Biosynthesis, Spectrophotometric Follow-Up, Characterization, and Variable Antimicrobial Activities of Ag Nanoparticles Prepared by Edible Macrofungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Mushroom Spawn and Other Chemicals
2.2. Myco-Synthesis and Purification of Ag NPs
2.3. Microorganism Source
2.4. Spectrophotometric Follow for the Ag NPs Formation
2.5. Characterization of the Ag NPs Formation
2.6. Antimicrobial Investigation
3. Results and Discussion
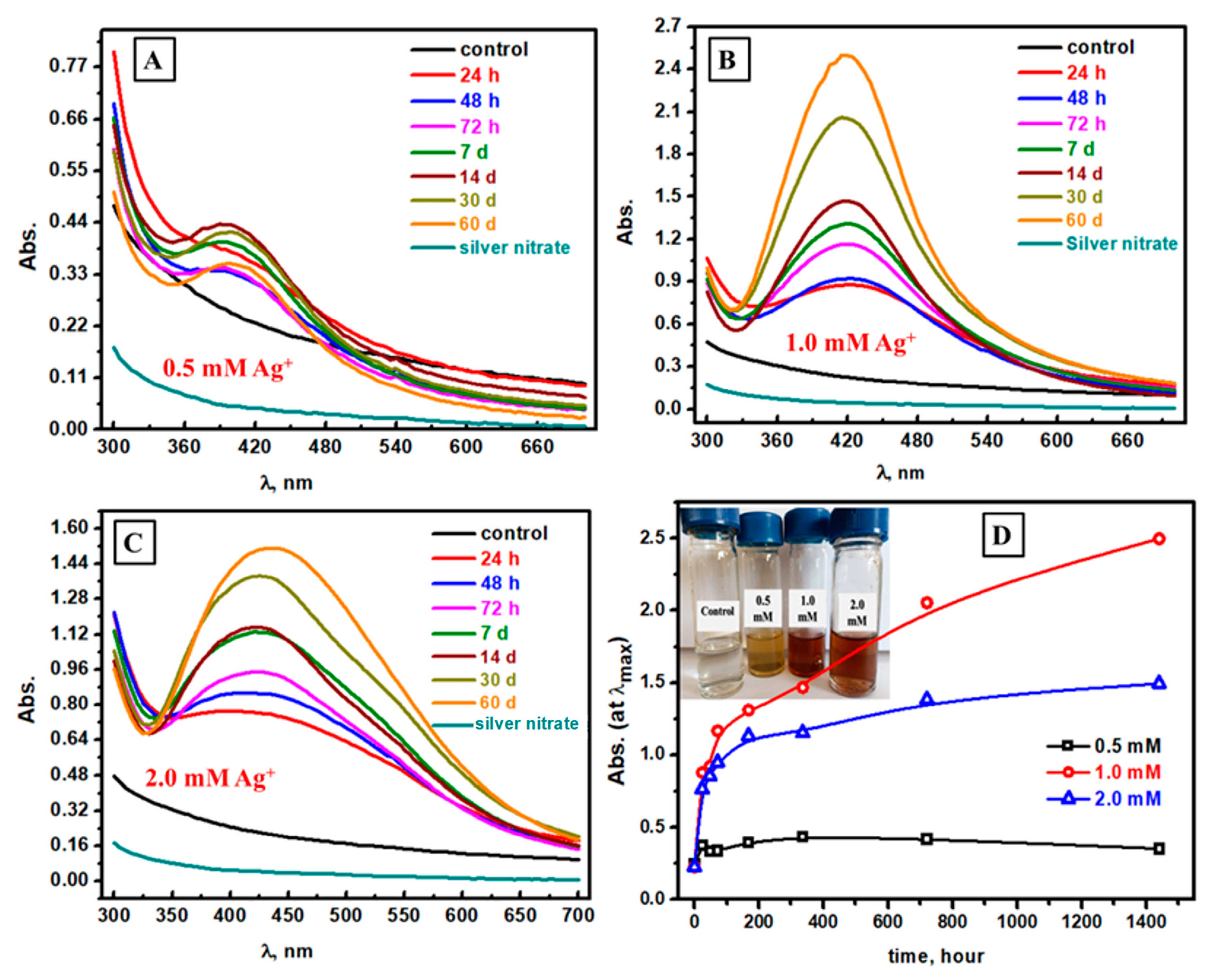
3.1. Spectrophotometric Follow-Up of Ag NPs Formation at Different AgNO3 Concentrations
3.2. Spectrophotometric Follow-Up of Ag NPs Formation at Different pH Values
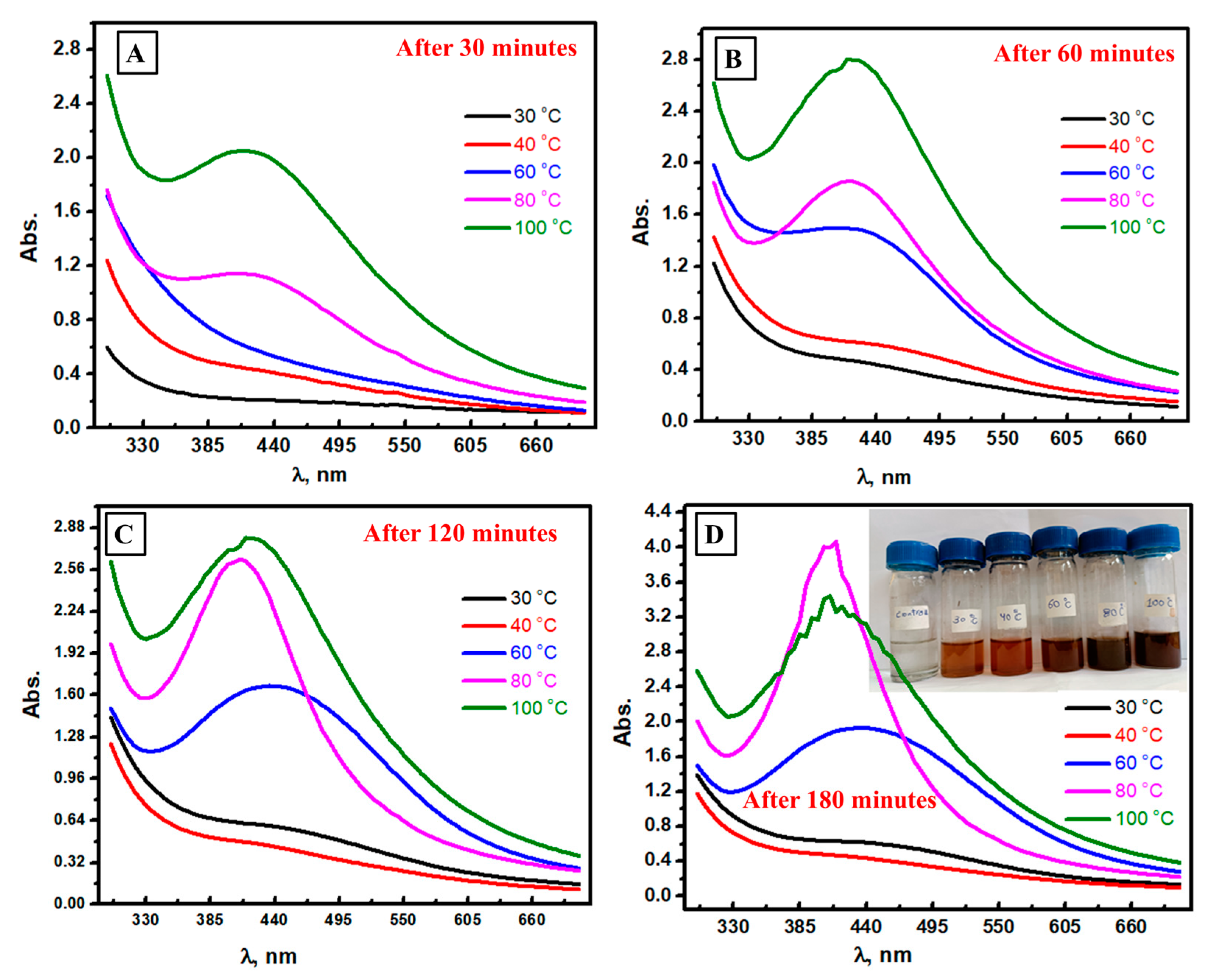
3.3. Spectrophotometric Follow-Up of Ag NPs Formation at Different Temperatures
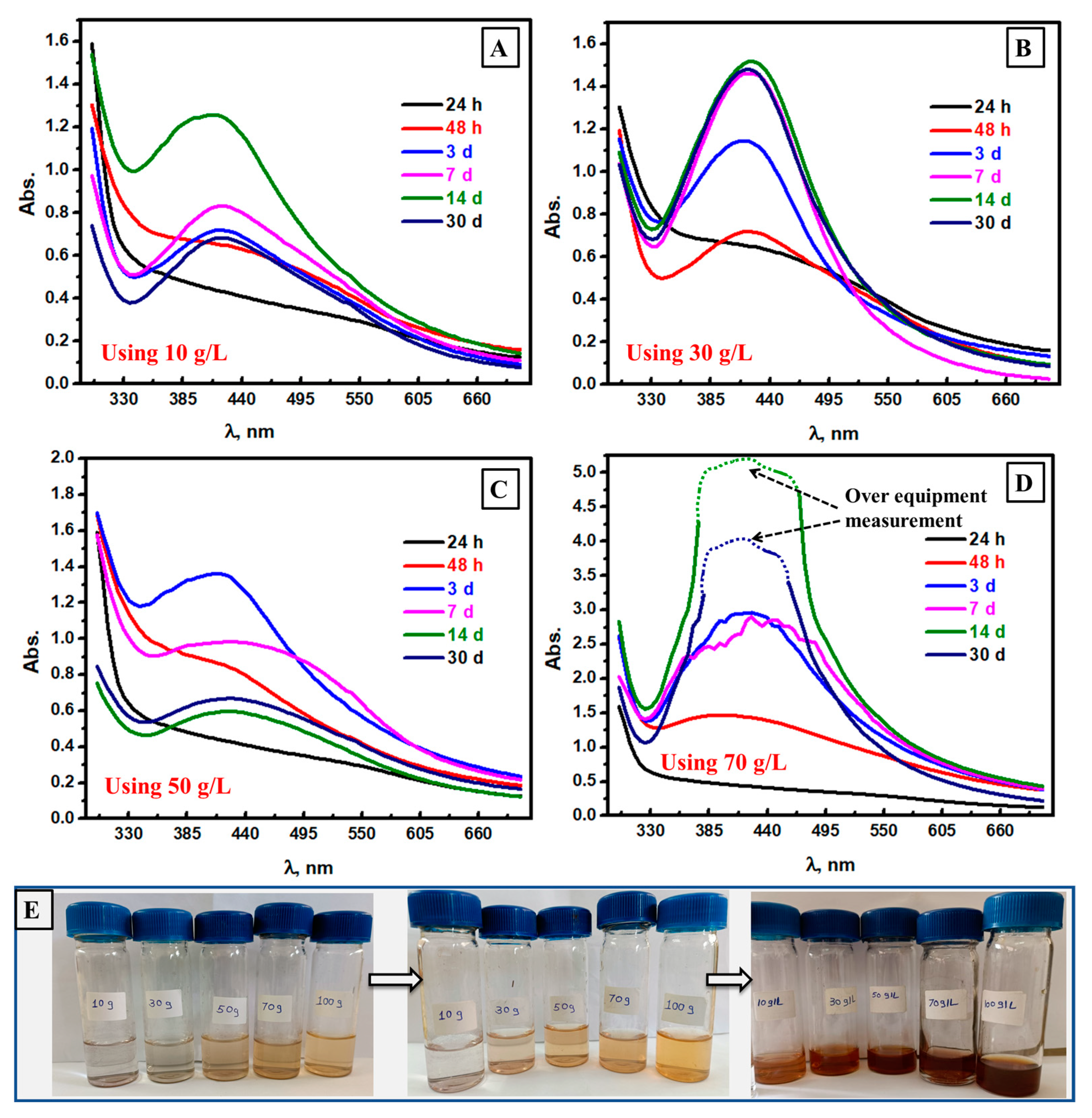
3.4. Spectrophotometric Follow of Ag NPs Formation at Different Mushroom Contents
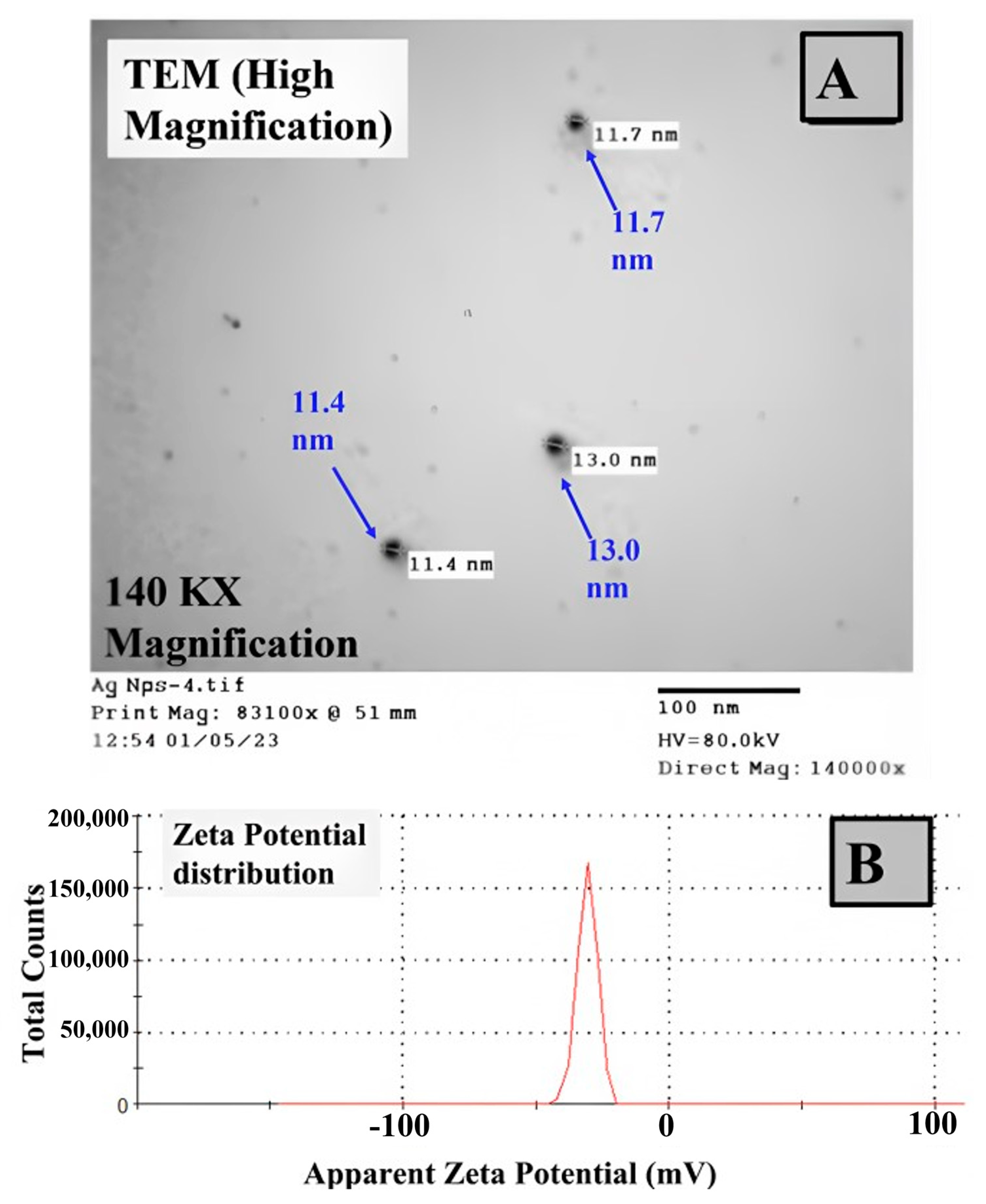
3.5. Characterization of the Synthesized Ag NPs by Edible Macrofungi
3.6. Antibacterial Activity of the Synthesized Ag NPs by Edible Macrofungi
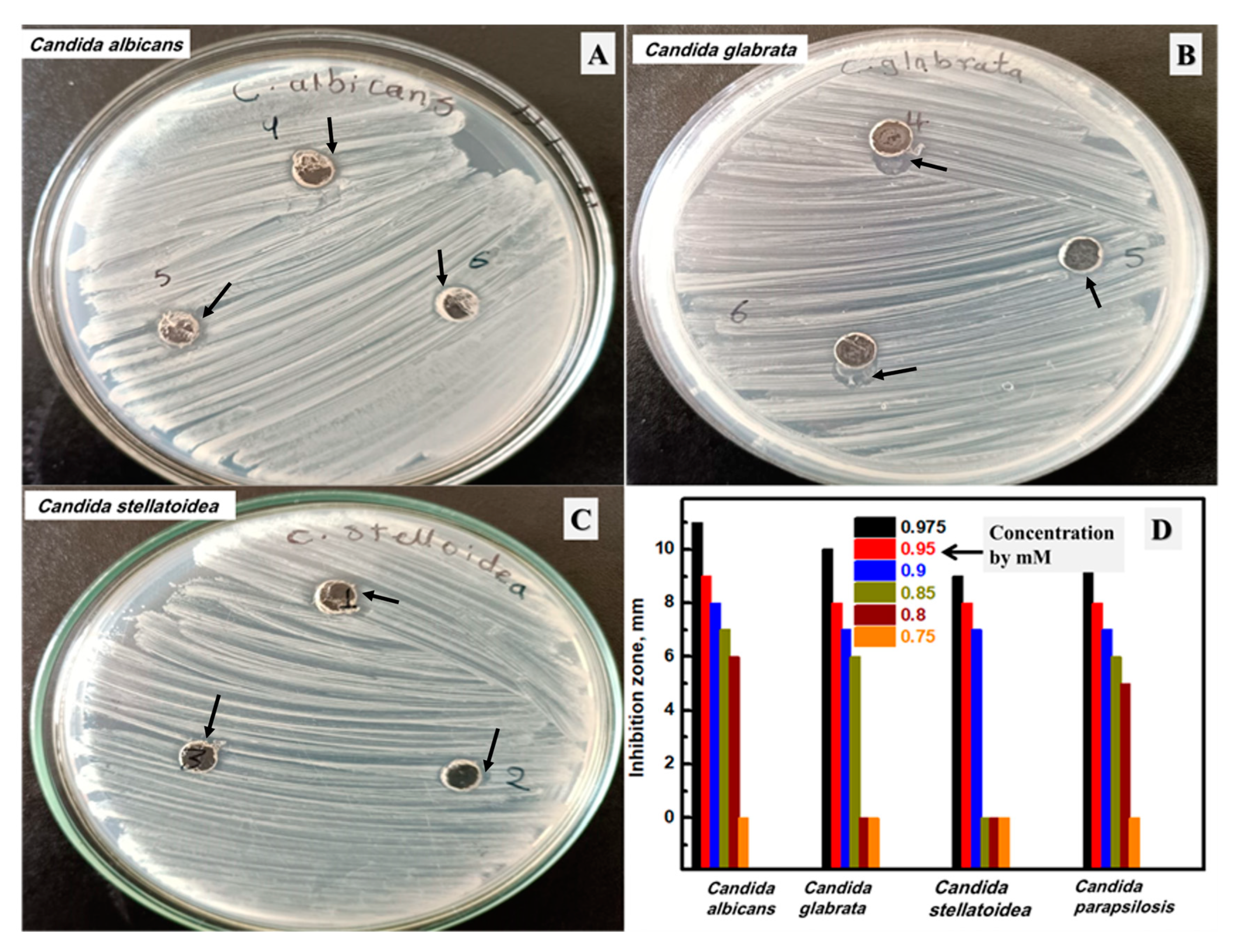
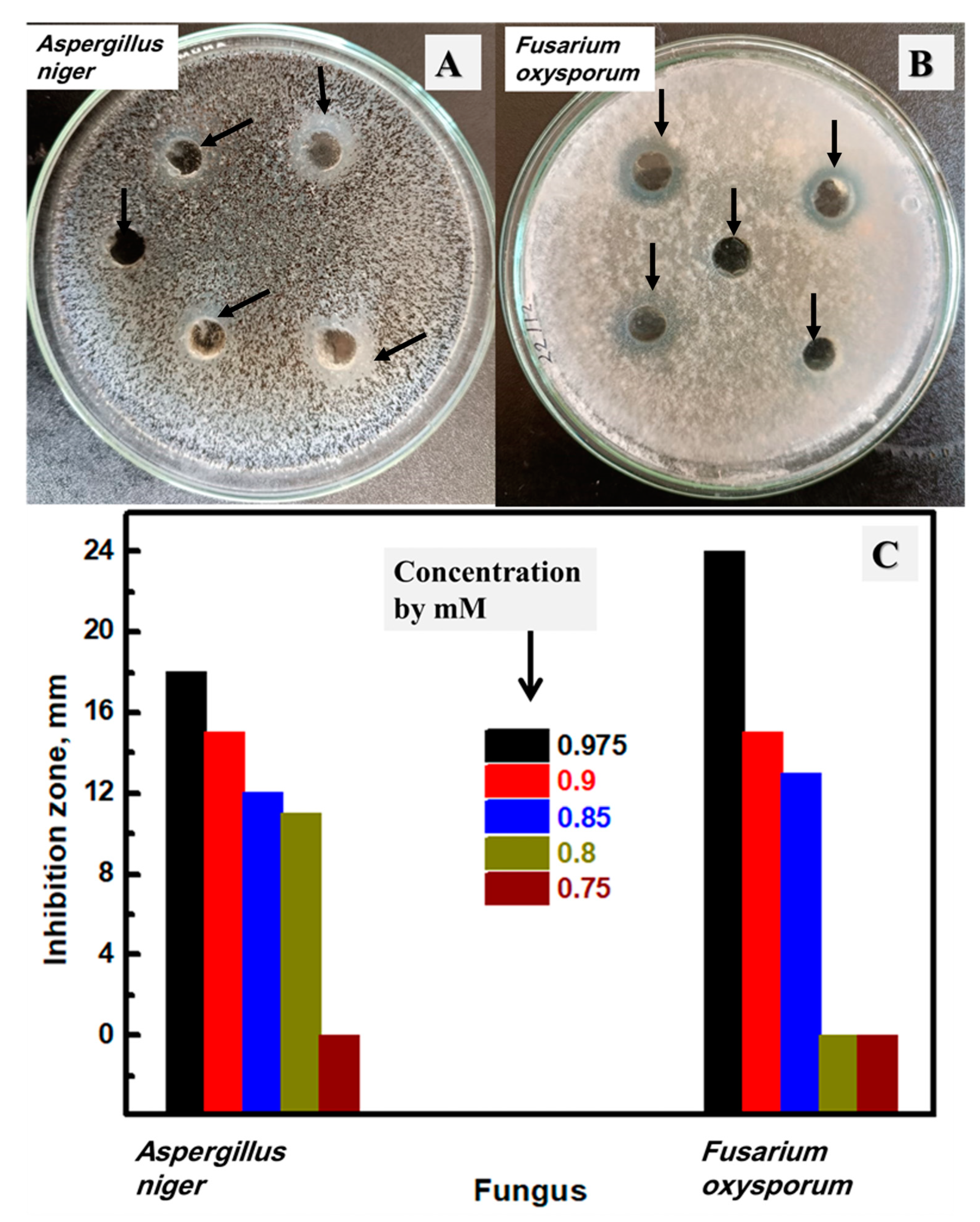
3.7. Antifungal Activity of the Synthesized Ag NPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, Y.; Zheng, W.; Tran, K.; Kamilar, E.; Bariwal, J.; Ma, H.; Liang, H. Hydrophilic nanoparticles that kill bacteria while sparing mammalian cells reveal the antibiotic role of nanostructures. Nat. Commun. 2022, 13, 197. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, R.; Nawaz, K.; Khan, A.K.; Hano, C.; Abbasi, B.H.; Anjum, S. An overview of the algae-mediated biosynthesis of nanoparticles and their biomedical applications. Biomolecules 2020, 10, 1498. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ezati, P.; Rhim, J.-W. Gelatin/carrageenan-based functional films with carbon dots from enoki mushroom for active food packaging applications. ACS Appl. Polym. Mater. 2021, 3, 6437–6445. [Google Scholar] [CrossRef]
- Papoutsis, K.; Grasso, S.; Menon, A.; Brunton, N.P.; Lyng, J.G.; Jacquier, J.-C.; Bhuyan, D.J. Recovery of ergosterol and vitamin D2 from mushroom waste—Potential valorization by food and pharmaceutical industries. Trends Food Sci. Technol. 2020, 99, 351–366. [Google Scholar] [CrossRef]
- González, A.; Cruz, M.; Losoya, C.; Nobre, C.; Loredo, A.; Rodríguez, R.; Contreras, J.; Belmares, R. Edible mushrooms as a novel protein source for functional foods. Food Funct. 2020, 11, 7400–7414. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Gao, M.; Wang, N.; Liu, S.; Wang, Q.; Sun, X. Lactic acid production from co-fermentation of food waste and spent mushroom substance with Aspergillus niger cellulase. Bioresour. Technol. 2021, 337, 125365. [Google Scholar] [CrossRef]
