Sunflower Hybrids and Inbred Lines Adopt Different Physiological Strategies and Proteome Responses to Cope with Water Deficit
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Proteomics
2.3. Genotyping Procedure
2.4. Statistical Analysis, Modelling and Data Integration
3. Results
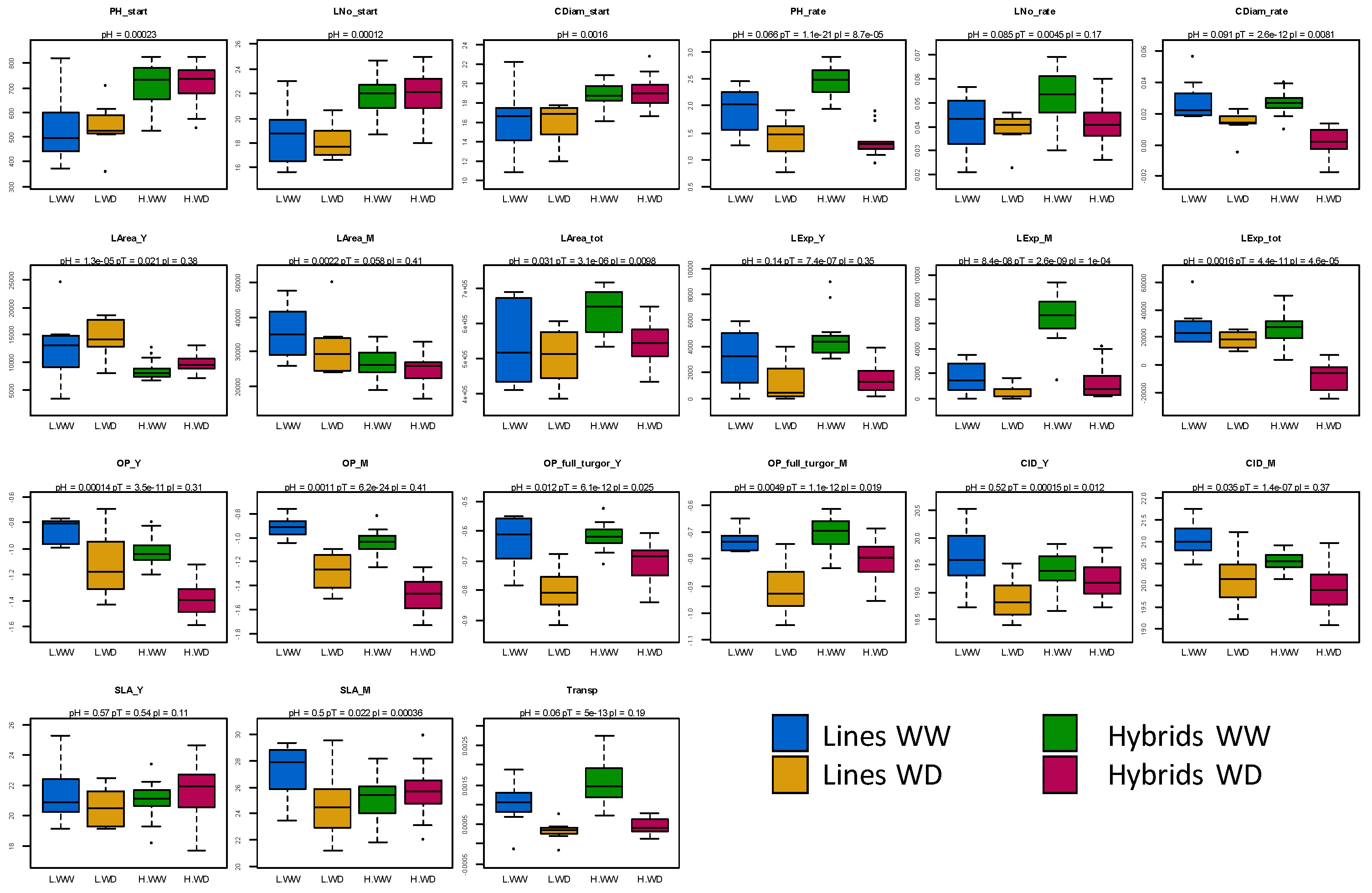
3.1. Large Effect of Water Deficit on Physiological and Growth Traits
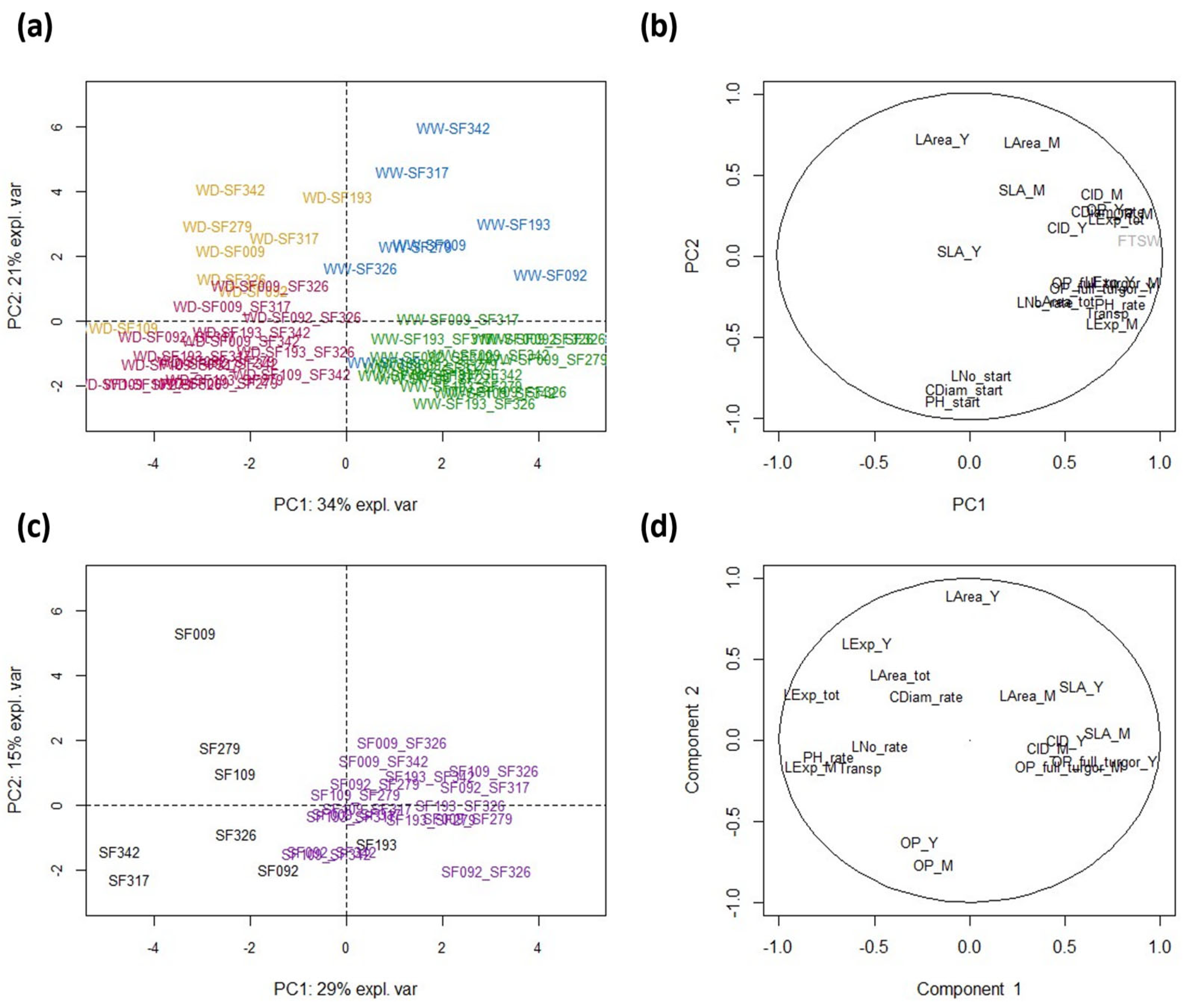
3.2. Hybrids Show Heterosis for Eco-Physiological Traits Measured before Treatment and Differ from Inbred Lines in Their Response to Water Deficit
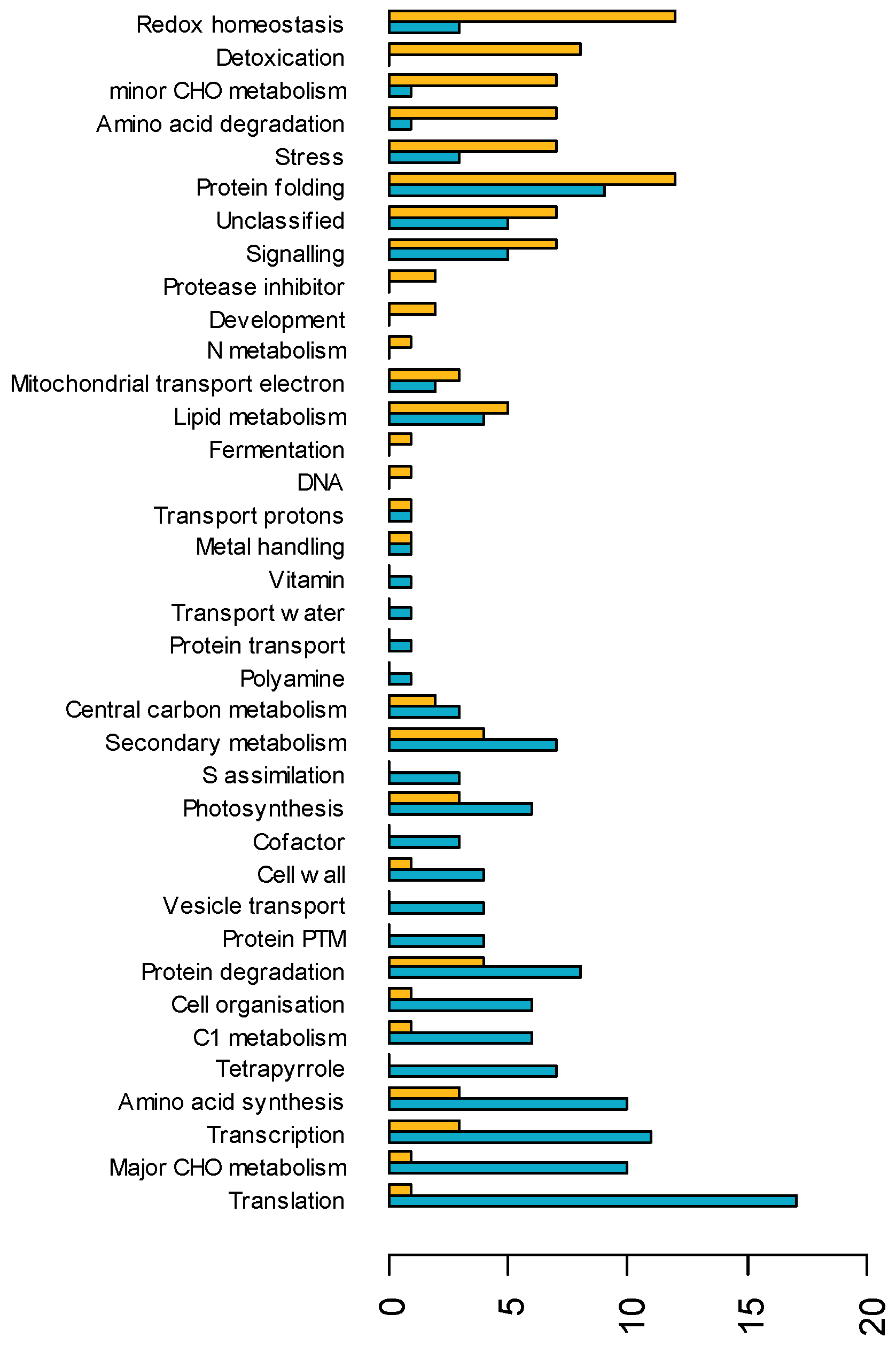
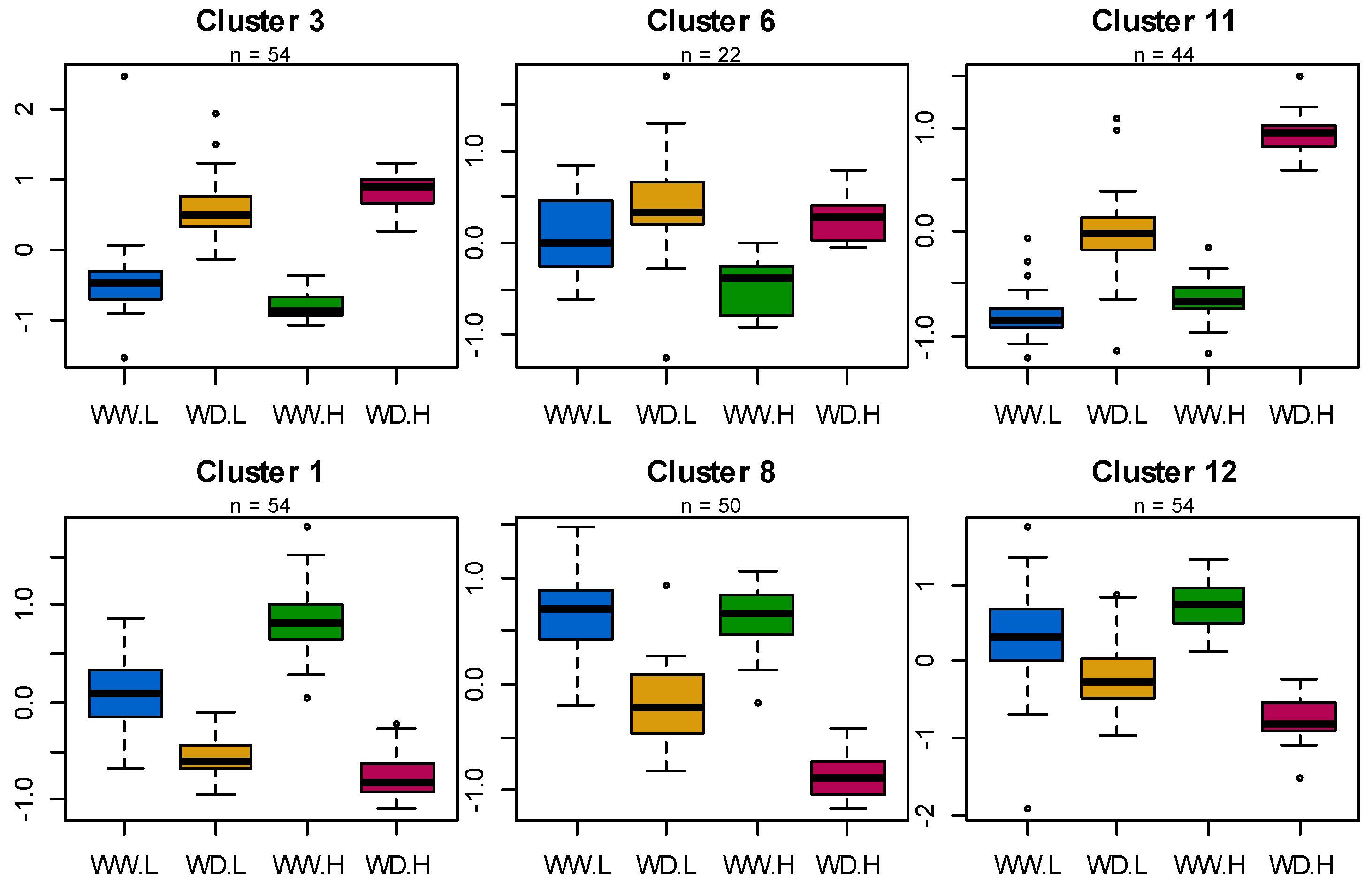
3.3. Distinct Populations of Proteins Are Up- and Downregulated in Sunflower Leaves in Response to Water Deficit
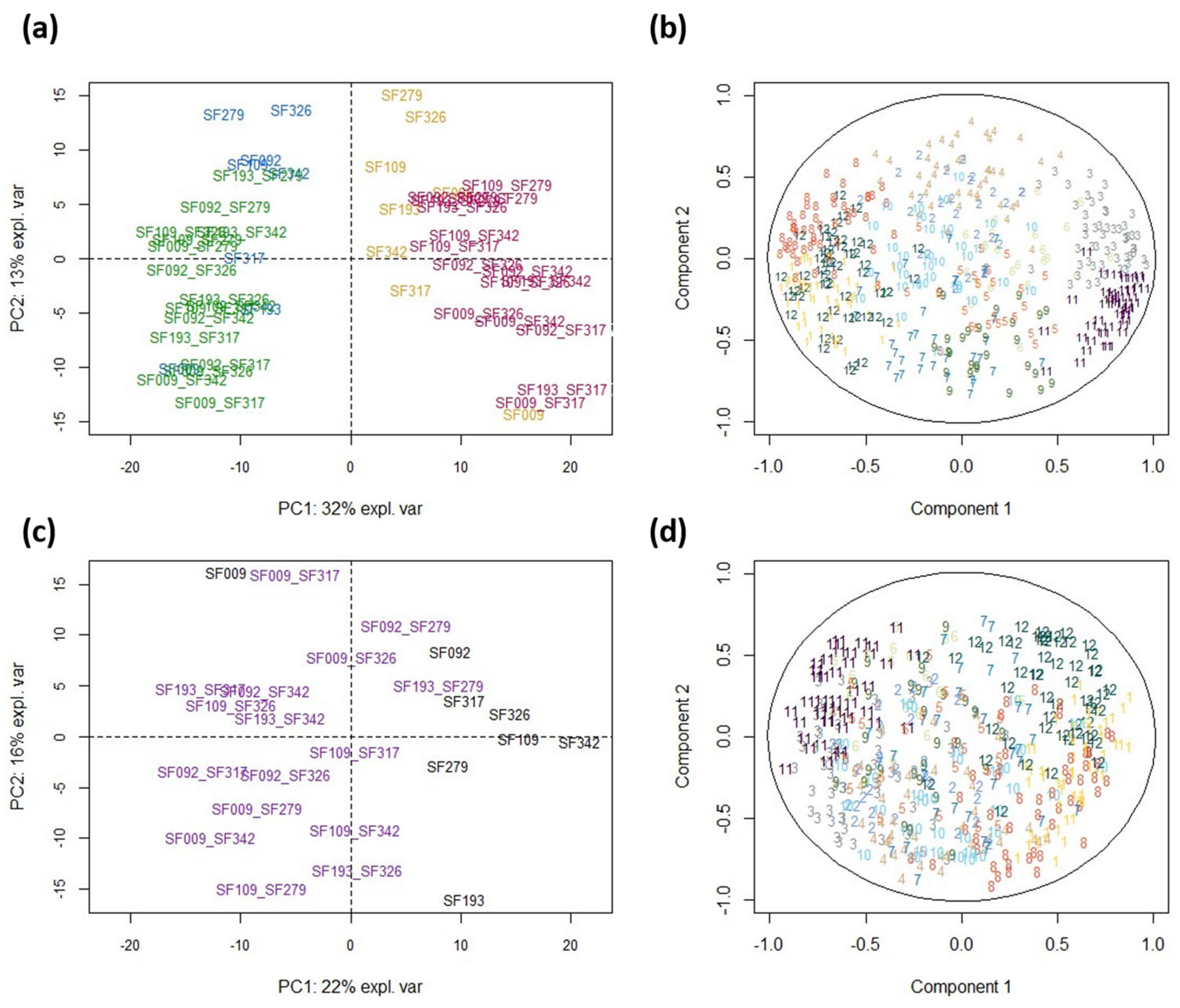
3.4. The Proteome Shows Constitutive and Water Status-Dependent Heterosis
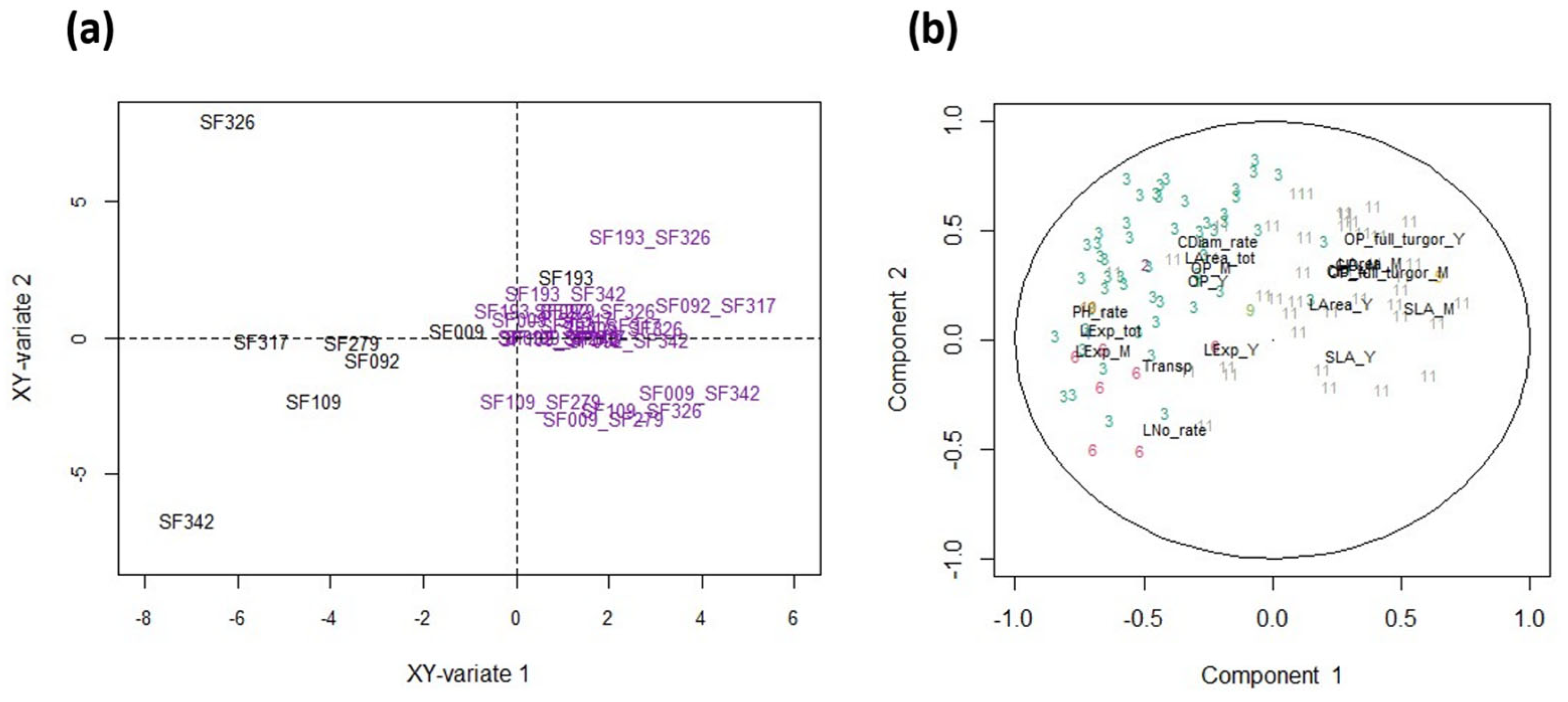
3.5. Co-Variation between Eco-Physiological Traits and Protein Abundances
3.6. The Response of Physiological Traits Is Partly Related to the Proteome under WW Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marcon, C.; Lamkemeyer, T.; Malik, W.A.; Ungrue, D.; Piepho, H.P.; Hochholdinger, F. Heterosis-associated proteome analyses of maize (Zea mays L.) seminal roots by quantitative label-free LC-MS. J. Proteom. 2013, 93, 295–302. [Google Scholar] [CrossRef]
- Blein-Nicolas, M.; Albertin, W.; da Silva, T.; Valot, B.; Balliau, T.; Masneuf-Pomarède, I.; Bely, M.; Marullo, P.; Sicard, D.; Dillmann, C.; et al. A systems approach to elucidate heterosis of protein abundances in yeast. Mol. Cell Proteom. 2015, 14, 2056–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botet, R.; Keurentjes, J.J.B. The role of transcriptional regulation in hybrid vigor. Front. Plant Sci. 2020, 11, 410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, R.C.; Törjék, O.; Becher, M.; Altmann, T. Heterosis of biomass production in arabidopsis. Establishment during early development. Plant Physiol. 2004, 134, 1813–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Coffey, L.; Garfin, J.; Miller, N.D.; White, M.R.; Spalding, E.P.; De Leon, N.; Kaeppler, S.M.; Schnable, P.S.; Springer, N.M.; et al. Genotype-by-environment interactions affecting heterosis in maize. PLoS ONE 2018, 14, e0219528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blum, A. Heterosis, stress, and the environment: A possible road map towards the general improvement of crop yield, J. Exp. Bot. 2013, 64, 4829–4837. [Google Scholar] [CrossRef] [Green Version]
