Regulation of Phosphoinositide Signaling by Scaffolds at Cytoplasmic Membranes
Abstract
:1. Introduction
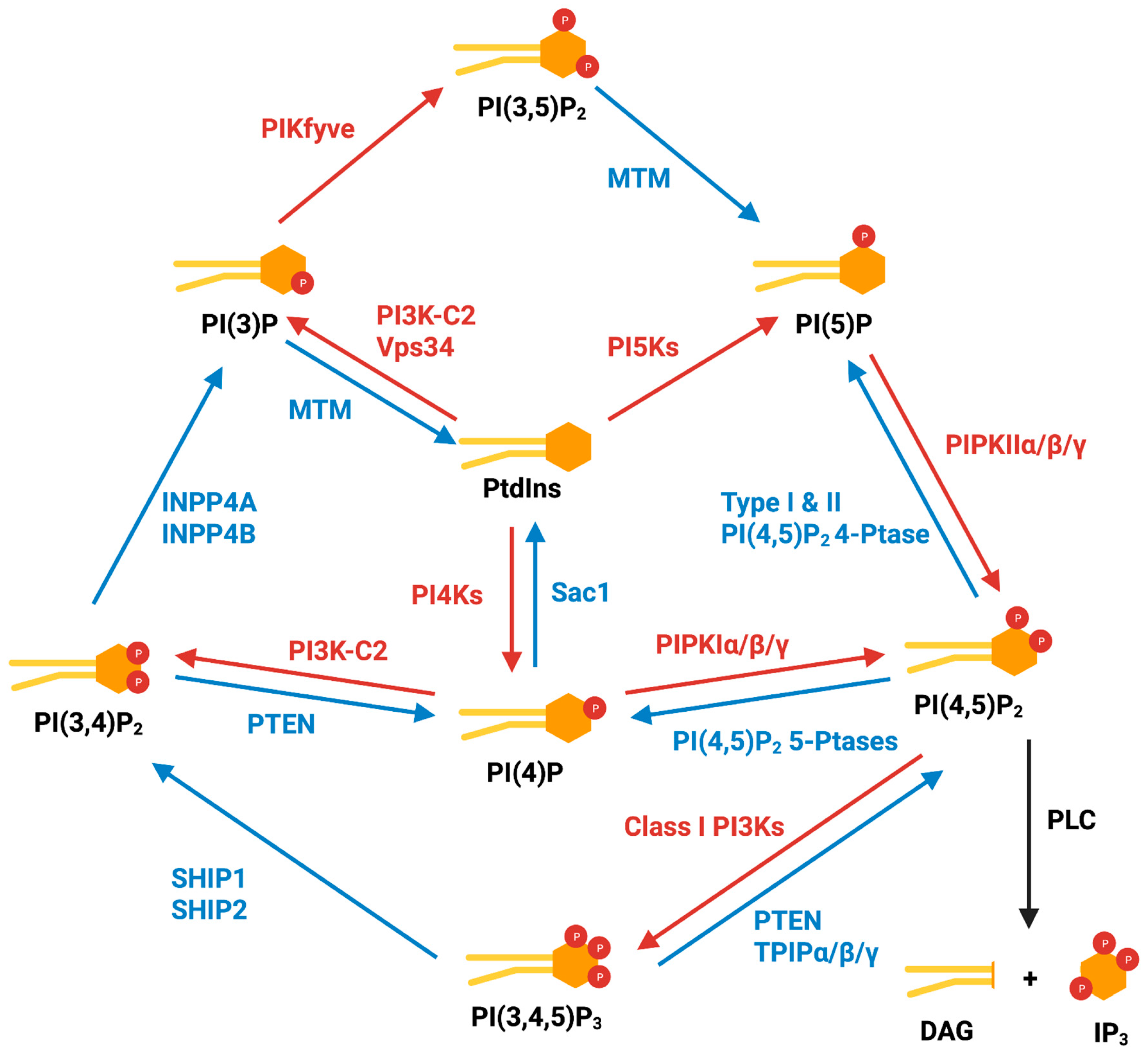
2. Cytoplasmic Phosphoinositides and Metabolism
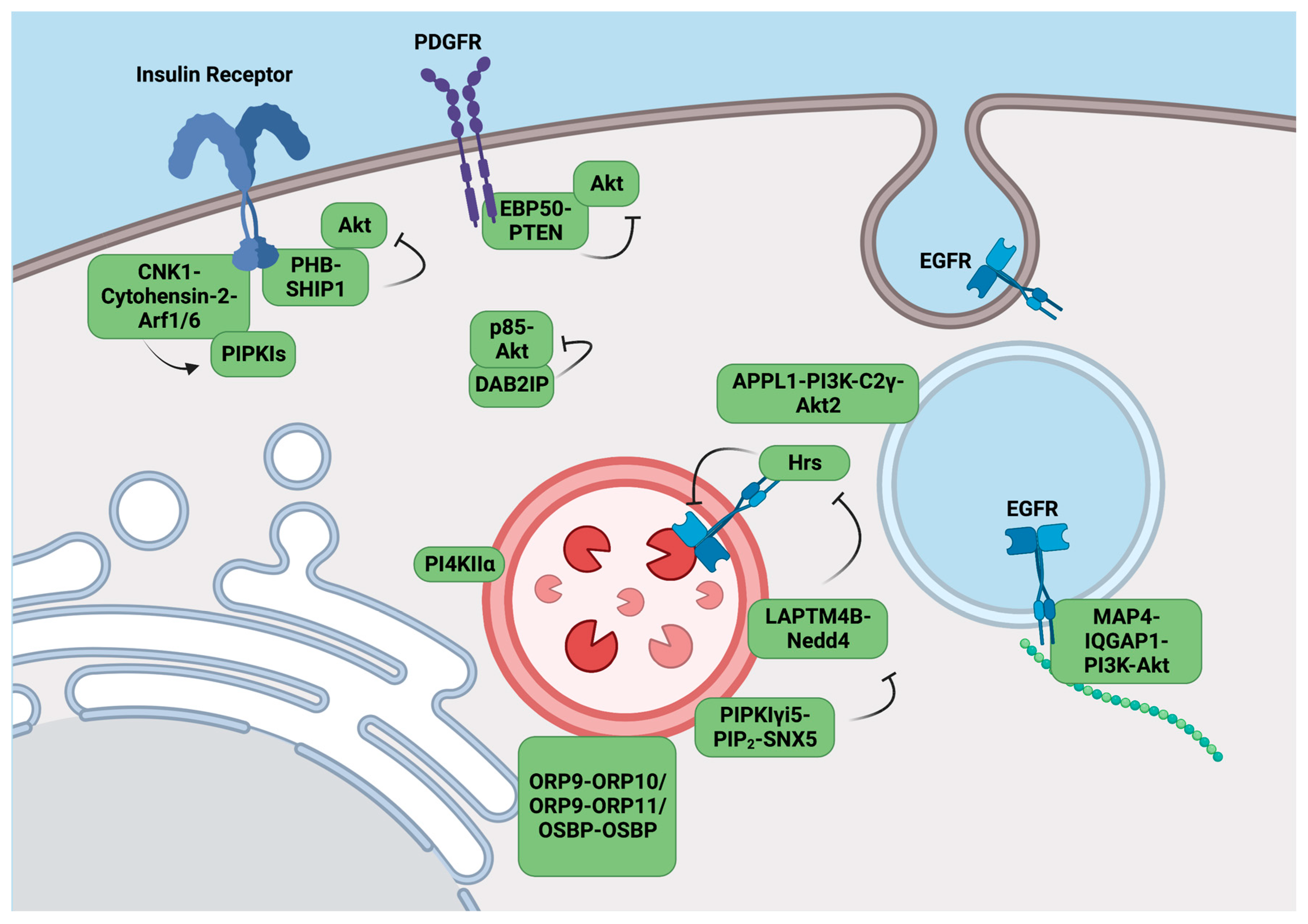
3. Scaffolding Complexes Target the PI3K-Akt Pathways to Specific Compartments
3.1. The CNK1 Scaffolding Complex Activates PIPKIs upon Insulin Stimulation
3.2. EBP50 Inhibits PI3K/Akt Signaling by Recruiting PTEN to the Membrane
3.3. PI3,4,5P3-Bound PHB Scaffolds SHIP1 to Inhibit Akt Activation
3.4. DAB2IP Blocks p85 and Akt Localization to the Membranes
3.5. APPL1 Scaffolds Akt2 on Endosomes for PI(3,4)P2 Activation
3.6. PI(4,5)P2 Mediated LAPTM4B Regulates EGFR Sorting and Degradation
3.7. IQGAP1 Scaffolds PI3,4,5P3 Synthesis at Membranes
3.8. MAP4 Scaffolds the PI3K-Akt Pathway Predominantly on Endosomal Membranes
4. PI Scaffolding Complexes in Autophagy
4.1. PI Complexes Regulate Autophagosome Maturation
4.2. PI Complexes Regulate Autolysosome Fusion
4.3. PI Complexes Regulate Autophagic Lysosome Reformation
5. The PI Scaffolding Complex Regulates Rapid Lysosome Repair
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sasaki, T.; Takasuga, S.; Sasaki, J.; Kofuji, S.; Eguchi, S.; Yamazaki, M.; Suzuki, A. Mammalian phosphoinositide kinases and phosphatases. Prog. Lipid Res. 2009, 48, 307–343. (In English) [Google Scholar] [CrossRef] [PubMed]
- Balla, T. Phosphoinositide-derived messengers in endocrine signaling. J. Endocrinol. 2006, 188, 135–153. (In English) [Google Scholar] [CrossRef]
- Falkenburger, B.H.; Jensen, J.B.; Dickson, E.J.; Suh, B.-C.; Hille, B. Phosphoinositides: Lipid regulators of membrane proteins. J. Physiol. 2010, 588, 3179–3185. (In English) [Google Scholar] [CrossRef] [PubMed]
- Kholodenko, B.N. Cell-signalling dynamics in time and space. Nat. Rev. Mol. Cell Biol. 2006, 7, 165–176. (In English) [Google Scholar] [CrossRef] [PubMed]
- Zeke, A.; Lukács, M.; Lim, W.A.; Reményi, A. Scaffolds: Interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009, 19, 364–374. (In English) [Google Scholar] [CrossRef] [PubMed]
- Thines, L.; Roushar, F.J.; Hedman, A.C.; Sacks, D.B. The IQGAP scaffolds: Critical nodes bridging receptor activation to cellular signaling. J. Cell Biol. 2023, 222, 5062. [Google Scholar] [CrossRef]
- Ande, S.R.; Gu, Y.; Nyomba, B.L.; Mishra, S. Insulin induced phosphorylation of prohibitin at tyrosine 114 recruits Shp1. Biochim. Biophys. Acta 2009, 1793, 1372–1378. (In English) [Google Scholar] [CrossRef]
- Dickson, E.J.; Hille, B. Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019, 476, 1–23. (In English) [Google Scholar] [CrossRef]
- Shisheva, A. Regulating Glut4 vesicle dynamics by phosphoinositide kinases and phosphoinositide phosphatases. Front. Biosci. 2003, 8, s945–s967. (In English) [Google Scholar] [CrossRef]
- Rameh, L.E.; Cantley, L.C. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 1999, 274, 8347–8350. (In English) [Google Scholar] [CrossRef]
- Liu, Y.; Bankaitis, V.A. Phosphoinositide phosphatases in cell biology and disease. Prog. Lipid Res. 2010, 49, 201–217. (In English) [Google Scholar] [CrossRef]
- Sarkes, D.; Rameh, L.E. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem. J. 2010, 428, 375–384. (In English) [Google Scholar] [CrossRef]
- Hokin, M.R.; Hokin, L.E. Enzyme secretion and the incorporation of P32 into phospholipides of pancreas slices. J. Biol. Chem. 1953, 203, 967–977. (In English) [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Guzman-Hernandez, M.L.; Balla, T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell 2011, 21, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.A.; Messa, M.; Sun, E.W.; Wheeler, H.; Torta, F.; Wenk, M.R.; De Camilli, P.; Reinisch, K.M. Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 2017, 355, aah6171. (In English) [Google Scholar] [CrossRef] [PubMed]
- Ashlin, T.G.; Blunsom, N.J.; Cockcroft, S. Courier service for phosphatidylinositol: PITPs deliver on demand. Biochim. Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2021, 1866, 158985. (In English) [Google Scholar] [CrossRef]
- Kim, Y.J.; Guzman-Hernandez, M.L.; Wisniewski, E.; Balla, T. Phosphatidylinositol-Phosphatidic Acid Exchange by Nir2 at ER-PM Contact Sites Maintains Phosphoinositide Signaling Competence. Dev. Cell 2015, 33, 549–561. (In English) [Google Scholar] [CrossRef]