- Banasik, A.; Kanellopoulos, A.; Bloemhof-Ruwaard, J.M.; Claassen, G. Accounting for uncertainty in eco-efficient agri-food supply chains: A case study for mushroom production planning. J. Clean. Prod. 2019, 216, 249–256. [Google Scholar] [CrossRef]
- Zhang, S.; Sugawara, Y.; Chen, S.; Beelman, R.B.; Tsuduki, T.; Tomata, Y.; Matsuyama, S.; Tsuji, I. Mushroom consumption and incident risk of prostate cancer in Japan: A pooled analysis of the Miyagi Cohort Study and the Ohsaki Cohort Study. Int. J. Cancer 2020, 146, 2712–2720. [Google Scholar] [CrossRef]
- Ajith, T.A.; Janardhanan, K.K. Indian medicinal mushrooms as a source of antioxidant and antitumor agents. J. Clin. Biochem. Nutr. 2007, 40, 157–162. [Google Scholar] [CrossRef]
- Wan-Mohtar, W.A.A.Q.I.; Halim-Lim, S.A.; Kamarudin, N.Z.; Rukayadi, Y.; Abd Rahim, M.H.; Jamaludin, A.A.; Ilham, Z. Fruiting-body-base flour from an Oyster mushroom waste in the development of antioxidative chicken patty. J. Food Sci. 2020, 85, 3124–3133. [Google Scholar] [CrossRef]
- Moussa, A.Y.; Fayez, S.; Xiao, H.; Xu, B. New insights into antimicrobial and antibiofilm effects of edible mushrooms. Food Res. Int. 2022, 162, 111982. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, R.V.; Oludemi, T.; Fernandes, Â.; Ferreira, I.C.; Barros, L. Bioactive Properties of Mushrooms with Potential Health Benefits. Edible Fungi Chem. Compos. Nutr. Health Eff. 2022, 161. [Google Scholar] [CrossRef]
- Kuang, Y.; Li, B.; Wang, Z.; Qiao, X.; Ye, M. Terpenoids from the medicinal mushroom Antrodia camphorata: Chemistry and medicinal potential. Nat. Prod. Rep. 2021, 38, 83–102. [Google Scholar] [CrossRef]
- Gong, P.; Wang, S.; Liu, M.; Chen, F.; Yang, W.; Chang, X.; Liu, N.; Zhao, Y.; Wang, J.; Chen, X. Extraction methods, chemical characterizations and biological activities of mushroom polysaccharides: A mini-review. Carbohydr. Res. 2020, 494, 108037. [Google Scholar] [CrossRef] [PubMed]
- Anthony, K.J.P.; Murugan, M.; Jeyaraj, M.; Rathinam, N.K.; Sangiliyandi, G. Synthesis of silver nanoparticles using pine mushroom extract: A potential antimicrobial agent against E. coli and B. subtilis. J. Ind. Eng. Chem. 2014, 20, 2325–2331. [Google Scholar] [CrossRef]
- Saravanan, A.; Kumar, P.S.; Karishma, S.; Vo, D.-V.N.; Jeevanantham, S.; Yaashikaa, P.; George, C.S. A review on biosynthesis of metal nanoparticles and its environmental applications. Chemosphere 2021, 264, 128580. [Google Scholar] [CrossRef]
- Qamar, S.U.R.; Ahmad, J.N. Nanoparticles: Mechanism of biosynthesis using plant extracts, bacteria, fungi, and their applications. J. Mol. Liq. 2021, 334, 116040. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, S.Y. Biosynthesis of inorganic nanomaterials using microbial cells and bacteriophages. Nat. Rev. Chem. 2020, 4, 638–656. [Google Scholar] [CrossRef]
- Janakiraman, V.; Govindarajan, K.; CR, M. Biosynthesis of silver nanoparticles from endophytic fungi, and its cytotoxic activity. BioNanoScience 2019, 9, 573–579. [Google Scholar] [CrossRef]
