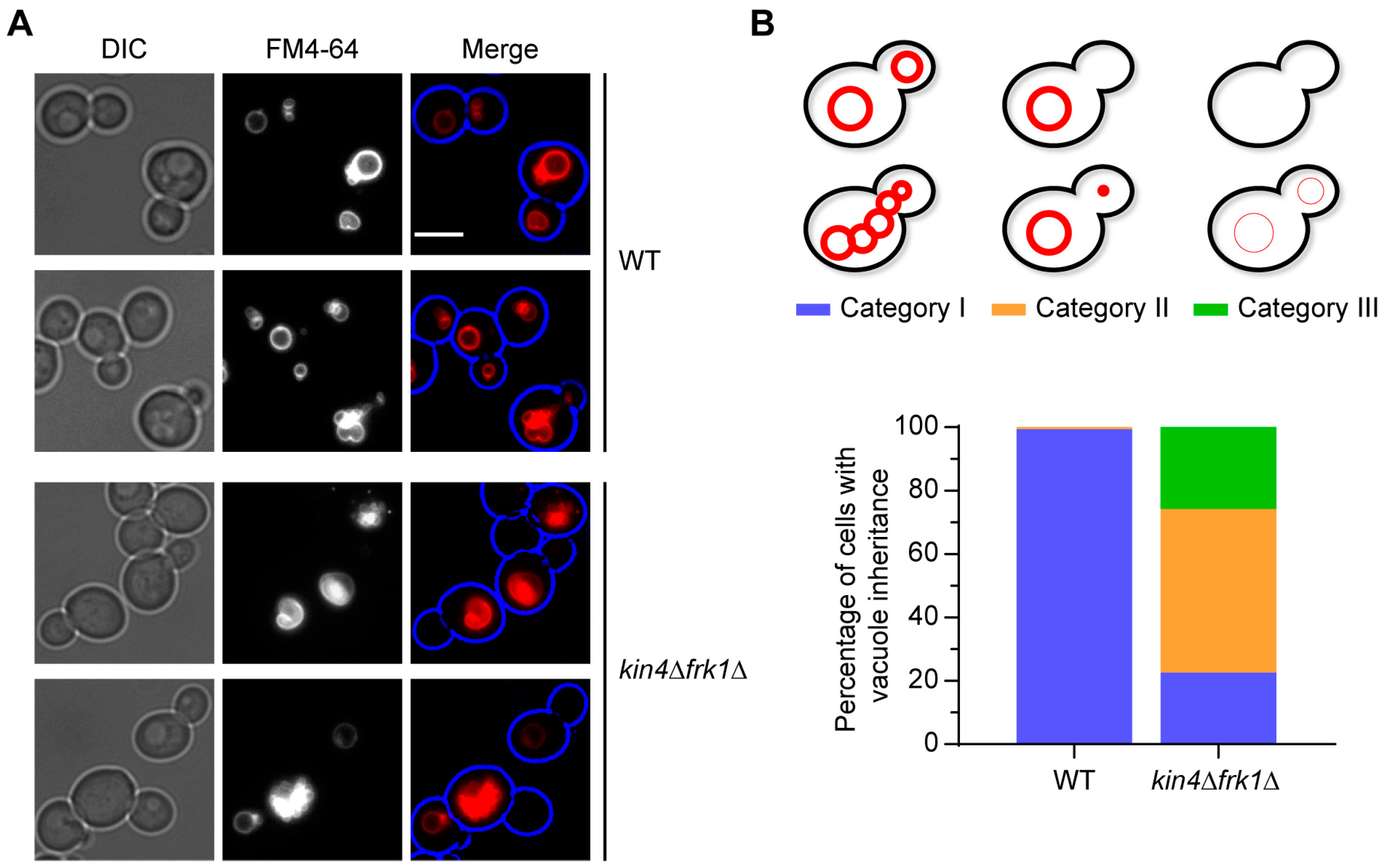
3.6. kin4Δfrk1Δ Cells That Fail to Inherit Peroxisomes Form Them De Novo
3.7. Kin4 Function in Peroxisome Inheritance Is Independent of Its Function in SPOC
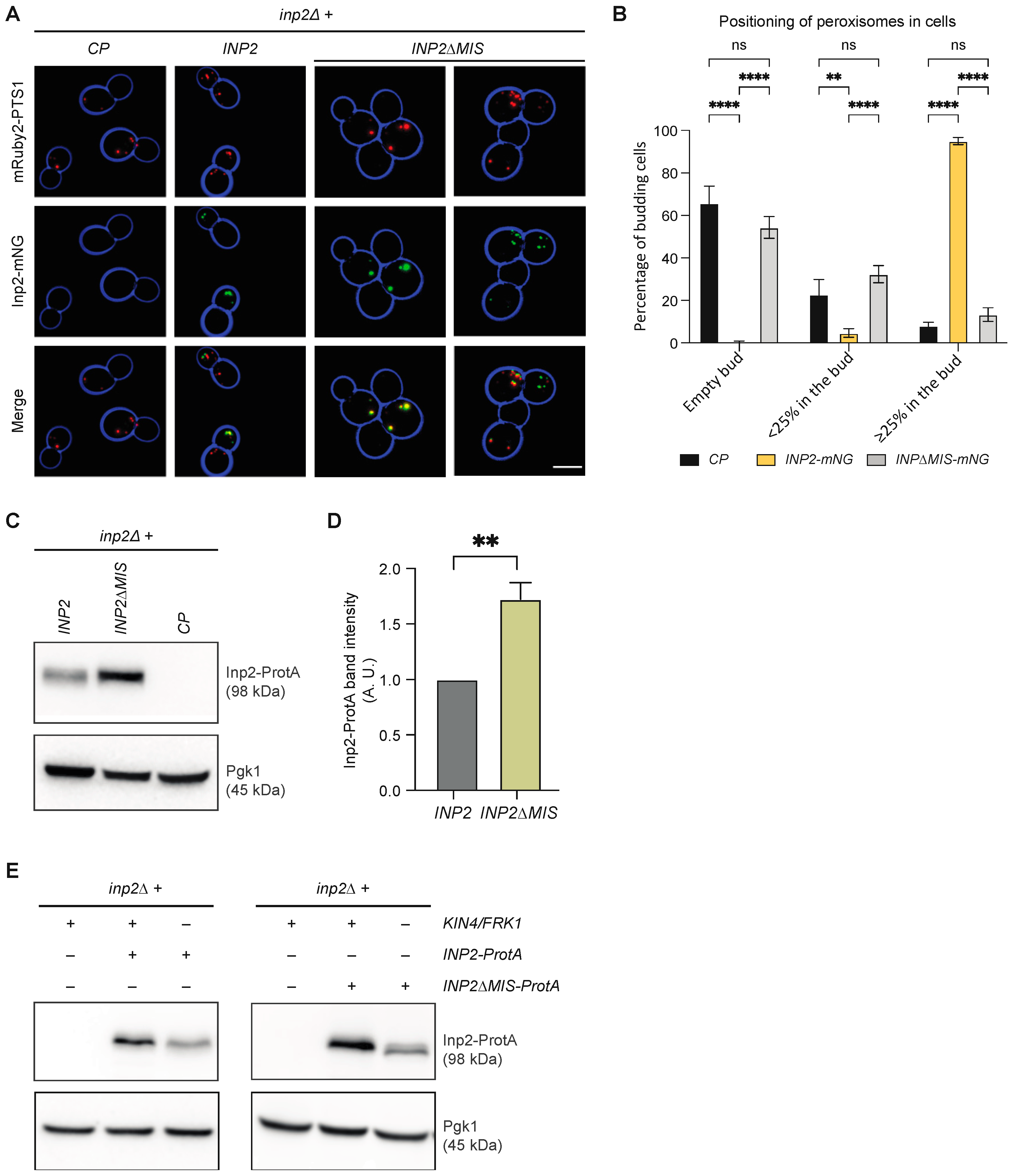
3.8. Kin4 and Frk1 Are Required to Maintain Inp2 Steady-State Levels
3.9. Kin4 and Frk1 Are Required to Maintain Elevated Inp2 Protein Levels in the Mother Cell
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wickner, G.W.A.W. Organelle Inheritance. Cell 1996, 84, 395–400. [Google Scholar]
- Knoblach, B.; Rachubinski, R.A. Sharing the cell’s bounty-organelle inheritance in yeast. J. Cell Sci. 2015, 128, 621–630. [Google Scholar] [CrossRef]
- Ménasché, G.; Pastural, E.; Feldmann, J.; Certain, S.; Ersoy, F.; Dupuis, S.; Wulffraat, N.; Bianchi, D.; Fischer, A.; Le Deist, F.; et al. Mutations in RAB27A cause Griscelli syndrome associated with haemophagocytic syndrome. Nat. Genet. 2000, 25, 173–176. [Google Scholar] [CrossRef]
- Li, K.W.; Lu, M.S.; Iwamoto, Y.; Drubin, D.G.; Pedersen, R.T.A. A preferred sequence for organelle inheritance during polarized cell growth. J. Cell Sci. 2021, 134, jcs258856. [Google Scholar] [CrossRef]
- Wong, S.; Weisman, L.S. Let it go: Mechanisms that detach myosin V from the yeast vacuole. Curr. Genet. 2021, 67, 865–869. [Google Scholar] [CrossRef]
- Yau, R.G.; Peng, Y.; Valiathan, R.R.; Birkeland, S.R.; Wilson, T.E.; Weisman, L.S. Release from myosin V via regulated recruitment of an E3 ubiquitin ligase controls organelle localization. Dev. Cell 2014, 28, 520–533. [Google Scholar] [CrossRef]
- Yau, R.G.; Wong, S.; Weisman, L.S. Spatial regulation of organelle release from myosin V transport by p21-activated kinases. J. Cell Biol. 2017, 216, 1557–1566. [Google Scholar] [CrossRef]
- Bartholomew, C.R.; Hardy, C.F. p21-activated kinases Cla4 and Ste20 regulate vacuole inheritance in Saccharomyces cerevisiae. Eukaryot. Cell 2009, 8, 560–572. [Google Scholar] [CrossRef][Green Version]
- Obara, K.; Yoshikawa, T.; Yamaguchi, R.; Kuwata, K.; Nakatsukasa, K.; Nishimura, K.; Kamura, T. Proteolysis of adaptor protein Mmr1 during budding is necessary for mitochondrial homeostasis in Saccharomyces cerevisiae. Nat. Commun. 2022, 13, 2005. [Google Scholar] [CrossRef]
- Yin, H.; Pruyne, D.; Huffaker, T.C.; Bretscher, A. Myosin V orientates the mitotic spindle in yeast. Nature 2000, 406, 1013–1015. [Google Scholar] [CrossRef]