- Chang, C.-L.; Hsieh, T.-S.; Yang, T.T.; Rothberg, K.G.; Azizoglu, D.B.; Volk, E.; Liao, J.-C.; Liou, J. Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 2013, 5, 813–825. (In English) [Google Scholar] [CrossRef]
- Kim, S.; Kedan, A.; Marom, M.; Gavert, N.; Keinan, O.; Selitrennik, M.; Laufman, O.; Lev, S. The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 2013, 14, 891–899. (In English) [Google Scholar] [CrossRef]
- Chang, C.L.; Liou, J. Phosphatidylinositol 4,5-Bisphosphate Homeostasis Regulated by Nir2 and Nir3 Proteins at Endoplasmic Reticulum-Plasma Membrane Junctions. J. Biol. Chem. 2015, 290, 14289–14301. (In English) [Google Scholar] [CrossRef]
- Marat, A.L.; Haucke, V. Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic. EMBO J. 2016, 35, 561–579. (In English) [Google Scholar] [CrossRef] [PubMed]
- Devereaux, K.; Dall’armi, C.; Alcazar-Roman, A.; Ogasawara, Y.; Zhou, X.; Wang, F.; Yamamoto, A.; De Camilli, P.; Di Paolo, G. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE 2013, 8, e76405. (In English) [Google Scholar] [CrossRef] [PubMed]
- Franco, I.; Gulluni, F.; Campa, C.C.; Costa, C.; Margaria, J.P.; Ciraolo, E.; Martini, M.; Monteyne, D.; De Luca, E.; Germena, G.; et al. PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev. Cell 2014, 28, 647–658. (In English) [Google Scholar] [CrossRef] [PubMed]
- Norris, F.A.; Auethavekiat, V.; Majerus, P.W. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J. Biol. Chem. 1995, 270, 16128–16133. (In English) [Google Scholar] [CrossRef]
- Norris, F.A.; Majerus, P.W. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J. Biol. Chem. 1994, 269, 8716–8720. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-W.; Hayashi, M.; Christoforidis, S.; Lacas-Gervais, S.; Hoepfner, S.; Wenk, M.R.; Modregger, J.; Uttenweiler-Joseph, S.; Wilm, M.; Nystuen, A.; et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 2005, 170, 607–618. [Google Scholar] [CrossRef]
- Nakatsu, F.; Baskin, J.M.; Chung, J.; Tanner, L.B.; Shui, G.; Lee, S.Y.; Pirruccello, M.; Hao, M.; Ingolia, N.T.; Wenk, M.R.; et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 2012, 199, 1003–1016. (In English) [Google Scholar] [CrossRef]
- De Matteis, M.; Godi, A.; Corda, D. Phosphoinositides and the golgi complex. Curr. Opin. Cell Biol. 2002, 14, 434–447. (In English) [Google Scholar] [CrossRef]
- Godi, A.; Pertile, P.; Meyers, R.; Marra, P.; Di Tullio, G.; Iurisci, C.; Luini, A.; Corda, D.; De Matteis, M.A. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol. 1999, 1, 280–287. (In English) [Google Scholar] [CrossRef]
- Wong, K.; Meyers, R.; Cantley, L.C. Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem. 1997, 272, 13236–13241. (In English) [Google Scholar] [CrossRef]
- Nakagawa, T.; Goto, K.; Kondo, H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 1996, 271, 12088–12094. (In English) [Google Scholar] [CrossRef]
- Manford, A.; Xia, T.; Saxena, A.K.; Stefan, C.; Hu, F.; Emr, S.D.; Mao, Y. Crystal structure of the yeast Sac1: Implications for its phosphoinositide phosphatase function. EMBO J. 2010, 29, 1489–1498. (In English) [Google Scholar] [CrossRef]
- Sbrissa, D.; Ikonomov, O.C.; Deeb, R.; Shisheva, A. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J. Biol. Chem. 2002, 277, 47276–47284. [Google Scholar] [CrossRef]
- Zolov, S.N.; Bridges, D.; Zhang, Y.; Lee, W.-W.; Riehle, E.; Verma, R.; Lenk, G.M.; Converso-Baran, K.; Weide, T.; Albin, R.L.; et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl. Acad. Sci. USA 2012, 109, 17472–17477. (In English) [Google Scholar] [CrossRef] [PubMed]
- Mason, D.; Mallo, G.V.; Terebiznik, M.R.; Payrastre, B.; Finlay, B.B.; Brumell, J.H.; Rameh, L.; Grinstein, S. Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J. Gen. Physiol. 2007, 129, 267–283. (In English) [Google Scholar] [CrossRef] [PubMed]
- Ungewickell, A.; Hugge, C.; Kisseleva, M.; Chang, S.-C.; Zou, J.; Feng, Y.; Galyov, E.E.; Wilson, M.; Majerus, P.W. The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc. Natl. Acad. Sci. USA 2005, 102, 18854–18859. (In English) [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Marjanovic, J.; Kisseleva, M.V.; Wilson, M.; Majerus, P.W. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 2007, 104, 16834–16839. (In English) [Google Scholar] [CrossRef]