- Alavi, M.; Nokhodchi, A. Synthesis and modification of bio-derived antibacterial Ag and ZnO nanoparticles by plants, fungi, and bacteria. Drug Discov. Today 2021, 26, 1953–1962. [Google Scholar] [CrossRef]
- Abu-Tahon, M.A.; Ghareib, M.; Abdallah, W.E. Environmentally benign rapid biosynthesis of extracellular gold nanoparticles using Aspergillus flavus and their cytotoxic and catalytic activities. Process Biochem. 2020, 95, 1–11. [Google Scholar] [CrossRef]
- Zhang, H.; Zhou, H.; Bai, J.; Li, Y.; Yang, J.; Ma, Q.; Qu, Y. Biosynthesis of selenium nanoparticles mediated by fungus Mariannaea sp. HJ and their characterization. Colloids Surf. A Physicochem. Eng. Asp. 2019, 571, 9–16. [Google Scholar] [CrossRef]
- Bunno, R.; Awakawa, T.; Mori, T.; Abe, I. Aziridine Formation by a FeII/α-Ketoglutarate Dependent Oxygenase and 2-Aminoisobutyrate Biosynthesis in Fungi. Angew. Chem. 2021, 133, 15961–15965. [Google Scholar] [CrossRef]
- Hashem, A.H.; Al Abboud, M.A.; Alawlaqi, M.M.; Abdelghany, T.M.; Hasanin, M. Synthesis of nanocapsules based on biosynthesized nickel nanoparticles and potato starch: Antimicrobial, antioxidant, and anticancer activity. Starch-Stärke 2022, 74, 2100165. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Shanmugam, S.; Varukattu, N.B.; MubarakAli, D.; Kathiresan, K.; Wang, M.-H. Biosynthesis and characterization of copper oxide nanoparticles from indigenous fungi and its effect of photothermolysis on human lung carcinoma. J. Photochem. Photobiol. B Biol. 2019, 190, 103–109. [Google Scholar] [CrossRef]
- Rehman, S.; Jermy, R.; Asiri, S.M.; Shah, M.A.; Farooq, R.; Ravinayagam, V.; Ansari, M.A.; Alsalem, Z.; Al Jindan, R.; Reshi, Z. Using Fomitopsis pinicola for bioinspired synthesis of titanium dioxide and silver nanoparticles, targeting biomedical applications. RSC Adv. 2020, 10, 32137–32147. [Google Scholar] [CrossRef]
- Chatterjee, S.; Mahanty, S.; Das, P.; Chaudhuri, P.; Das, S. Biofabrication of iron oxide nanoparticles using manglicolous fungus Aspergillus niger BSC-1 and removal of Cr (VI) from aqueous solution. Chem. Eng. J. 2020, 385, 123790. [Google Scholar] [CrossRef]
- Huang, Y.; Bai, L.; Yang, Y.; Yin, Z.; Guo, B. Biodegradable gelatin/silver nanoparticle composite cryogel with excellent antibacterial and antibiofilm activity and hemostasis for Pseudomonas aeruginosa-infected burn wound healing. J. Colloid Interface Sci. 2022, 608, 2278–2289. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, X.; Li, M.; Lu, Y.; Ai, C.; Jiang, C.; Liu, Y.; Pan, Z.; Shi, J. Antifungal activity of silver nanoparticles synthesized by iturin against Candida albicans in vitro and in vivo. Appl. Microbiol. Biotechnol. 2021, 105, 3759–3770. [Google Scholar] [CrossRef]
- Majeed, S.; Bakhtiar, N.F.B.; Danish, M.; Ibrahim, M.M.; Hashim, R. Green approach for the biosynthesis of silver nanoparticles and its antibacterial and antitumor effect against osteoblast MG-63 and breast MCF-7 cancer cell lines. Sustain. Chem. Pharm. 2019, 12, 100138. [Google Scholar] [CrossRef]