- Debaeke, P.; Casadebaig, P.; Flenet, F.; Langlade, N. Sunflower crop and climate change: Vulnerability, adaptation, and mitigation potential from case-studies in Europe. OCL 2017, 24, D102. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.M.; Rauf, S.; Riaz, M.A.; Al-Khayri, J.M.; Monneveux, P. Determination of drought tolerance related traits in Helianthus argophillus, Helianthus annuus and their hybrids. Breed. Sci. 2017, 67, 257–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- USDA. Oilseeds: World Markets and Trade. [WWW Document] 2020. Available online: https://apps.fas.usda.gov/psdonline/circulars/oilseeds.pdf (accessed on 3 July 2020).
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef] [Green Version]
- Volaire, F. A unified framework of plant adaptive strategies to drought: Crossing scales and disciplines. Glob. Change Biol. 2018, 24, 2929–2938. [Google Scholar] [CrossRef] [Green Version]
- Tardieu, F.; Simonneau, T. Variability among species of stomatal control under fluctuating soil water status and evaporative demand: Modelling isohydric and anisohydric behaviours. J. Exp. Bot. 1998, 49, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Maseda, P.H.; Fernandez, R.J. Stay wet or else: Three ways in which plants can adjust hydraulically to their environment. J. Exp. Bot. 2006, 57, 3963–3977. [Google Scholar] [CrossRef]
- Kiani, S.P.; Grieu, P.; Maury, P.; Hewezi, T.; Gentzbittel, L.; Sarrafi, A. Genetic variability for physiological traits under drought conditions and differential expression of water stress-associated genes in sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2007, 114, 193–207. [Google Scholar] [CrossRef]
- Fulda, S.; Mikkat, S.; Stegmann, H.; Horn, R. Physiology and proteomics of drought stress acclimation in sunflower (Helianthus annuus L.). Plant Biol. 2011, 13, 632–642. [Google Scholar] [CrossRef] [PubMed]
- Rengel, D.; Arribat, S.; Maury, P.; Martin-Magniette, M.L.; Hourlier, T.; Laporte, M.; Varès, D.; Carrère, S.; Grieu, P.; Balzergue, S.; et al. A gene-phenotype network based on genetic variability for drought responses reveals key physiological processes in controlled and natural environments. PLoS ONE 2012, 7, e45249. [Google Scholar] [CrossRef] [PubMed]
