Effect of Metformin Treatment on Serum Metabolic Profile Changes in Lean and Obese Zucker Rat Model for Fatty Liver Disease
Abstract
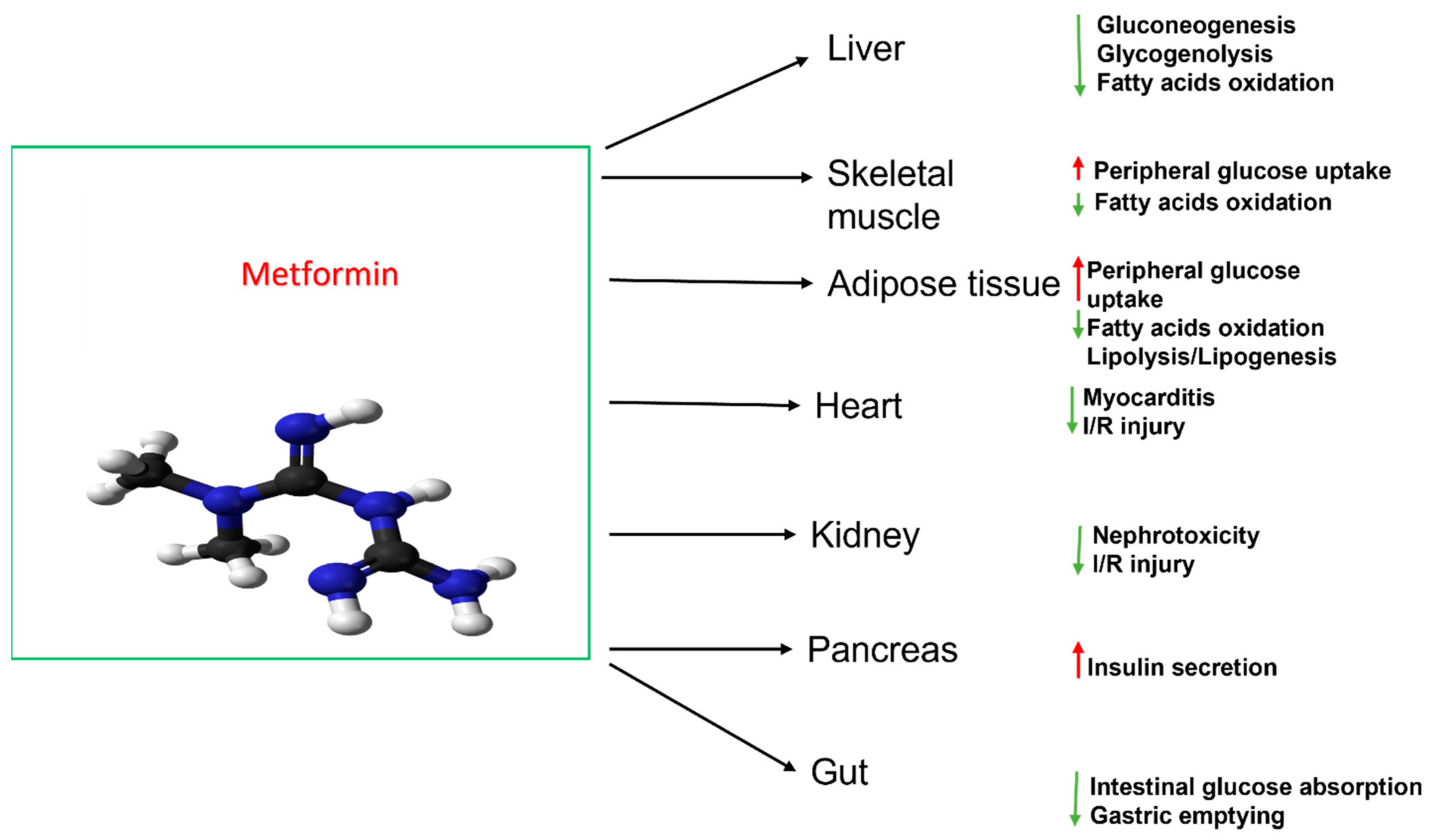
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Preparation for Measurement of Serum Metabolites
2.3. HPLC and LC-MS Methods
2.3.1. HPLC-ECD
2.3.2. HPLC-UV
2.3.3. LC-MS
2.4. Data Processing and Statistical Analysis
3. Results
3.1. Body Weight
3.2. Untargeted Metabolomics Data
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lustig, R.H.; Collier, D.; Kassotis, C.; Roepke, T.A.; Kim, M.J.; Blanc, E.; Barouki, R.; Bansal, A.; Cave, M.C.; Chatterjee, S.; et al. Obesity I: Overview and molecular and biochemical mechanisms. Biochem. Pharmacol. 2022, 199, 115012. [Google Scholar] [CrossRef] [PubMed]
- Engin, A. The Definition and Prevalence of Obesity and Metabolic Syndrome. Adv. Exp. Med. Biol. 2017, 960, 1–17. [Google Scholar] [CrossRef]
- Ala, M.; Ala, M. Metformin for Cardiovascular Protection, Inflammatory Bowel Disease, Osteoporosis, Periodontitis, Polycystic Ovarian Syndrome, Neurodegeneration, Cancer, Inflammation and Senescence: What Is Next? ACS Pharmacol. Transl. Sci. 2021, 4, 1747–1770. [Google Scholar] [CrossRef] [PubMed]
- Guh, D.P.; Zhang, W.; Bansback, N.; Amarsi, Z.; Birmingham, C.L.; Anis, A.H. The incidence of co-morbidities related to obesity and overweight: A systematic review and meta-analysis. BMC Public Health 2009, 9, 88. [Google Scholar] [CrossRef] [Green Version]
- Solis-Herrera, C.; Triplitt, C.; Cersosimo, E.; DeFronzo, R.A. Pathogenesis of Type 2 Diabetes Mellitus. In Endotext; Internet; MDText.com, Inc.: South Dartmouth, MA, USA, 2021. [Google Scholar]
- Aras, M.; Tchang, B.G.; Pape, J. Obesity and Diabetes. Nurs. Clin. N. Am. 2021, 56, 527–541. [Google Scholar] [CrossRef]
- Apovian, C.M. Obesity: Definition, comorbidities, causes, and burden. Am. J. Manag. Care 2016, 22 (Suppl. S7), S176–S185. [Google Scholar] [PubMed]
- Khodadadi, M.; Jafari-Gharabaghlou, D.; Zarghami, N. An update on mode of ac-tion of metformin in modulation of meta-inflammation and inflammaging. Pharmacol. Rep. 2022, 74, 310–322. [Google Scholar] [CrossRef]
- Clish, C.B. Metabolomics: An emerging but powerful tool for precision medicine. Cold Spring Harb. Mol. Case Stud. 2015, 1, a000588. [Google Scholar] [CrossRef] [Green Version]
- Costanzo, M.; Caterino, M.; Sotgiu, G.; Ruoppolo, M.; Franconi, F.; Campesi, I. Sex differences in the human metabolome. Biol. Sex Differ. 2022, 13, 30. [Google Scholar] [CrossRef]