- Chi, N.T.L.; Narayanan, M.; Chinnathambi, A.; Govindasamy, C.; Subramani, B.; Brindhadevi, K.; Pimpimon, T.; Pikulkaew, S. Fabrication, characterization, anti-inflammatory, and anti-diabetic activity of silver nanoparticles synthesized from Azadirachta indica kernel aqueous extract. Environ. Res. 2022, 208, 112684. [Google Scholar] [CrossRef]
- Yadav, R.; Saini, H.; Kumar, D.; Pasi, S.; Agrawal, V. Bioengineering of Piper longum L. extract mediated silver nanoparticles and their potential biomedical applications. Mater. Sci. Eng. C 2019, 104, 109984. [Google Scholar] [CrossRef] [PubMed]
- Parmar, S.; Kaur, H.; Singh, J.; Matharu, A.S.; Ramakrishna, S.; Bechelany, M. Recent advances in green synthesis of Ag NPs for extenuating antimicrobial resistance. Nanomaterials 2022, 12, 1115. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.; Goshisht, M.K. Recent advances and mechanistic insights into antibacterial activity, antibiofilm activity, and cytotoxicity of silver nanoparticles. ACS Appl. Bio Mater. 2022, 5, 1391–1463. [Google Scholar] [CrossRef] [PubMed]
- Das, T.K.; Ganguly, S.; Bhawal, P.; Remanan, S.; Mondal, S.; Das, N. Mussel inspired green synthesis of silver nanoparticles-decorated halloysite nanotube using dopamine: Characterization and evaluation of its catalytic activity. Appl. Nanosci. 2018, 8, 173–186. [Google Scholar] [CrossRef]
- Elegbede, J.A.; Lateef, A.; Azeez, M.A.; Asafa, T.B.; Yekeen, T.A.; Oladipo, I.C.; Adebayo, E.A.; Beukes, L.S.; Gueguim-Kana, E.B. Fungal xylanases-mediated synthesis of silver nanoparticles for catalytic and biomedical applications. IET Nanobiotechnol. 2018, 12, 857–863. [Google Scholar] [CrossRef]
- Priyadarshni, K.C.; Krishnamoorthi, R.; Mumtha, C.; Mahalingam, P.U. Biochemical analysis of cultivated mushroom, Pleurotus florida and synthesis of silver nanoparticles for enhanced antimicrobial effects on clinically important human pathogens. Inorg. Chem. Commun. 2022, 142, 109673. [Google Scholar] [CrossRef]
- Panjikkaran, S.T.; Mathew, D. An environmentally friendly and cost effective technique for the commercial cultivation of oyster mushroom [Pleurotus florida (Mont.) Singer]. J. Sci. Food Agric. 2013, 93, 973–976. [Google Scholar] [CrossRef]
- Guo, H.; Han, F.; Shang, H.; Xiong, S.; Huynh, M.; Thistle, L.; Meng, L.; He, L.; Xing, B. New insight into naturally formed nanosilver particles: Role of plant root exudates. Environ. Sci. Nano 2021, 8, 1580–1592. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, X.; Chen, L.; Zhou, X.; Fan, X.; Hu, Y.; Niu, X.; Xu, X.; Zhou, G.; Ullah, N. Antibacterial aerogels with nano-silver reduced in situ by carboxymethyl cellulose for fresh meat preservation. Int. J. Biol. Macromol. 2022, 213, 621–630. [Google Scholar] [CrossRef]