- Gupta, A.; Toscano, S.; Trivedi, D.; Jones, D.R.; Mathre, S.; Clarke, J.H.; Divecha, N.; Raghu, P. Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc. Natl. Acad. Sci. USA 2013, 110, 5963–5968. (In English) [Google Scholar] [CrossRef]
- Emerling, B.M.; Hurov, J.B.; Poulogiannis, G.; Tsukazawa, K.S.; Choo-Wing, R.; Wulf, G.M.; Bell, E.L.; Shim, H.-S.; Lamia, K.A.; Rameh, L.E.; et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 2013, 155, 844–857. (In English) [Google Scholar] [CrossRef]
- Viaud, J.; Boal, F.; Tronchère, H.; Gaits-Iacovoni, F.; Payrastre, B. Phosphatidylinositol 5-phosphate: A nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. Bioessays 2013, 36, 260–272. (In English) [Google Scholar] [CrossRef]
- Misawa, H.; Ohtsubo, M.; Copeland, N.G.; Gilbert, D.J.; Jenkins, N.A.; Yoshimura, A. Cloning and characterization of a novel class II phosphoinositide 3-kinase containing C2 domain. Biochem. Biophys. Res. Commun. 1998, 244, 531–539. (In English) [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-L.; Wang, Z.-G.; Hu, Y.; Xin, Y.; Singaram, I.; Gorai, S.; Zhou, X.; Shim, Y.; Min, J.-H.; Gong, L.-W.; et al. Quantitative Lipid Imaging Reveals a New Signaling Function of Phosphatidylinositol-3,4-Bisphophate: Isoform- and Site-Specific Activation of Akt. Mol. Cell 2018, 71, 1092–1104.e5. (In English) [Google Scholar] [CrossRef] [PubMed]
- Stephens, L.R.; Hughes, K.T.; Irvine, R.F. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature 1991, 351, 33–39. (In English) [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.T.; Jackson, T.R.; Stephens, L.R. Platelet-derived growth factor stimulates synthesis of PtdIns(3,4,5)P3 by activating a PtdIns(4,5)P2 3-OH kinase. Nature 1992, 358, 157–159. (In English) [Google Scholar] [CrossRef]
- Sasaki, J.; Kofuji, S.; Itoh, R.; Momiyama, T.; Takayama, K.; Murakami, H.; Chida, S.; Tsuya, Y.; Takasuga, S.; Eguchi, S.; et al. The PtdIns(3,4)P(2) phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 2010, 465, 497–501. (In English) [Google Scholar] [CrossRef]
- Malek, M.; Kielkowska, A.; Chessa, T.; Anderson, K.E.; Barneda, D.; Pir, P.; Nakanishi, H.; Eguchi, S.; Koizumi, A.; Sasaki, J.; et al. PTEN Regulates PI(3,4)P. Mol. Cell 2017, 68, 566–580.e10. (In English) [Google Scholar] [CrossRef]
- Shisheva, A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol. Int. 2008, 32, 591–604. (In English) [Google Scholar] [CrossRef]
- Ikonomov, O.C.; Sbrissa, D.; Shisheva, A. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J. Biol. Chem. 2001, 276, 26141–26147. [Google Scholar] [CrossRef]
- Hong, N.H.; Qi, A.; Weaver, A.M. PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions. J. Cell Biol. 2015, 210, 753–769. (In English) [Google Scholar] [CrossRef]
- Loijens, J.C.; Anderson, R.A. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J. Biol. Chem. 1996, 271, 32937–32943. [Google Scholar] [CrossRef]
- Doughman, R.L.; Firestone, A.J.; Anderson, R.A. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 2003, 194, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yuan, Q.; Kang, X.; Qin, Y.; Li, L.; Ha, Y.; Wu, D. Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled. Nat. Commun. 2015, 6, 8205. (In English) [Google Scholar] [CrossRef] [PubMed]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. (In English) [Google Scholar] [CrossRef]
- Foster, F.M.; Traer, C.J.; Abraham, S.M.; Fry, M.J. The phosphoinositide (PI) 3-kinase family. J. Cell Sci. 2003, 116, 3037–3040. (In English) [Google Scholar] [CrossRef] [PubMed]
- Milne, S.B.; Ivanova, P.T.; DeCamp, D.; Hsueh, R.C.; Brown, H.A. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J. Lipid Res. 2005, 46, 1796–1802. (In English) [Google Scholar] [CrossRef] [PubMed]
- Clark, J.; Anderson, K.E.; Juvin, V.; Smith, T.S.; Karpe, F.; Wakelam, M.J.O.; Stephens, L.R.; Hawkins, P.T. Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 2011, 8, 267–272. (In English) [Google Scholar] [CrossRef]