- Adames, N.R.; Cooper, J.A. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000, 149, 863–874. [Google Scholar] [CrossRef]
- Huffaker, T.C.; Thomas, J.H.; Botstein, D. Diverse effects of beta-tubulin mutations on microtubule formation and function. J. Cell Biol. 1988, 106, 1997–2010. [Google Scholar] [CrossRef]
- Yeh, E.; Skibbens, R.V.; Cheng, J.W.; Salmon, E.D.; Bloom, K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995, 130, 687–700. [Google Scholar] [CrossRef]
- Bardin, A.J.; Visintin, R.; Amon, A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 2000, 102, 21–31. [Google Scholar] [CrossRef]
- Bloecher, A.; Venturi, G.M.; Tatchell, K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat. Cell Biol. 2000, 2, 556–558. [Google Scholar] [CrossRef]
- Lee, S.E.; Frenz, L.M.; Wells, N.J.; Johnson, A.L.; Johnston, L.H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001, 11, 784–788. [Google Scholar] [CrossRef][Green Version]
- Mohl, D.A.; Huddleston, M.J.; Collingwood, T.S.; Annan, R.S.; Deshaies, R.J. Dbf2-Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 2009, 184, 527–539. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Pereira, G. SPOC alert--when chromosomes get the wrong direction. Exp. Cell Res. 2012, 318, 1421–1427. [Google Scholar] [CrossRef]
- Geymonat, M.; Spanos, A.; Smith, S.J.; Wheatley, E.; Rittinger, K.; Johnston, L.H.; Sedgwick, S.G. Control of mitotic exit in budding yeast.In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 2002, 277, 28439–28445. [Google Scholar] [CrossRef]
- Pereira, G.; Höfken, T.; Grindlay, J.; Manson, C.; Schiebel, E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell 2000, 6, 1–10. [Google Scholar] [CrossRef]
- Pereira, G.; Schiebel, E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell 2005, 19, 209–221. [Google Scholar] [CrossRef]
- Maekawa, H.; Priest, C.; Lechner, J.; Pereira, G.; Schiebel, E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 2007, 179, 423–436. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Kurtulmus, B.; Orrico, M.I.; Hofmann, A.; Ibrahim, B.; Pereira, G. Elm1 kinase activates the spindle position checkpoint kinase Kin4. J. Cell Biol. 2010, 190, 975–989. [Google Scholar] [CrossRef]
- Moore, J.K.; Chudalayandi, P.; Heil-Chapdelaine, R.A.; Cooper, J.A. The spindle position checkpoint is coordinated by the Elm1 kinase. J. Cell Biol. 2010, 191, 493–503. [Google Scholar] [CrossRef]