- Moschen, S.; Di Rienzo, J.A.; Higgins, J.; Tohge, T.; Watanabe, M.; González, S.; Rivarola, M.; García-García, F.; Dopazo, J.; Hopp, H.E.; et al. Integration of transcriptomic and metabolic data reveals hub transcription factors involved in drought stress response in sunflower (Helianthus annuus L.). Plant Mol. Biol. 2017, 94, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.; Toorchi, M.; Valizadeh, M.; Shakiba, M. Proteomic prospects for tolerance of sunflower (Helianthus annuus) to drought stress during the flowering stage. Crop Pasture Sci. 2017, 68, 457. [Google Scholar] [CrossRef]
- Liang, C.; Wang, W.; Wang, J.; Ma, J.; Li, C.; Zhou, F.; Zhang, S.; Yu, Y.; Zhang, L.; Li, W.; et al. Identification of differentially expressed genes in sunflower (Helianthus annuus) leaves and roots under drought stress by RNA sequencing. Bot. Stud. 2017, 58, 42. [Google Scholar] [CrossRef]
- Fernandez, O.; Urrutia, M.; Berton, T.; Bernillon, S.; Deborde, C.; Jacob, D.; Maucourt, M.; Maury, P.; Duruflé, H.; Gibon, Y.; et al. Metabolomic characterization of sunflower leaf allows discriminating genotype groups or stress levels with a minimal set of metabolic markers. Metabolomics 2019, 15, 56. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, Y.; Shi, H.; Hu, H.; Yi, L.; Hou, J. Time-course transcriptome and WGCNA analysis revealed the drought response mechanism of two sunflower inbred lines. PLoS ONE 2022, 17, e0265447. [Google Scholar] [CrossRef]
- Mojayad, F.; Planchon, C. Stomatal and photosynthetic adjustment to water deficit as the expression of heterosis in sunflower. Crop Sci. 1994, 34, 103–107. [Google Scholar] [CrossRef]
- Cadic, E.; Coque, M.; Vear, F.; Grezes-Besset, B.; Pauquet, J.; Piquemal, J.; Lippi, Y.; Blanchard, P.; Romestant, M.; Pouilly, N.; et al. Combined linkage and association mapping of flowering time in Sunflower (Helianthus annuus L.). Theor. Appl. Genet. 2013, 126, 1337–1356. [Google Scholar] [CrossRef] [PubMed]
- Mangin, B.; Casadebaig, P.; Cadic, E.; Blanchet, N.; Boniface, M.C.; Carrère, S.; Gouzy, J.; Legrand, L.; Mayjonade, B.; Pouilly, N.; et al. Genetic control of plasticity of oil yield for combined abiotic stresses using a joint approach of crop modelling and genome-wide association. Plant Cell Environ. 2017, 40, 2276–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanchet, N.; Casadebaig, P.; Debaeke, P.; Duruflé, H.; Gody, L.; Gosseau, F.; Langlade, N.B.; Maury, P. Data describing the eco-physiological responses of twenty-four sunflower genotypes to water deficit. Data Brief 2018, 21, 1296–1301. [Google Scholar] [CrossRef] [PubMed]