- Kim, H.W. Metabolomic Approaches to Investigate the Effect of Metformin: An Overview. Int. J. Mol. Sci. 2021, 22, 10275. [Google Scholar] [CrossRef]
- Xie, B.; Waters, M.J.; Schirra, H.J. Investigating Potential Mechanisms of Obesity by Metabolomics. J. Biomed. Biotechnol. 2012, 805683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazza, A.; Fruci, B.; Garinis, G.A.; Giuliano, S.; Malaguarnera, R.; Belfiore, A. The role of metformin in the management of NAFLD. Exp. Diabetes Res. 2012, 2012, 716404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, K.H.; Lee, C.H.; Cheng, Y.D.; Gau, S.Y.; Tsai, T.H.; Chung, N.J.; Lee, C.Y. Correlation between long-term use of metformin and incidence of NAFLD among patients with type 2 diabetes mellitus: A real-world cohort study. Front. Endocrinol. 2022, 13, 1027484. [Google Scholar] [CrossRef] [PubMed]
- Hakkak, R.; Rose, S.; Spray, B.; Kozaczek, M.; Korourian, S. Effects of Obesity and 10 Weeks of Metformin Treatment on Liver Steatosis. J. Biomed. Rep. 2021, 14, 49. [Google Scholar] [CrossRef]
- Robeson, M.; Manna, K.; Randolph, C.; Byrum, S.; Hakkak, R. Short-term Metformin Treatment En-riches Bacteroides dorei in Obese Liver Steatosis Zucker Rat Model. Front. Microbiol. 2021, 13, 834776. [Google Scholar] [CrossRef]
- Zucker, T.F.; Zucker, L.M. Fat accretion and growth in the rat. J. Nutr. 1963, 80, 6–19. [Google Scholar] [CrossRef]
- Zucker, L.M.; Zucker, T.F. Fatty, a new mutation in the rat. J. Hered. 1961, 52, 275–278. [Google Scholar] [CrossRef]
- Chua, S.C.; Chung, W.K.; Wu-Peng, X.S.; Zhang, Y.; Liu, S.M.; Tartaglia, L.; Leibel, R.L. Phenotypes of mouse ‘diabetes’ and rat ‘fatty’ due to mutations in the OB (leptin) receptor. Science 1996, 271, 994–996. [Google Scholar] [CrossRef]
- Tartaglia, L.A.; Dembski, M.; Weng, X.; Deng, N.; Culpepper, J.; Devos, R.; Richards, G.J.; Campfield, L.A.; Clark, F.T.; Deeds, J.; et al. Identification and expression cloning of a leptin receptor, OB-R. Cell 1995, 83, 1263–1271. [Google Scholar] [CrossRef] [Green Version]
- Zucker, L.M.; Antoniades, H.N. Insulin and obesity in the Zucker genetically obese rat “fatty”. Endocrinology 1971, 90, 1320–1330. [Google Scholar] [CrossRef]
- Melnyk, S.; Hakkak, R. Metabolic Status of Lean and Obese Zucker Rats Based on Untarget-ed and Targeted Metabolomics Analysis of Serum. Biomedicines 2022, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, S.; Pogribna, M.; Pogribny, I.; Hine, R.J.; James, S.J. A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection. J. Nutr. Biochem. 1999, 10, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Melnyk, S.; Pogribna, M.; Pogribny, I.P.; Yi, P.; James, S.J. Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: Alter-ations with plasma homocysteine and pyridoxal 5′-phosphate concentrations. Clin. Chem. 2000, 46, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Chen, Y.T.; Yang, Y.X.; Shou, D.; Li, C.Y. Urinary Metabolomic Profiling in Zucker Diabetic Fatty Rats with Type 2 Diabetes Mellitus Treated with Glimepiride, Metformin, and Their Combination. Molecules 2016, 21, 1446. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Teng, Y.; Li, S.; Li, Y.; Li, H.; Jiao, L.; Wu, W. UHPLC-MS-Based Serum and Urine Metabolomics Reveals the Anti-Diabetic Mechanism of Ginsenoside Re in Type 2 Diabetic Rats. Molecules 2021, 26, 6657. [Google Scholar] [CrossRef]
- Sambe, T.; Mason, R.P.; Dawoud, H.; Bhatt, D.L.; Malinski, T. Metformin treatment decreases nitroxidative stress, restores nitric oxide bioavailability and endothelial function beyond glucose control. Biomed. Pharmacother. 2018, 98, 149–156. [Google Scholar] [CrossRef]
- Shoghi, K.I.; Finck, B.N.; Schechtman, K.B.; Sharp, T.; Herrero, P.; Gropler, R.J.; Welch, M.J. In vivo metabolic phenotyping of myocardial substrate metabolism in rodents: Differential efficacy of metformin and rosig-litazone monotherapy. Circ. Cardiovasc. Imaging 2009, 2, 373–381. [Google Scholar] [CrossRef] [Green Version]
- Triggle, C.R.; Mohammed, I.; Bshesh, K.; Marei, I.; Ye, K.; Ding, H.; MacDonald, R.; Hollenberg, M.D.; Hill, M.A. Metformin: Is it a drug for all reasons and diseases? Metabolism 2022, 133, 155223. [Google Scholar] [CrossRef]
- Sui, Y.; Kong, X.; Fan, R.; Ye, Y.; Mai, H.; Zhuo, S.; Lu, W.; Ruan, P.; Fang, S.; Yang, T. Long-term treatment with metformin in the prevention of fatty liver in Zucker diabetic fatty rats. Diabetol. Metab. Syndr. 2019, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Cwynar-Zając, Ł. Metformin—A new approach. Pediatr. Endocrinol. Diabetes Metab. 2021, 27, 134–140. [Google Scholar] [CrossRef]
- Zhou, J.; Massey, S.; Story, D.; Li, L. Metformin: An Old Drug with New Applications. Int. J. Mol. Sci. 2018, 19, 2863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.P.; Go, Y.-M.; Anderson, C.L.; Ziegler, T.R.; Kinkade, J.M., Jr.; Kirlin, W.G. Cysteine/cystine couple is a newly recognized node in the circuitry for biologic redox signaling and control. FASEB J. 2004, 18, 1246–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
 Statistically higher (p < 0.05);
Statistically higher (p < 0.05);  Statistically lower (p < 0.05);
Statistically lower (p < 0.05);  Marginal (0.1 > p > 0.05);
Marginal (0.1 > p > 0.05);  No difference (p > 0.05).