- Bertazzi, D.T.; Kurtulmus, B.; Pereira, G. The cortical protein Lte1 promotes mitotic exit by inhibiting the spindle position checkpoint kinase Kin4. J. Cell Biol. 2011, 193, 1033–1048. [Google Scholar] [CrossRef]
- Falk, J.E.; Chan, L.Y.; Amon, A. Lte1 promotes mitotic exit by controlling the localization of the spindle position checkpoint kinase Kin4. Proc. Natl. Acad. Sci. USA 2011, 108, 12584–12590. [Google Scholar] [CrossRef]
- Falk, J.E.; Campbell, I.W.; Joyce, K.; Whalen, J.; Seshan, A.; Amon, A. LTE1 promotes exit from mitosis by multiple mechanisms. Mol. Biol. Cell 2016, 27, 3991–4001. [Google Scholar] [CrossRef]
- Baro, B.; Queralt, E.; Monje-Casas, F. Regulation of Mitotic Exit in Saccharomyces cerevisiae. Methods Mol. Biol. 2017, 1505, 3–17. [Google Scholar]
- Segal, M. Mitotic exit control: A space and time odyssey. Curr. Biol. 2011, 21, R857–R859. [Google Scholar] [CrossRef]
- Caydasi, A.K.; Khmelinskii, A.; Duenas-Sanchez, R.; Kurtulmus, B.; Knop, M.; Pereira, G. Temporal and compartment-specific signals coordinate mitotic exit with spindle position. Nat. Commun. 2017, 8, 14129. [Google Scholar] [CrossRef]
- Hoepfner, D.; Berg, M.V.D.; Philippsen, P.; Tabak, H.F.; Hettema, E.H. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2001, 155, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Rachubinski, R.; Mast, F.D.; Jm, M.; Am, M.; Jm, N.; Eh, H. A dual function for Pex3p in peroxisome formation and inheritance. J. Cell Biol. 2009, 187, 463–471. [Google Scholar]
- Knoblach, B.; Sun, X.; Coquelle, N.; Fagarasanu, A.; Poirier, R.L.; Rachubinski, R.A. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013, 32, 2439–2453. [Google Scholar] [CrossRef]
- Fagarasanu, M.; Fagarasanu, A.; Tam, Y.Y.C.; Aitchison, J.D.; Rachubinski, R.A. Inp1p is a peroxisomal membrane protein required for peroxisome inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2005, 169, 765–775. [Google Scholar] [CrossRef]
- Hulmes, G.E.; Hutchinson, J.D.; Dahan, N.; Nuttall, J.M.; Allwood, E.G.; Ayscough, K.R.; Hettema, E.H. The Pex3-Inp1 complex tethers yeast peroxisomes to the plasma membrane. J. Cell Biol. 2020, 219, e201906021. [Google Scholar] [CrossRef]
- Krikken, A.M.; Wu, H.; de Boer, R.; Devos, D.P.; Levine, T.P.; van der Klei, I.J. Peroxisome retention involves Inp1-dependent peroxisome-plasma membrane contact sites in yeast. J. Cell Biol. 2020, 219, e201906023. [Google Scholar] [CrossRef] [PubMed]