- Gosseau, F.; Blanchet, N.; Varès, D.; Burger, P.; Campergue, D.; Colombet, C.; Gody, L.; Liévin, J.F.; Mangin, B.; Tison, G.; et al. Heliaphen, an outdoor high-throughput phenotyping platform for genetic studies and crop modeling. Front. Plant Sci. 2019, 9, 1908. [Google Scholar] [CrossRef] [Green Version]
- Terzić, S.; Boniface, M.C.; Marek, L.; Alvarez, D.; Baumann, K.; Gavrilova, V.; Joita-Pacureanu, M.; Sujatha, M.; Valkova, D.; Velasco, L.; et al. Gene banks for wild and cultivated sunflower genetic resources. OCL 2020, 27, 9. [Google Scholar] [CrossRef] [Green Version]
- Leclercq, P. Une stérilité mâle cytoplasmique chez le tournesol. Ann. Amelior. Plantes 1969, 19, 99–106. [Google Scholar]
- Schneiter, A.A.; Miller, J.F. Description of sunflower growth stages. Crop Sci. 1981, 21, 901–903. [Google Scholar] [CrossRef]
- Marchand, G.; Mayjonade, B.; Varès, D.; Blanchet, N.; Boniface, M.C.; Maury, P.; Andrianasolo, F.N.; Burger, P.; Debaeke, P.; Casadebaig, P.; et al. A biomarker based on gene expression indicates plant water status in controlled and natural environments. Plant Cell Environ. 2013, 36, 2175–2189. [Google Scholar] [CrossRef] [PubMed]
- Andrianasolo, F.N.; Casadebaig, P.; Langlade, N.; Debaeke, P.; Maury, P. Effects of plant growth stage and leaf againg on the response of transpiration and photosynthesis to water deficit in sunflower. Funct. Plant Biol. 2016, 43, 797–805. [Google Scholar] [CrossRef]
- Sinclair, T.R. Theoretical analysis of soil and plant traits influencing daily plant water flux on drying soils. Agron. J. 2005, 97, 1148–1152. [Google Scholar] [CrossRef]
- Balliau, T.; Blein-Nicolas, M.; Zivy, M. Evaluation of Optimized Tube-Gel Methods of Sample Preparation for Large-Scale Plant Proteomics. Proteomes 2018, 6, 6. [Google Scholar] [CrossRef] [Green Version]
- Balliau, T.; Duruflé, H.; Blanchet, N.; Blein-Nicolas, M.; Langlade, N.B.; Zivy, M. Proteomic data from leaves of twenty-four sunflower genotypes underwater deficit. OCL 2021, 28, 12. [Google Scholar] [CrossRef]
- Méchin, V.; Damerval, C.; Zivy, M. Total protein extraction with TCA-acetone. Methods Mol. Biol. 2007, 355, 1–8. [Google Scholar] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [Green Version]