No difference (p > 0.05).
 Statistically higher (p < 0.05);
Statistically higher (p < 0.05);  Statistically lower (p < 0.05);
Statistically lower (p < 0.05);  Marginal (0.1 > p > 0.05);
Marginal (0.1 > p > 0.05);  No difference (p > 0.05).
No difference (p > 0.05).
 p < 0.05,
p < 0.05,  0.1 > p > 0.05,
0.1 > p > 0.05,  p > 0.05.
p > 0.05.
 p < 0.05,
p < 0.05,  0.1 > p > 0.05,
0.1 > p > 0.05,  p > 0.05.
p > 0.05.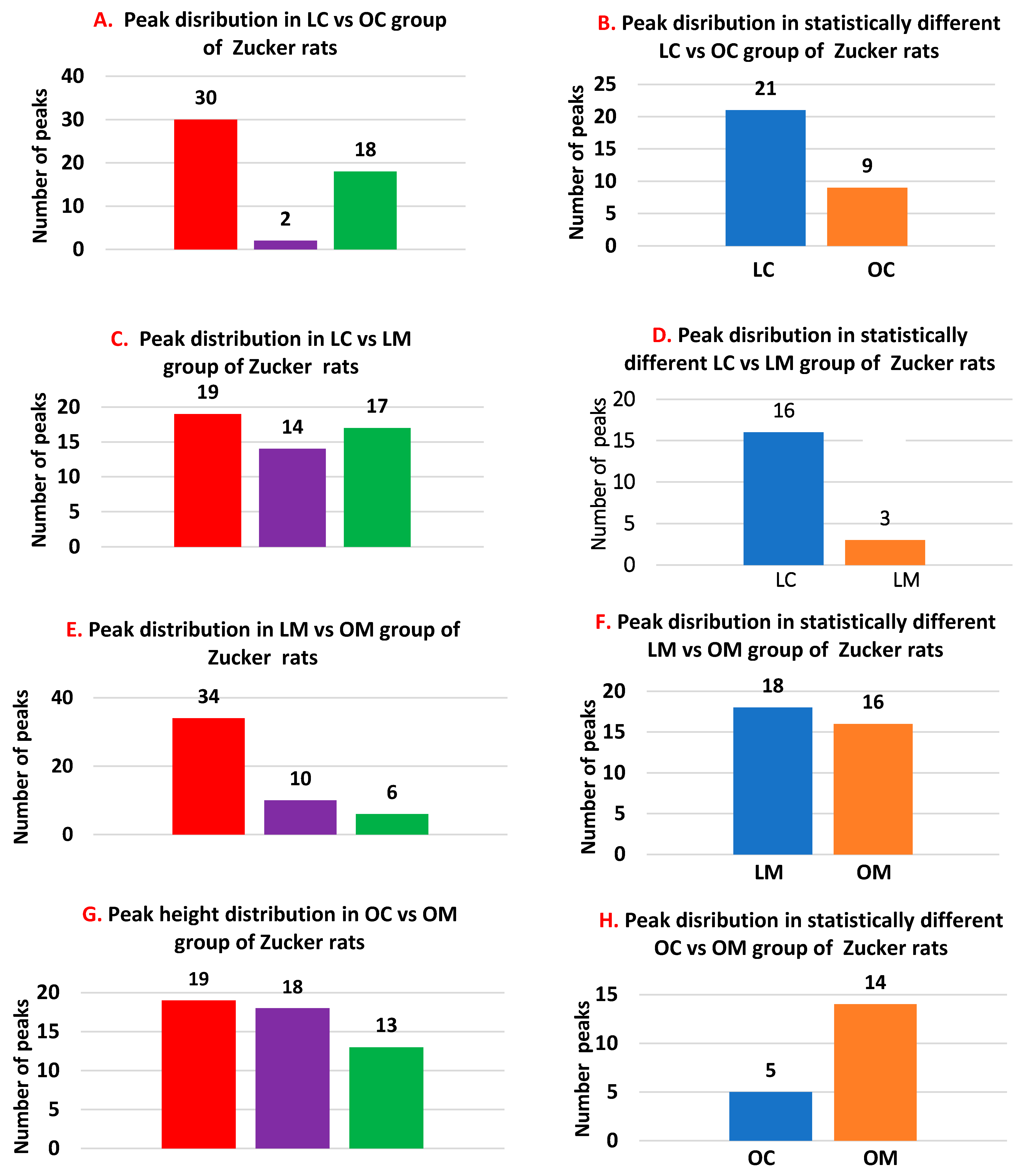
 Peaks more pronounced in obese animals.
Peaks more pronounced in obese animals.  Peaks more pronounced in lean animals.
Peaks more pronounced in lean animals.  Peak that are similar among lean and obese animals.
Peak that are similar among lean and obese animals.
 Peaks more pronounced in obese animals.
Peaks more pronounced in obese animals.  Peaks more pronounced in lean animals.
Peaks more pronounced in lean animals.  Peak that are similar among lean and obese animals.