- Fagarasanu, A.; Fagarasanu, M.; Eitzen, G.A.; Aitchison, J.D.; Rachubinski, R.A. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 2006, 10, 587–600. [Google Scholar] [CrossRef]
- Saraya, R.; Cepińska, M.N.; Kiel, J.A.; Veenhuis, M.; van der Klei, I.J. A conserved function for Inp2 in peroxisome inheritance. Biochim. Biophys. Acta 2010, 1803, 617–622. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Otzen, M.; Rucktäschel, R.; Thoms, S.; Emmrich, K.; Krikken, A.M.; Erdmann, R.; van der Klei, I.J. Pex19p contributes to peroxisome inheritance in the association of peroxisomes to Myo2p. Traffic 2012, 13, 947–959. [Google Scholar] [CrossRef]
- Hettema, E.H.; Girzalsky, W.; van den Berg, M.; Erdmann, R.; Distel, B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000, 19, 223–233. [Google Scholar] [CrossRef]
- Fagarasanu, A.; Mast, F.D.; Knoblach, B.; Jin, Y.; Brunner, M.J.; Logan, M.R.; Glover, J.M.; Eitzen, G.A.; Aitchison, J.D.; Weisman, L.S.; et al. Myosin-driven peroxisome partitioning in S. cerevisiae. J. Cell Biol. 2009, 186, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Kuravi, K.; Nagotu, S.; Krikken, A.M.; Sjollema, K.; Deckers, M.; Erdmann, R.; Veenhuis, M.; van der Klei, I.J. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 2006, 119, 3994–4001. [Google Scholar] [CrossRef] [PubMed]
- Motley, A.M.; Hettema, E.H. Yeast peroxisomes multiply by growth and division. J. Cell Biol. 2007, 178, 399–410. [Google Scholar] [CrossRef]
- Huber, A.; Koch, J.; Kragler, F.; Brocard, C.; Hartig, A. A subtle interplay between three Pex11 proteins shapes de novo formation and fission of peroxisomes. Traffic 2012, 13, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Motley, A.M.; Galvin, P.C.; Ekal, L.; Nuttall, J.M.; Hettema, E.H. Reevaluation of the role of Pex1 and dynamin-related proteins in peroxisome membrane biogenesis. J. Cell Biol. 2015, 211, 1041–1056. [Google Scholar] [CrossRef]
- Ekal, L.; Hettema, E. Targeted Modifications of the Yeast Genome to Study Peroxisomes. Methods Mol. Biol. 2023, 2643, 217–232. [Google Scholar]
- Goldstein, A.L.; McCusker, J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999, 15, 1541–1553. [Google Scholar] [CrossRef]
- Shaner, N.C.; Lambert, G.G.; Chammas, A.; Ni, Y.; Cranfill, P.J.; Baird, M.A.; Sell, B.R.; Allen, J.R.; Day, R.N.; Israelsson, M.; et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 2013, 10, 407–409. [Google Scholar] [CrossRef]