- Bahari, N.; Hashim, N.; Abdan, K.; Md Akim, A.; Maringgal, B.; Al-Shdifat, L. Role of Honey as a Bifunctional Reducing and Capping/Stabilizing Agent: Application for Silver and Zinc Oxide Nanoparticles. Nanomaterials 2023, 13, 1244. [Google Scholar] [CrossRef] [PubMed]
- Hebbalalu, D.; Lalley, J.; Nadagouda, M.N.; Varma, R.S. Greener techniques for the synthesis of silver nanoparticles using plant extracts, enzymes, bacteria, biodegradable polymers, and microwaves. ACS Sustain. Chem. Eng. 2013, 1, 703–712. [Google Scholar] [CrossRef]
- Mohammadi, F.M.; Ghasemi, N. Influence of temperature and concentration on biosynthesis and characterization of zinc oxide nanoparticles using cherry extract. J. Nanostruct. Chem. 2018, 8, 93–102. [Google Scholar] [CrossRef]
- Abdel-Kareem, M.; Ahmed Zohri, A.-N. Inhibition of three toxigenic fungal strains and their toxins production using selenium nanoparticles. Czech Mycol. 2017, 69, 193–204. [Google Scholar] [CrossRef]
- Abdel-Kareem, M.; Zohri, A.-N.; Rasmey, A.-H. Biosynthesis of silver nanoparticles by Aspergillus sakultaensis and its antibacterial activity against human pathogens. Egypt. J. Microbiol. 2021, 56, 11–24. [Google Scholar] [CrossRef]
- Stamplecoskie, K.G.; Scaiano, J.C. Light emitting diode irradiation can control the morphology and optical properties of silver nanoparticles. J. Am. Chem. Soc. 2010, 132, 1825–1827. [Google Scholar] [CrossRef]
- Kassem, M.A.; El-Feky, H.H. The Impact of Ultrasonication on SDS-stabilized Silver Nanoparticles as a Probe for Trace Determination of Diaveridine. Plasmonics 2023, 18, 395–405. [Google Scholar] [CrossRef]
- Barabadi, H.; Noqani, H.; Ashouri, F.; Prasad, A.; Jounaki, K.; Mobaraki, K.; Mohanta, Y.K.; Mostafavi, E. Nanobiotechnological approaches in anticoagulant therapy: The role of bioengineered silver and gold nanomaterials. Talanta 2023, 256, 124279. [Google Scholar] [CrossRef]
- Xie, X.; Sun, T.; Xue, J.; Miao, Z.; Yan, X.; Fang, W.; Li, Q.; Tang, R.; Lu, Y.; Tang, L. Ag nanoparticles cluster with pH-triggered reassembly in targeting antimicrobial applications. Adv. Funct. Mater. 2020, 30, 2000511. [Google Scholar] [CrossRef]
- Salleh, A.; Naomi, R.; Utami, N.D.; Mohammad, A.W.; Mahmoudi, E.; Mustafa, N.; Fauzi, M.B. The potential of silver nanoparticles for antiviral and antibacterial applications: A mechanism of action. Nanomaterials 2020, 10, 1566. [Google Scholar] [CrossRef]
- Sanghi, R.; Verma, P. Biomimetic synthesis and characterisation of protein capped silver nanoparticles. Bioresour. Technol. 2009, 100, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kalishwaralal, K.; Vaidyanathan, R.; Venkataraman, D.; Pandian, S.R.K.; Muniyandi, J.; Hariharan, N.; Eom, S.H. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf. B Biointerfaces 2009, 74, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Biswal, A.K.; Misra, P.K. Biosynthesis and characterization of silver nanoparticles for prospective application in food packaging and biomedical fields. Mater. Chem. Phys. 2020, 250, 123014. [Google Scholar] [CrossRef]