- King, D.; Yeomanson, D.; Bryant, H.E. PI3King the lock: Targeting the PI3K/Akt/mTOR pathway as a novel therapeutic strategy in neuroblastoma. J. Pediatr. Hematol. Oncol. 2015, 37, 245–251. (In English) [Google Scholar] [CrossRef]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009, 4, 127–150. (In English) [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. (In English) [Google Scholar] [CrossRef]
- Kolch, W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005, 6, 827–837. (In English) [Google Scholar] [CrossRef]
- Fritz, R.D.; Varga, Z.; Radziwill, G. CNK1 is a novel Akt interaction partner that promotes cell proliferation through the Akt-FoxO signalling axis. Oncogene 2010, 29, 3575–3582. (In English) [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Weber, W.; Warscheid, B.; Radziwill, G. AKT-dependent phosphorylation of the SAM domain induces oligomerization and activation of the scaffold protein CNK1. Biochim. Biophys. Acta. Mol. Cell Res. 2017, 1864, 89–100. (In English) [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Hao, P.; An, S.; Yang, Y.; Liu, Y.; Hao, Q.; Ejaz, M.; Guo, X.-X.; Xu, T.-R. Akt scaffold proteins: The key to controlling specificity of Akt signaling. Am. J. Physiol. Physiol. 2021, 321, C429–C442. (In English) [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Morales, F.C.; Kreimann, E.L.; Georgescu, M.-M. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 2006, 25, 910–920. (In English) [Google Scholar] [CrossRef]
- Wang, B.; Yang, Y.; Friedman, P.A. Na/H exchange regulatory factor 1, a novel AKT-associating protein, regulates extracellular signal-regulated kinase signaling through a B-Raf-mediated pathway. Mol. Biol. Cell 2008, 19, 1637–1645. (In English) [Google Scholar] [CrossRef]
- Molina, J.R.; Agarwal, N.K.; Morales, F.C.; Hayashi, Y.; Aldape, K.D.; Cote, G.; Georgescu, M.-M. PTEN, NHERF1 and PHLPP form a tumor suppressor network that is disabled in glioblastoma. Oncogene 2012, 31, 1264–1274. (In English) [Google Scholar] [CrossRef]
- Molina, J.R.; Morales, F.C.; Hayashi, Y.; Aldape, K.D.; Georgescu, M.M. Loss of PTEN binding adapter protein NHERF1 from plasma membrane in glioblastoma contributes to PTEN inactivation. Cancer Res 2010, 70, 6697–6703. (In English) [Google Scholar] [CrossRef]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef]
- Ande, S.R.; Mishra, S. Prohibitin interacts with phosphatidylinositol 3,4,5-triphosphate (PIP3) and modulates insulin signaling. Biochem. Biophys. Res. Commun. 2009, 390, 1023–1028. (In English) [Google Scholar] [CrossRef]
- Sun, L.; Liu, L.; Yang, X.J.; Wu, Z. Akt binds prohibitin 2 and relieves its repression of MyoD and muscle differentiation. J. Cell Sci. 2004, 117 Pt 4, 3021–3029. (In English) [Google Scholar] [CrossRef]
- Xie, D.; Gore, C.; Zhou, J.; Pong, R.-C.; Zhang, H.; Yu, L.; Vessella, R.L.; Min, W.; Hsieh, J.-T. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc. Natl. Acad. Sci. USA 2009, 106, 19878–19883. (In English) [Google Scholar] [CrossRef]
- Dai, X.; North, B.J.; Inuzuka, H. Negative regulation of DAB2IP by Akt and SCFFbw7 pathways. Oncotarget 2014, 5, 3307–3315. (In English) [Google Scholar] [CrossRef]
- Mitsuuchi, Y.; Johnson, S.W.; Sonoda, G.; Tanno, S.; Golemis, E.A.; Testa, J.R. Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene 1999, 18, 4891–4898. (In English) [Google Scholar] [CrossRef] [PubMed]
- Bae, G.-U.; Lee, J.-R.; Kim, B.-G.; Han, J.-W.; Leem, Y.-E.; Ho, S.-M.; Hahn, M.-J.; Kang, J.-S.; Lee, H.-J.; Choi, H.-K.; et al. Cdo interacts with APPL1 and activates Akt in myoblast differentiation. Mol. Biol. Cell 2010, 21, 2399–2411. (In English) [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.K.; Iglesias, M.A.; Lam, K.S.; Wang, Y.; Sweeney, G.; Zhu, W.; Vanhoutte, P.M.; Kraegen, E.W.; Xu, A. APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metab. 2009, 9, 417–427. (In English) [Google Scholar] [CrossRef] [PubMed]
- Lin, D.C.; Quevedo, C.; Brewer, N.E.; Bell, A.; Testa, J.R.; Grimes, M.L.; Miller, F.D.; Kaplan, D.R. APPL1 associates with TrkA and GIPC1 and is required for nerve growth factor-mediated signal transduction. Mol. Cell Biol. 2006, 26, 8928–8941. (In English) [Google Scholar] [CrossRef]