- Craig, R.; Cortens, J.P.; Beavis, R.C. Open source system for analyzing, validating, and storing protein identification data. J. Proteome Res. 2004, 3, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Langella, O.; Valot, B.; Balliau, T.; Blein-Nicolas, M.; Bonhomme, L.; Zivy, M. X!TandemPipeline: A tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res. 2017, 16, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Valot, B.; Langella, O.; Nano, E.; Zivy, M. MassChroQ: A versatile tool for mass spectrometry quantification. Proteomics 2011, 11, 3572–3577. [Google Scholar] [CrossRef] [Green Version]
- Millan-Oropeza, A.; Henry, C.; Blein-Nicolas, M.; Aubert-Frambourg, A.; Moussa, F.; Bleton, J.; Virolle, M.J. Quantitative proteomics analysis confirmed oxidative metabolism predominates in Streptomyces coelicolor versus glycolytic metabolism in Streptomyces lividans. J. Proteome Res. 2017, 16, 2597–2613. [Google Scholar] [CrossRef]
- Belouah, I.; Blein-Nicolas, M.; Balliau, T.; Gibon, Y.; Zivy, M.; Colombié, S. Peptide filtering differently affects the performances of XIC-based quantification methods. J. Proteom. 2019, 193, 131–141. [Google Scholar] [CrossRef]
- Pecrix, Y.; Penouilh-Suzette, C.; Muños, S.; Vear, F.; Godiard, L. Ten broad spectrum resistances to downy mildew physically mapped on the sunflower genome. Front. Plant Sci. 2018, 9, 1780. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, X.; Xiong, W.; Ye, T.; Wu, Y. Overexpression of the aspartic protease ASPG1 gene confers drought avoidance in Arabidopsis. J. Exp. Bot. 2012, 63, 2579–2593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bown, A.W.; Shelp, B.J. Plant GABA: Not just a metabolite. Trends Plant Sci. 2016, 21, 811–813. [Google Scholar] [CrossRef]
- Deuschle, K.; Funck, D.; Hellmann, H.; Däschner, K.; Binder, S.; Frommer, W.B. A nuclear gene encoding mitochondrial Δ1-pyrroline-5-carboxylate dehydrogenase and its potential role in protection from proline toxicity. Plant J. 2001, 27, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Orians, C.M.; Schweiger, R.; Dukes, J.S.; Scott, E.R.; Müller, C. Combined impacts of prolonged drought and warming on plant size and foliar chemistry. Ann. Bot. 2019, 124, 41–52. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, J.; Li, C.; Zhang, Z.; Ma, F.; Li, M. Response of sugar metabolism in apple leaves subjected to short-term drought stress. Plant Physiol. Biochem. 2019, 141, 164–171. [Google Scholar] [CrossRef]