- Singh, R.; Hano, C.; Nath, G.; Sharma, B. Green biosynthesis of silver nanoparticles using leaf extract of Carissa carandas L. and their antioxidant and antimicrobial activity against human pathogenic bacteria. Biomolecules 2021, 11, 299. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.R.; Qiu, P.; Zhao, K.; Shi, X.; Chen, L. Ag2Q-Based (Q = S, Se, Te) Silver Chalcogenide Thermoelectric Materials. Adv. Mater. 2023, 35, 2110236. [Google Scholar] [CrossRef]
- Vigneshwaran, N.; Kathe, A.A.; Varadarajan, P.V.; Nachane, R.P.; Balasubramanya, R.H. Silver-protein (core-shell) nanoparticle production using spent mushroom substrate. Langmuir 2007, 23, 7113–7117. [Google Scholar] [CrossRef]
- Sen, I.K.; Maity, K.; Islam, S.S. Green synthesis of gold nanoparticles using a glucan of an edible mushroom and study of catalytic activity. Carbohydr. Polym. 2013, 91, 518–528. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, Y.; Wang, H.; Wu, F.; Zhao, Y.; Liu, X.; Wu, H.; Wang, L.; Su, H. Green synthesis of silver nanoparticles using mushroom Flammulina velutipes Extract and their antibacterial activity against aquatic pathogens. Food Bioprocess Technol. 2020, 13, 1908–1917. [Google Scholar] [CrossRef]
- Roy, S.; Rhim, J.-W. Gelatin/cellulose nanofiber-based functional films added with mushroom-mediated sulfur nanoparticles for active packaging applications. J. Nanostruct. Chem. 2022, 12, 979–990. [Google Scholar] [CrossRef]
- Rasmussen, M.K.; Pedersen, J.N.; Marie, R. Size and surface charge characterization of nanoparticles with a salt gradient. Nat. Commun. 2020, 11, 2337. [Google Scholar] [CrossRef]
- Bhatnagar, S.; Kobori, T.; Ganesh, D.; Ogawa, K.; Aoyagi, H. Biosynthesis of silver nanoparticles mediated by extracellular pigment from Talaromyces purpurogenus and their biomedical applications. Nanomaterials 2019, 9, 1042. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.S.; Ashok, A.; Edis, Z.; Bloukh, S.H.; Gaikwad, M.; Patil, R.; Pandey, B.; Bhagat, N. Meta-Analysis of Cytotoxicity Studies Using Machine Learning Models on Physical Properties of Plant Extract-Derived Silver Nanoparticles. Int. J. Mol. Sci. 2023, 24, 4220. [Google Scholar] [CrossRef] [PubMed]
- Gul, A.; Shaheen, A.; Ahmad, I.; Khattak, B.; Ahmad, M.; Ullah, R.; Bari, A.; Ali, S.S.; Alobaid, A.; Asmari, M.M. Green synthesis, characterization, enzyme inhibition, antimicrobial potential, and cytotoxic activity of plant mediated silver nanoparticle using Ricinus communis leaf and root extracts. Biomolecules 2021, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Song, M.; Chai, Z.; Wang, X. Enhanced photocatalytic activity of Ag/Ag2Ta4O11/g-C3N4 under wide-spectrum-light irradiation: H2 evolution from water reduction without co-catalyst. J. Colloid Interface Sci. 2019, 550, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Li, T.; Li, J.; Liu, H.; Wang, Y. Geographic identification of Boletus mushrooms by data fusion of FT-IR and UV spectroscopies combined with multivariate statistical analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 198, 257–263. [Google Scholar] [CrossRef]