- Yang, L.; Lin, H.-K.; Altuwaijri, S.; Xie, S.; Wang, L.; Chang, C. APPL suppresses androgen receptor transactivation via potentiating Akt activity. J. Biol. Chem. 2003, 278, 16820–16827. (In English) [Google Scholar] [CrossRef]
- Braccini, L.; Ciraolo, E.; Campa, C.C.; Perino, A.; Longo, D.L.; Tibolla, G.; Pregnolato, M.; Cao, Y.; Tassone, B.; Damilano, F.; et al. PI3K-C2γ is a Rab5 effector selectively controlling endosomal Akt2 activation downstream of insulin signalling. Nat. Commun. 2015, 6, 7400. (In English) [Google Scholar] [CrossRef]
- Shao, G.-Z.; Zhou, R.-L.; Zhang, Q.-Y.; Zhang, Y.; Liu, J.-J.; Rui, J.-A.; Wei, X.; Ye, D.-X. Molecular cloning and characterization of LAPTM4B, a novel gene upregulated in hepatocellular carcinoma. Oncogene 2003, 22, 5060–5069. [Google Scholar] [CrossRef]
- Li, Y.; Zou, L.; Li, Q.; Haibe-Kains, B.; Tian, R.; Li, Y.; Desmedt, C.; Sotiriou, C.; Szallasi, Z.; Iglehart, J.D.; et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat. Med. 2010, 16, 214–218. [Google Scholar] [CrossRef]
- Kang, Y.; Yin, M.; Jiang, W.; Zhang, H.; Xia, B.; Xue, Y.; Huang, Y. Overexpression of LAPTM4B-35 is associated with poor prognosis in colorectal carcinoma. Am. J. Surg. 2012, 204, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shan, Y.; Yang, H.; Zhang, S.; Lin, M.; Zhu, P.; Chen, X.-Y.; Yi, J.; Mcnutt, M.A.; Shao, G.-Z.; et al. Upregulation of LAPTM4B-35 promotes malignant transformation and tumorigenesis in L02 human liver cell line. Anat. Rec. 2011, 294, 1135–1142. (In English) [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xiong, F.; Wei, X.; Yang, Y.; McNutt, M.A.; Zhou, R. Overexpression of LAPTM4B-35 promotes growth and metastasis of hepatocellular carcinoma in vitro and in vivo. Cancer Lett. 2010, 294, 236–244. [Google Scholar] [CrossRef]
- Tan, X.; Sun, Y.; Thapa, N.; Liao, Y.; Hedman, A.C.; Anderson, R.A. LAPTM4B is a PtdIns(4,5)P2 effector that regulates EGFR signaling, lysosomal sorting, and degradation. EMBO J. 2015, 34, 475–490. [Google Scholar] [CrossRef]
- Mendelsohn, J.; Baselga, J. Epidermal growth factor receptor targeting in cancer. Semin. Oncol. 2006, 33, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Wiley, H.S. Trafficking of the ErbB receptors and its influence on signaling. Exp. Cell Res. 2003, 284, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, A.; Goh, L.K. Endocytosis and intracellular trafficking of ErbBs. Exp. Cell Res. 2008, 314, 3094–3106. [Google Scholar] [CrossRef]
- Eden, E.R.; White, I.J.; Futter, C.E. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem. Soc. Trans. 2009, 37 Pt 1, 173–177. (In English) [Google Scholar] [CrossRef]
- Williams, R.L.; Urbe, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 2007, 8, 355–368. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Katz, M.; Shtiegman, K.; Tal-Or, P.; Yakir, L.; Mosesson, Y.; Harari, D.; Machluf, Y.; Asao, H.; Jovin, T.; Sugamura, K.; et al. Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 2002, 3, 740–751. [Google Scholar] [CrossRef]
- Hoeller, D.; Crosetto, N.; Blagoev, B.; Raiborg, C.; Tikkanen, R.; Wagner, S.; Kowanetz, K.; Breitling, R.; Mann, M.; Stenmark, H.; et al. Regulation of ubiquitin-binding proteins by monoubiquitination. Nature 2006, 8, 163–169. [Google Scholar] [CrossRef]
- Persaud, A.; Alberts, P.; Amsen, E.M.; Xiong, X.; Wasmuth, J.; Saadon, Z.; Fladd, C.; Parkinson, J.; Rotin, D. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Mol. Syst. Biol. 2009, 5, 333. [Google Scholar] [CrossRef]
- Tan, X.; Thapa, N.; Sun, Y.; Anderson, R.A. A kinase-independent role for EGF receptor in autophagy initiation. Cell 2015, 160, 145–160. [Google Scholar] [CrossRef]
- Smith, J.M.; Hedman, A.C.; Sacks, D.B. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015, 25, 171–184. (In English) [Google Scholar] [CrossRef]
- Hedman, A.C.; Smith, J.M.; Sacks, D.B. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep. 2015, 16, 427–446. (In English) [Google Scholar] [CrossRef]
- Ren, J.-G.; Li, Z.; Sacks, D.B. IQGAP1 modulates activation of B-Raf. Proc. Natl. Acad. Sci. USA 2007, 104, 10465–10469. (In English) [Google Scholar] [CrossRef] [PubMed]