- Thomas, B.R.; Inouhe, M.; Simmons, C.R.; Nevins, D.J. Endo-1,3;1,4-beta-glucanase from coleoptiles of rice and maize: Role in the regulation of plant growth. Int. J. Biol. Macromol. 2000, 27, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Orians, G.H.; Solbrig, O.T. A cost-income model of leaves and roots with special reference to arid and semiarid areas. Am. Nat. 1977, 111, 677–690. [Google Scholar] [CrossRef]
- Hernández, E.I.; Pausas, J.G.; Vilagrosa, A. Leaf physiological traits in relation to resprouter ability in the Mediterranean Basin. Plant Ecol. 2011, 212, 1959–1966. [Google Scholar] [CrossRef]
- Moreno-Gutiérrez, C.; Dawson, T.E.; Nicolás, E.; Querejeta, J.I. Isotopes reveal contrasting water use strategies among coexisting plant species in a mediterranean ecosystem. New Phytol. 2012, 196, 489–496. [Google Scholar] [CrossRef]
- Schultz, H.R. Differences in hydraulic architecture account for near-isohydric and anisohydric behaviour of two field-grown Vitis vinifera L. cultivars during drought. Plant Cell Environ. 2003, 26, 1393–1405. [Google Scholar] [CrossRef]
- Attia, Z.; Domec, J.C.; Oren, R.; Way, D.A.; Moshelion, M. Growth and physiological responses of isohydric and anisohydric poplars to drought. J. Exp. Bot. 2015, 66, 4373–4381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birdseye, D.; de Boer, L.A.; Bai, H.; Zhou, P.; Shen, Z.; Schmelz, E.A.; Springer, N.M.; Briggs, S.P. Plant height heterosis is quantitatively associated with expression levels of plastid ribosomal proteins. Proc. Natl. Acad. Sci. USA 2021, 118, e2109332118. [Google Scholar] [CrossRef] [PubMed]
- Mohayeji, M.; Capriotti, A.L.; Cavaliere, C.; Piovesana, S.; Samperi, R.; Stampachiacchiere, S.; Toorchi, M.; Lagana, A. Heterosis profile of sunflower leaves: A label free proteomics approach. J. Proteom. 2014, 99, 101–110. [Google Scholar] [CrossRef]
- Xing, J.; Sun, Q.; Ni, Z. Proteomic patterns associated with heterosis. Biochim. Biophys. Acta Proteins Proteom. 2016, 1864, 908–915. [Google Scholar] [CrossRef]
- Rockenbach, M.F.; Corrêa, C.C.G.; Heringer, A.S.; Freitas, I.L.J.; Santa-Catarina, C.; Do Amaral, A.T.; Silveira, V. Differentially abundant proteins associated with heterosis in the primary roots of popcorn. PLoS ONE 2018, 13, e0197114. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef]