- Dias, C.; Ayyanar, M.; Amalraj, S.; Khanal, P.; Subramaniyan, V.; Das, S.; Gandhale, P.; Biswa, V.; Ali, R.; Gurav, N. Biogenic synthesis of zinc oxide nanoparticles using mushroom fungus Cordyceps militaris: Characterization and mechanistic insights of therapeutic investigation. J. Drug Deliv. Sci. Technol. 2022, 73, 103444. [Google Scholar] [CrossRef]
- Mousa, H.M.; Hussein, K.H.; Sayed, M.M.; El-Aassar, M.R.; Mohamed, I.M.A.; Kwak, H.-H.; Woo, H.-M.; Abdal-hay, A. Development of biocompatible tri-layered nanofibers patches with endothelial cells for cardiac tissue engineering. Eur. Polym. J. 2020, 129, 109630. [Google Scholar] [CrossRef]
- Mohamed, I.M.A.; Yasin, A.S.; Liu, C. Synthesis, surface characterization and electrochemical performance of ZnO@ activated carbon as a supercapacitor electrode material in acidic and alkaline electrolytes. Ceram. Int. 2020, 46, 3912–3920. [Google Scholar] [CrossRef]
- Rai, M.K.; Deshmukh, S.; Ingle, A.; Gade, A. Silver nanoparticles: The powerful nanoweapon against multidrug-resistant bacteria. J. Appl. Microbiol. 2012, 112, 841–852. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Ivanova, K.; Hoyo, J.; Perelshtein, I.; Owen, G.; Haegert, A.; Lin, Y.-Y.; LeBihan, S.; Gedanken, A.; Hafeli, U.O. Novel lignin-capped silver nanoparticles against multidrug-resistant bacteria. ACS Appl. Mater. Interfaces 2021, 13, 22098–22109. [Google Scholar] [CrossRef]
- Mateo, E.M.; Jiménez, M. Silver Nanoparticle-Based Therapy: Can It Be Useful to Combat Multi-Drug Resistant Bacteria? Antibiotics 2022, 11, 1205. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, P.; Yu, R.; Jiang, J.; Liang, R.; Liu, G. Cost-efficient collagen fibrous aerogel cross-linked by Fe (III)/silver nanoparticle complexes for simultaneously degrading antibiotics, eliminating antibiotic-resistant bacteria, and adsorbing heavy metal ions from wastewater. Sep. Purif. Technol. 2022, 303, 122209. [Google Scholar] [CrossRef]
- More, P.R.; Pandit, S.; Filippis, A.D.; Franci, G.; Mijakovic, I.; Galdiero, M. Silver nanoparticles: Bactericidal and mechanistic approach against drug resistant pathogens. Microorganisms 2023, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yu, Z.; Lin, M.; Mustapha, A. Toxicity of silver nanoparticle incorporated-bacterial nanocellulose to human cells and intestinal bacteria. Int. J. Biol. Macromol. 2023, 241, 124705. [Google Scholar] [CrossRef]
- Ślosarczyk, A.; Klapiszewska, I.; Skowrońska, D.; Janczarek, M.; Jesionowski, T.; Klapiszewski, Ł. A comprehensive review of building materials modified with metal and metal oxide nanoparticles against microbial multiplication and growth. Chem. Eng. J. 2023, 466, 143276. [Google Scholar] [CrossRef]