- Jameson, K.L.; Mazur, P.K.; Zehnder, A.M.; Zhang, J.; Zarnegar, B.; Sage, J.; A Khavari, P. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat. Med. 2013, 19, 626–630. (In English) [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Thapa, N.; Hedman, A.C.; Li, Z.; Sacks, D.B.; Anderson, R.A. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013, 32, 2617–2630. (In English) [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; McNulty, D.E.; Marler, K.J.M.; Lim, L.; Hall, C.; Annan, R.S.; Sacks, D.B. IQGAP1 promotes neurite outgrowth in a phosphorylation-dependent manner. J. Biol. Chem. 2005, 280, 13871–13878. (In English) [Google Scholar] [CrossRef] [PubMed]
- Fukata, M.; Watanabe, T.; Noritake, J.; Nakagawa, M.; Yamaga, M.; Kuroda, S.; Matsuura, Y.; Iwamatsu, A.; Perez, F.; Kaibuchi, K. Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 2002, 109, 873–885. (In English) [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Wang, S.; Noritake, J.; Sato, K.; Fukata, M.; Takefuji, M.; Nakagawa, M.; Izumi, N.; Akiyama, T.; Kaibuchi, K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell 2004, 7, 871–883. (In English) [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Anderson, R.A. IQGAP1 is a phosphoinositide effector and kinase scaffold. Adv. Biol. Regul. 2016, 60, 29–35. (In English) [Google Scholar] [CrossRef]
- Le Clainche, C.; Schlaepfer, D.; Ferrari, A.; Klingauf, M.; Grohmanova, K.; Veligodskiy, A.; Didry, D.; Le, D.; Egile, C.; Carlier, M.-F.; et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 2007, 282, 426–435. (In English) [Google Scholar] [CrossRef]
- Rohatgi, R.; Ho, H.-Y.H.; Kirschner, M.W. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 2000, 150, 1299–1310. (In English) [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. (In English) [Google Scholar] [CrossRef]
- Kelly, K.L.; Ruderman, N.B.; Chen, K.S. Phosphatidylinositol-3-kinase in isolated rat adipocytes. Activation by insulin and subcellular distribution. J. Biol. Chem. 1992, 267, 3423–3428. (In English) [Google Scholar] [CrossRef]
- Sato, M.; Ueda, Y.; Takagi, T.; Umezawa, Y. Production of PtdInsP3 at endomembranes is triggered by receptor endocytosis. Nature 2003, 5, 1016–1022. (In English) [Google Scholar] [CrossRef]
- Naguib, A.; Bencze, G.; Cho, H.; Zheng, W.; Tocilj, A.; Elkayam, E.; Faehnle, C.R.; Jaber, N.; Pratt, C.P.; Chen, M.; et al. PTEN functions by recruitment to cytoplasmic vesicles. Mol. Cell 2015, 58, 255–268. (In English) [Google Scholar] [CrossRef]
- Choi, S.; Hedman, A.C.; Sayedyahossein, S.; Thapa, N.; Sacks, D.B.; Anderson, R.A. Agonist-stimulated phosphatidylinositol-3,4,5-trisphosphate generation by scaffolded phosphoinositide kinases. Nat. Cell Biol. 2016, 18, 1324–1335. (In English) [Google Scholar] [CrossRef]
- Tekletsadik, Y.K.; Sonn, R.; Osman, M.A. A conserved role of IQGAP1 in regulating TOR complex 1. J. Cell Sci. 2012, 125, 2041–2052. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhu, H.-H.; Zhou, L.-F.; Wu, S.-S.; Wang, J.; Chen, Z. IQGAP1 is overexpressed in hepatocellular carcinoma and promotes cell proliferation by Akt activation. Exp. Mol. Med. 2010, 42, 477–483. (In English) [Google Scholar] [CrossRef] [PubMed]
- Rameh, L.E.; Mackey, A.M. IQGAP1 makes PI(3)K signalling as easy as PIP, PIP. Nat. Cell Biol. 2016, 18, 1263–1265. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Choi, S.; Jung, O.; Wen, T.; Baum, C.; Thapa, N.; Lambert, P.F.; Rapraeger, A.C.; Anderson, R.A. The Specificity of EGF-Stimulated IQGAP1 Scaffold Towards the PI3K-Akt Pathway is Defined by the IQ3 motif. Sci. Rep. 2019, 9, 9126. (In English) [Google Scholar] [CrossRef]
- Jethwa, N.; Chung, G.H.C.; Lete, M.G.; Alonso, A.; Byrne, R.D.; Calleja, V.; Larijani, B. Endomembrane PtdIns(3,4,5)P3 activates the PI3K-Akt pathway. J. Cell Sci. 2015, 128, 3456–3465. (In English) [Google Scholar] [CrossRef]
- Fields, I.C.; King, S.M.; Shteyn, E.; Kang, R.S.; Fölsch, H. Phosphatidylinositol 3,4,5-trisphosphate localization in recycling endosomes is necessary for AP-1B-dependent sorting in polarized epithelial cells. Mol. Biol. Cell 2010, 21, 95–105. (In English) [Google Scholar] [CrossRef]
- Thapa, N.; Chen, M.; Horn, H.T.; Choi, S.; Wen, T.; Anderson, R.A. Phosphatidylinositol-3-OH kinase signalling is spatially organized at endosomal compartments by microtubule-associated protein 4. Nat. Cell Biol. 2020, 22, 1357–1370. (In English) [Google Scholar] [CrossRef]