- Todesco, M.; Balasubramanian, S.; Hu, T.T.; Traw, M.B.; Horton, M.; Epple, P.; Kuhns, C.; Sureshkumar, S.; Schwartz, C.; Lanz, C.; et al. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature 2010, 465, 632–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajpai, P.K.; Reichelt, M.; Augustine, R.; Gershenzon, J.; Bisht, N.C. Heterotic patterns of primary and secondary metabolites in the oilseed crop Brassica juncea. Heredity 2019, 123, 318–336. [Google Scholar] [CrossRef] [PubMed]
- Groszmann, M.; Gonzalez-Bayon, R.; Lyons, R.L.; Greaves, I.K.; Kazan, K.; Peacock, W.J.; Dennis, E.S. Hormone-regulated defense and stress response networks contribute to heterosis in Arabidopsis F1 hybrids. Proc. Natl. Acad. Sci. USA 2015, 112, E6397–E6406. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.; Song, Q.; Shi, X.; Juenger, T.E.; Chen, Z.J. Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat. Comm. 2015, 6, 7453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiévet, J.B.; Nidelet, T.; Dillmann, C.; de Vienne, D. Heterosis is a systemic property emerging from non-linear genotype-phenotype relationships: Evidence from in vitro genetics and computer simulations. Front. Genet. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasseur, F.; Fouqueau, L.; De Vienne, D.; Nidelet, T.; Violle, C.; Weigel, D. Nonlinear phenotypic variation uncovers the emergence of heterosis in Arabidopsis thaliana. PLoS Biol. 2019, 17, 3000214. [Google Scholar] [CrossRef] [Green Version]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 22, 2105–2112. [Google Scholar] [CrossRef] [Green Version]
- Goff, S.A. A unifying theory for general multigenic heterosis: Energy efficiency, protein metabolism, and implications for molecular breeding. New Phytol. 2011, 189, 923–937. [Google Scholar] [CrossRef]
- Lee, J.S.; Jahani, M.; Huang, K.; Mandel, J.R.; Marek, L.F.; Burke, J.M.; Langlade, N.B.; Owens, G.L.; Rieseberg, L.H. Expression complementation of gene presence/absence polymorphisms in hybrids contributes importantly to heterosis in sunflower. J. Adv. Res. 2022, 42, 83–98. [Google Scholar] [CrossRef]

| Category | Level of Organisation | Trait | Abbreviation | Unit | Measurement Time |
|---|---|---|---|---|---|
| Water deficit experienced by the plant | Plant | Fraction of transpirable soil water | FTSW | (N/A) | End of exp. |
| Morphology | Plant | Plant height | PH_start | (mm. plant−1) | 37 DAG |
| Number of leaves | LNo_start | (plant−1) | 37 DAG | ||
| Collar diameter | CDiam_start | (mm. plant−1) | 37 DAG | ||
| Leaf area | LArea_tot | (mm2. plant−1) | End of exp. | ||
| Leaf | Mature−leaf area | LArea_M | (mm2) | End of exp. | |