- Liu, L.; Liang, Z.; Zhou, Y.; Pan, H.; Liu, H. Effect of Amphotericin B on the thermodynamic stability, aggregation state, hemolysis and antifungal activity of Amphotericin B-nonionic surfactant micellar system. J. Mol. Liq. 2023, 376, 121486. [Google Scholar] [CrossRef]
- Panáček, A.; Kolář, M.; Večeřová, R.; Prucek, R.; Soukupová, J.; Kryštof, V.; Hamal, P.; Zbořil, R.; Kvítek, L. Antifungal activity of silver nanoparticles against Candida spp. Biomaterials 2009, 30, 6333–6340. [Google Scholar] [CrossRef]
- Sun, A.; Wang, W.-X. A yeast-based biosensor for silver nanoparticle accumulation and cellular dissolution. Biosens. Bioelectron. 2022, 205, 114082. [Google Scholar] [CrossRef]
- Bonilla, J.J.A.; Guerrero, D.J.P.; Sáez, R.G.T.; Ishida, K.; Fonseca, B.B.; Rozental, S.; López, C.C.O. Green synthesis of silver nanoparticles using maltose and cysteine and their effect on cell wall envelope shapes and microbial growth of Candida spp. J. Nanosci. Nanotechnol. 2017, 17, 1729–1739. [Google Scholar] [CrossRef]
- Rónavári, A.; Bélteky, P.; Boka, E.; Zakupszky, D.; Igaz, N.; Szerencsés, B.; Pfeiffer, I.; Kónya, Z.; Kiricsi, M. Polyvinyl-Pyrrolidone-Coated Silver Nanoparticles—The Colloidal, Chemical, and Biological Consequences of Steric Stabilization under Biorelevant Conditions. Int. J. Mol. Sci. 2021, 22, 8673. [Google Scholar] [CrossRef]
- Kvítek, L.; Panáček, A.; Soukupova, J.; Kolář, M.; Večeřová, R.; Prucek, R.; Holecová, M.; Zbořil, R. Effect of surfactants and polymers on stability and antibacterial activity of silver nanoparticles (NPs). J. Phys. Chem. C 2008, 112, 5825–5834. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Youssef, M.S.; Ahmed, S.I.; Mohamed, I.M.A.; Abdel-Kareem, M.M. Biosynthesis, Spectrophotometric Follow-Up, Characterization, and Variable Antimicrobial Activities of Ag Nanoparticles Prepared by Edible Macrofungi. Biomolecules 2023, 13, 1102. https://doi.org/10.3390/biom13071102
Youssef MS, Ahmed SI, Mohamed IMA, Abdel-Kareem MM. Biosynthesis, Spectrophotometric Follow-Up, Characterization, and Variable Antimicrobial Activities of Ag Nanoparticles Prepared by Edible Macrofungi. Biomolecules. 2023; 13(7):1102. https://doi.org/10.3390/biom13071102
Chicago/Turabian StyleYoussef, Mohamed S., Sanaa Ibrahim Ahmed, Ibrahim M. A. Mohamed, and Marwa M. Abdel-Kareem. 2023. "Biosynthesis, Spectrophotometric Follow-Up, Characterization, and Variable Antimicrobial Activities of Ag Nanoparticles Prepared by Edible Macrofungi" Biomolecules 13, no. 7: 1102. https://doi.org/10.3390/biom13071102
APA StyleYoussef, M. S., Ahmed, S. I., Mohamed, I. M. A., & Abdel-Kareem, M. M. (2023). Biosynthesis, Spectrophotometric Follow-Up, Characterization, and Variable Antimicrobial Activities of Ag Nanoparticles Prepared by Edible Macrofungi. Biomolecules, 13(7), 1102. https://doi.org/10.3390/biom13071102