- Murphy, J.E.; Padilla, B.E.; Hasdemir, B.; Cottrell, G.S.; Bunnett, N.W. Endosomes: A legitimate platform for the signaling train. Proc. Natl. Acad. Sci. USA 2009, 106, 17615–17622. (In English) [Google Scholar] [CrossRef]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. (In English) [Google Scholar] [CrossRef]
- Goh, L.K.; Sorkin, A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a017459. [Google Scholar] [CrossRef] [PubMed]
- Ebner, M.; Lucic, I.; Leonard, T.A.; Yudushkin, I. PI(3,4,5)P3 Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 2017, 65, 416–431.e6. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; He, C.; Wu, A.; Zhou, J.; Wu, J. Microtubule-Associated Protein 4 Is a Prognostic Factor and Promotes Tumor Progression in Lung Adenocarcinoma. Dis. Markers 2018, 2018, 8956072. (In English) [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Zheng, X.; Gao, Y.; Shu, M.; Leng, T.; Li, Y.; Yin, W.; Zhu, W.; Huang, Y.; Zhou, Y.; et al. Activation of cyclic AMP/PKA pathway inhibits bladder cancer cell invasion by targeting MAP4-dependent microtubule dynamics. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 47.e21–47.e28. (In English) [Google Scholar] [CrossRef]
- Palamiuc, L.; Ravi, A.; Emerling, B.M. Phosphoinositides in autophagy: Current roles and future insights. FEBS J. 2019, 287, 222–238. (In English) [Google Scholar] [CrossRef]
- Tan, X.; Thapa, N.; Choi, S.; Anderson, R.A. Emerging roles of PtdIns(4,5)P2--beyond the plasma membrane. J. Cell Sci. 2015, 128, 4047–4056. (In English) [Google Scholar] [CrossRef]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. (In English) [Google Scholar] [CrossRef]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. (In English) [Google Scholar] [CrossRef]
- Shi, C.S.; Kehrl, J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010, 3, ra42. (In English) [Google Scholar] [CrossRef]
- Debnath, J.; Gammoh, N.; Ryan, K.M. Autophagy and autophagy-related pathways in cancer. Nat. Rev. Mol. Cell Biol. 2023, 24, 560–575. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–5372. (In English) [Google Scholar] [CrossRef] [PubMed]
- Baskaran, S.; Carlson, L.-A.; Stjepanovic, G.; Young, L.N.; Kim, D.J.; Grob, P.; Stanley, R.E.; Nogales, E.; Hurley, J.H. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife 2014, 3, e05115. (In English) [Google Scholar] [CrossRef] [PubMed]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Araki, Y.; Ku, W.-C.; Akioka, M.; May, A.I.; Hayashi, Y.; Arisaka, F.; Ishihama, Y.; Ohsumi, Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 2013, 203, 299–313. [Google Scholar] [CrossRef]
- Hitomi, K.; Kotani, T.; Noda, N.N.; Kimura, Y.; Nakatogawa, H. The Atg1 complex, Atg9, and Vac8 recruit PI3K complex I to the pre-autophagosomal structure. J. Cell Biol. 2023, 222, 10017. [Google Scholar] [CrossRef]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef]
- Fujioka, Y.; Suzuki, S.W.; Yamamoto, H.; Kondo-Kakuta, C.; Kimura, Y.; Hirano, H.; Akada, R.; Inagaki, F.; Ohsumi, Y.; Noda, N.N. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat. Struct. Mol. Biol. 2014, 21, 513–521. [Google Scholar] [CrossRef]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Cheung, P.C.; Trinkle-Mulcahy, L.; Cohen, P.; Lucocq, J.M. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 2001, 355 Pt 1, 113–121. (In English) [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. (In English) [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. (In English) [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Itoh, T.; Omori, H.; Fukuda, M.; Noda, T.; Yoshimori, T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 2008, 19, 2092–2100. (In English) [Google Scholar] [CrossRef] [PubMed]
- Hanada, T.; Noda, N.N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007, 282, 37298–37302. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. (In English) [Google Scholar] [CrossRef] [PubMed]
- Judith, D.; Jefferies, H.B.J.; Boeing, S.; Frith, D.; Snijders, A.P.; Tooze, S.A. ATG9A shapes the forming autophagosome through Arfaptin 2 and phosphatidylinositol 4-kinase IIIβ. J. Cell Biol. 2019, 218, 1634–1652. (In English) [Google Scholar] [CrossRef]