- Gietz, R.D.; Sugino, A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 1988, 74, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Falcón, A.A.; Aris, J.P. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 41607–41617. [Google Scholar] [CrossRef]
- Orr-Weaver, T.L.; Szostak, J.W. Yeast recombination: The association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 1983, 80, 4417–4421. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.N.; Chang, A.C.; Boyer, H.W.; Helling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 1973, 70, 3240–3244. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.; Boone, C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 2006, 313, 171–192. [Google Scholar] [PubMed]
- Cohen, Y.; Schuldiner, M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 2011, 781, 127–159. [Google Scholar]
- Breker, M.; Gymrek, M.; Schuldiner, M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 2013, 200, 839–850. [Google Scholar] [CrossRef]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Breslow, D.K.; Cameron, D.M.; Collins, S.R.; Schuldiner, M.; Stewart-Ornstein, J.; Newman, H.W.; Braun, S.; Madhani, H.D.; Krogan, N.J.; Weissman, J.S. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 2008, 5, 711–718. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Vida, T.A.; Emr, S.D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 1995, 128, 779–792. [Google Scholar] [CrossRef]
- Ekal, L.; Alqahtani, A.M.S.; Hettema, E.H. The dynamin-related protein Vps1 and the peroxisomal membrane protein Pex27 function together during peroxisome fission. J. Cell Sci. 2023, 136, jcs246348. [Google Scholar] [CrossRef]
- Bi, E.; Maddox, P.; Lew, D.J.; Salmon, E.; McMillan, J.N.; Yeh, E.; Pringle, J.R. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J. Cell Biol. 1998, 142, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, J.B.; Drubin, D.G.; Barnes, G. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J. Cell Biol. 2010, 191, 795–808. [Google Scholar] [CrossRef]
- Lockshon, D.; Surface, L.E.; Kerr, E.O.; Kaeberlein, M.; Kennedy, B.K. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics 2007, 175, 77–91. [Google Scholar] [CrossRef]