| Young−leaf area | LArea_Y | (mm2) | End of exp. | ||
| Mature Specific leaf area | SLA_M | (m2. kg−1) | End of exp. | ||
| Young Specific leaf area | SLA_Y | (m2. kg−1) | End of exp. | ||
| Growth | Plant | Plant height growth rate | PH_rate | (mm. °Cd−1) | From 35 DAG to the end of exp. |
| Rate of leaf appearance | LNo_rate | (leaves. °Cd−1) | From 35 DAG to the end of exp. | ||
| Collar diameter growth rate | CDiam_rate | (mm. °Cd−1) | From 35 DAG to the end of exp. | ||
| Leaf expansion rate | LExp_tot | (mm2. d−1) | Last 24 h of the exp | ||
| Leaf | Mature-leaf expansion rate | LExp_M | (mm2. d−1) | Last 24 h of the exp. | |
| Young-leaf expansion rate | LExp_Y | (mm2. d−1) | Last 24 h of the exp. | ||
| Physiology | Plant | Plant transpiration rate | Transp | (g. mm−2. d−1) | Last 24 h of the exp. |
| Leaf | Osmotic potential of mature leaf | OP_M | (MPa) | End of exp. | |
| Osmotic potential of young leaf | OP_Y | (MPa) | End of exp. | ||
| Osmotic potential at full turgor of mature leaf | OP_full_turgor_M | (MPa) | End of exp. | ||
| Osmotic potential at full turgor of young leaf | OP_full_turgor_Y | (MPa) | End of exp. | ||
| Carbon isotope discrimination of mature leaf | CID_M | (‰) | End of exp. | ||
| Carbon isotope discrimination of young leaf | CID_Y | (‰) | End of exp. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duruflé, H.; Balliau, T.; Blanchet, N.; Chaubet, A.; Duhnen, A.; Pouilly, N.; Blein-Nicolas, M.; Mangin, B.; Maury, P.; Langlade, N.B.; et al. Sunflower Hybrids and Inbred Lines Adopt Different Physiological Strategies and Proteome Responses to Cope with Water Deficit. Biomolecules 2023, 13, 1110. https://doi.org/10.3390/biom13071110
Duruflé H, Balliau T, Blanchet N, Chaubet A, Duhnen A, Pouilly N, Blein-Nicolas M, Mangin B, Maury P, Langlade NB, et al. Sunflower Hybrids and Inbred Lines Adopt Different Physiological Strategies and Proteome Responses to Cope with Water Deficit. Biomolecules. 2023; 13(7):1110. https://doi.org/10.3390/biom13071110
Chicago/Turabian StyleDuruflé, Harold, Thierry Balliau, Nicolas Blanchet, Adeline Chaubet, Alexandra Duhnen, Nicolas Pouilly, Mélisande Blein-Nicolas, Brigitte Mangin, Pierre Maury, Nicolas Bernard Langlade, and et al. 2023. "Sunflower Hybrids and Inbred Lines Adopt Different Physiological Strategies and Proteome Responses to Cope with Water Deficit" Biomolecules 13, no. 7: 1110. https://doi.org/10.3390/biom13071110