Peak that are similar among lean and obese animals.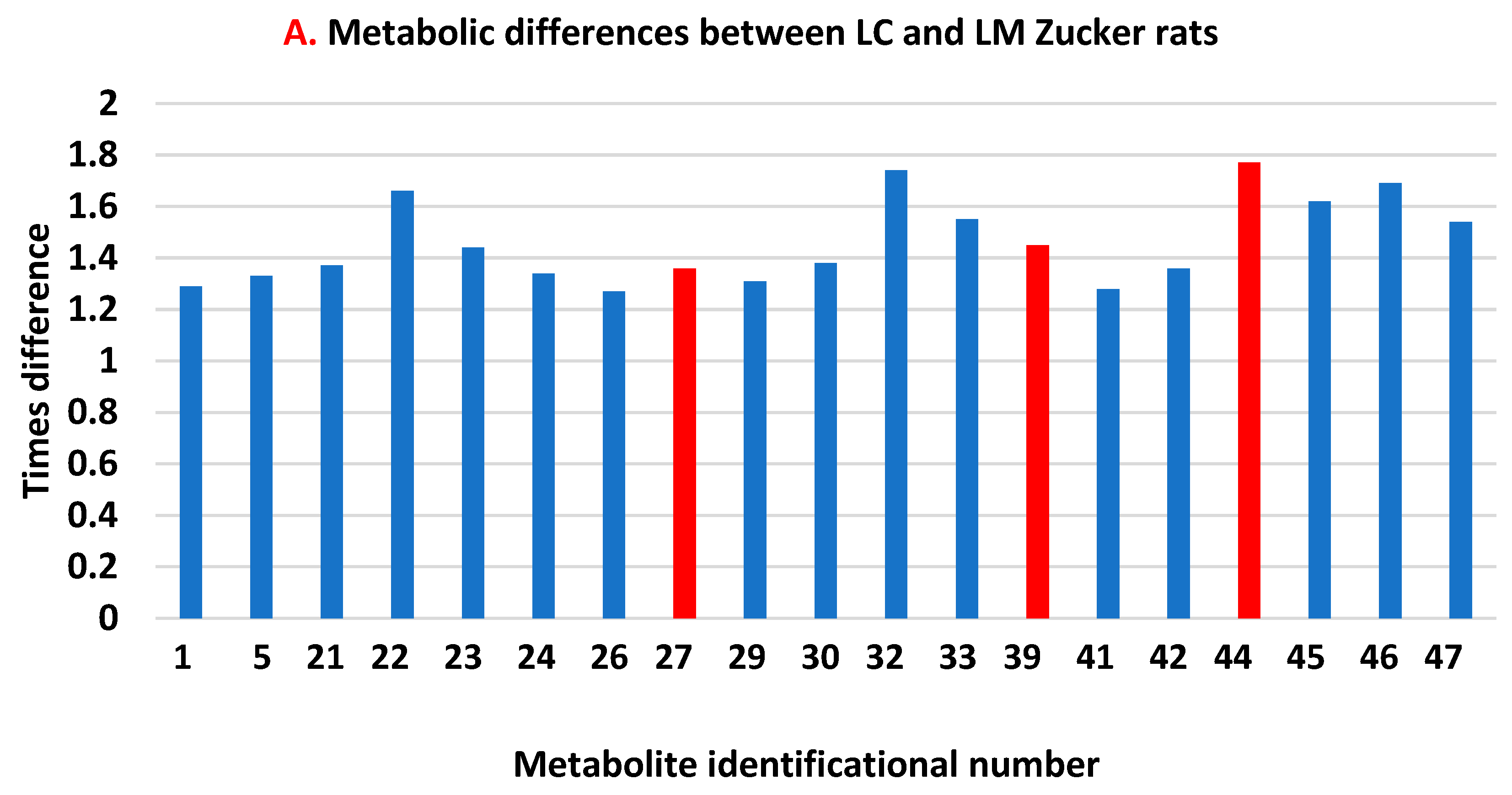

 Peaks higher in the lean group.
Peaks higher in the lean group.  Peaks higher in the obese group.
Peaks higher in the obese group.
 Peaks higher in the lean group.
Peaks higher in the lean group.  Peaks higher in the obese group.
Peaks higher in the obese group.
| OC | OM | p-Value | |
|---|---|---|---|
| Methionine, nmol/mL | 36.4 ± 5.72 | 34.4 ± 10.18 | 0.34 |
| Cysteine, nmol/mL | 10.1 ± 1.34 | 12.74 ± 2.61 | 0.024 |
| Cystine, nmol/mL | 14.7 ± 2.71 | 18.69 ± 5.75 | 0.071 |
| Cysteine/Cystine | 0.71 ± 0.168 | 0.74 ± 0.317 | 0.42 |
| Tryptophan, nmol/mL | 39.1 ± 9.14 | 48.2 ± 6.28 | 0.029 |
| Kynurenic acid, nmol/mL | 1.66 ± 0.332 | 2.27 ± 0.576 | 0.023 |
| Tryptophane/Kynurenic acid | 24.6 ± 8.02 | 22.7 ± 7.04 | 0.32 |
| Tyrosine, nmol/mL | 62.2 ± 10.83 | 66.5 ± 8.21 | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melnyk, S.; Hakkak, R. Effect of Metformin Treatment on Serum Metabolic Profile Changes in Lean and Obese Zucker Rat Model for Fatty Liver Disease. Biomolecules 2023, 13, 1234. https://doi.org/10.3390/biom13081234
Melnyk S, Hakkak R. Effect of Metformin Treatment on Serum Metabolic Profile Changes in Lean and Obese Zucker Rat Model for Fatty Liver Disease. Biomolecules. 2023; 13(8):1234. https://doi.org/10.3390/biom13081234
Chicago/Turabian StyleMelnyk, Stepan, and Reza Hakkak. 2023. "Effect of Metformin Treatment on Serum Metabolic Profile Changes in Lean and Obese Zucker Rat Model for Fatty Liver Disease" Biomolecules 13, no. 8: 1234. https://doi.org/10.3390/biom13081234
APA StyleMelnyk, S., & Hakkak, R. (2023). Effect of Metformin Treatment on Serum Metabolic Profile Changes in Lean and Obese Zucker Rat Model for Fatty Liver Disease. Biomolecules, 13(8), 1234. https://doi.org/10.3390/biom13081234