- Smith, J.J.; Marelli, M.; Christmas, R.H.; Vizeacoumar, F.J.; Dilworth, D.J.; Ideker, T.; Galitski, T.; Dimitrov, K.; Rachubinski, R.A.; Aitchison, J.D. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 2002, 158, 259–271. [Google Scholar] [CrossRef]
- Dean, M.; Allikmets, R.; Gerrard, B.; Stewart, C.; Kistler, A.; Shafer, B.; Michaelis, S.; Strathern, J. Mapping and sequencing of two yeast genes belonging to the ATP-binding cassette superfamily. Yeast 1994, 10, 377–383. [Google Scholar] [CrossRef]
- Miller, R.K.; Rose, M.D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 1998, 140, 377–390. [Google Scholar] [CrossRef]
- Beach, D.L.; Thibodeaux, J.; Maddox, P.; Yeh, E.; Bloom, K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 2000, 10, 1497–1506. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Li, Y.; Yu, C.; Wei, Z. Structural mechanism for versatile cargo recognition by the yeast class V myosin Myo2. J. Biol. Chem. 2019, 294, 5896–5906. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kauffman, E.J.; Novak, J.L.; Nau, J.J.; Catlett, N.L.; Weisman, L.S. Regulated degradation of a class V myosin receptor directs movement of the yeast vacuole. Nature 2003, 422, 87–92. [Google Scholar] [CrossRef]
- D’Aquino, K.E.; Monje-Casas, F.; Paulson, J.; Reiser, V.; Charles, G.M.; Lai, L. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell 2005, 19, 223–234. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekal, L.; Alqahtani, A.M.S.; Schuldiner, M.; Zalckvar, E.; Hettema, E.H.; Ayscough, K.R. Spindle Position Checkpoint Kinase Kin4 Regulates Organelle Transport in Saccharomyces cerevisiae. Biomolecules 2023, 13, 1098. https://doi.org/10.3390/biom13071098
Ekal L, Alqahtani AMS, Schuldiner M, Zalckvar E, Hettema EH, Ayscough KR. Spindle Position Checkpoint Kinase Kin4 Regulates Organelle Transport in Saccharomyces cerevisiae. Biomolecules. 2023; 13(7):1098. https://doi.org/10.3390/biom13071098
Chicago/Turabian StyleEkal, Lakhan, Abdulaziz M. S. Alqahtani, Maya Schuldiner, Einat Zalckvar, Ewald H. Hettema, and Kathryn R. Ayscough. 2023. "Spindle Position Checkpoint Kinase Kin4 Regulates Organelle Transport in Saccharomyces cerevisiae" Biomolecules 13, no. 7: 1098. https://doi.org/10.3390/biom13071098
APA StyleEkal, L., Alqahtani, A. M. S., Schuldiner, M., Zalckvar, E., Hettema, E. H., & Ayscough, K. R. (2023). Spindle Position Checkpoint Kinase Kin4 Regulates Organelle Transport in Saccharomyces cerevisiae. Biomolecules, 13(7), 1098. https://doi.org/10.3390/biom13071098