- Karanasios, E.; Walker, S.A.; Okkenhaug, H.; Manifava, M.; Hummel, E.; Zimmermann, H.; Ahmed, Q.; Domart, M.-C.; Collinson, L.; Ktistakis, N.T. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 2016, 7, 12420. (In English) [Google Scholar] [CrossRef] [PubMed]
- Vicinanza, M.; Korolchuk, V.I.; Ashkenazi, A.; Puri, C.; Menzies, F.M.; Clarke, J.H.; Rubinsztein, D.C. PI(5)P regulates autophagosome biogenesis. Mol. Cell 2015, 57, 219–234. (In English) [Google Scholar] [CrossRef] [PubMed]
- Knævelsrud, H.; Søreng, K.; Raiborg, C.; Håberg, K.; Rasmuson, F.; Brech, A.; Liestøl, K.; Rusten, T.E.; Stenmark, H.; Neufeld, T.P.; et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J. Cell Biol. 2013, 202, 331–349. (In English) [Google Scholar] [CrossRef]
- Tan, X.; Thapa, N.; Liao, Y.; Choi, S.; Anderson, R.A. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc. Natl. Acad. Sci. USA 2016, 113, 10896–10901. (In English) [Google Scholar] [CrossRef]
- Olsvik, H.L.; Lamark, T.; Takagi, K.; Larsen, K.B.; Evjen, G.; Øvervatn, A.; Mizushima, T.; Johansen, T. FYCO1 Contains a C-terminally Extended, LC3A/B-preferring LC3-interacting Region (LIR) Motif Required for Efficient Maturation of Autophagosomes during Basal Autophagy. J. Biol. Chem. 2015, 290, 29361–29374. (In English) [Google Scholar] [CrossRef] [PubMed]
- Pankiv, S.; Alemu, E.A.; Brech, A.; Bruun, J.-A.; Lamark, T.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 2010, 188, 253–269. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Fan, W.; Lu, Y.; Ding, X.; Chen, S.; Zhong, Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol. Cell 2012, 45, 629–641. (In English) [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, H.-Q.; Zhu, X.; Zhang, L.; Albanesi, J.; Levine, B.; Yin, H. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc. Natl. Acad. Sci. USA 2015, 112, 7015–7020. (In English) [Google Scholar] [CrossRef]
- McEwan, D.G.; Popovic, D.; Gubas, A.; Terawaki, S.; Suzuki, H.; Stadel, D.; Coxon, F.P.; de Stegmann, D.M.; Bhogaraju, S.; Maddi, K.; et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell 2015, 57, 39–54. (In English) [Google Scholar] [CrossRef]
- Baba, T.; Toth, D.J.; Sengupta, N.; Kim, Y.J.; Balla, T. Phosphatidylinositol 4,5-bisphosphate controls Rab7 and PLEKHM1 membrane cycling during autophagosome-lysosome fusion. EMBO J. 2019, 38, e100312. (In English) [Google Scholar] [CrossRef]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. (In English) [Google Scholar] [CrossRef]
- Sridhar, S.; Patel, B.; Aphkhazava, D.; Macian, F.; Santambrogio, L.; Shields, D.; Cuervo, A.M. The lipid kinase PI4KIIIβ preserves lysosomal identity. EMBO J. 2012, 32, 324–339. (In English) [Google Scholar] [CrossRef]
- Rong, Y.; Liu, M.; Ma, L.; Du, W.; Zhang, H.; Tian, Y.; Cao, Z.; Li, Y.; Ren, H.; Zhang, C.; et al. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat. Cell Biol. 2012, 14, 924–934. (In English) [Google Scholar] [CrossRef]
- Du, W.; Su, Q.P.; Chen, Y.; Zhu, Y.; Jiang, D.; Rong, Y.; Zhang, S.; Zhang, Y.; Ren, H.; Zhang, C.; et al. Kinesin 1 Drives Autolysosome Tubulation. Dev. Cell 2016, 37, 326–336. (In English) [Google Scholar] [CrossRef]
- Yang, H.; Tan, J.X. Lysosomal quality control: Molecular mechanisms and therapeutic implications. Trends Cell Biol. 2023, 33, 749–764. (In English) [Google Scholar] [CrossRef]
- Hung, Y.-H.; Chen, L.M.-W.; Yang, J.-Y.; Yang, W.Y. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 2013, 4, 2111. (In English) [Google Scholar] [CrossRef] [PubMed]
- Maejima, I.; Takahashi, A.; Omori, H.; Kimura, T.; Takabatake, Y.; Saitoh, T.; Yamamoto, A.; Hamasaki, M.; Noda, T.; Isaka, Y.; et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013, 32, 2336–2347. (In English) [Google Scholar] [CrossRef]
- Skowyra, M.L.; Schlesinger, P.H.; Naismith, T.V.; Hanson, P.I. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science 2018, 360, aar5078. (In English) [Google Scholar] [CrossRef] [PubMed]
- Tan, J.X.; Finkel, T. A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature 2022, 609, 815–821. (In English) [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wen, T.; Horn, H.T.; Chandrahas, V.K.; Thapa, N.; Choi, S.; Cryns, V.L.; Anderson, R.A. The nuclear phosphoinositide response to stress. Cell Cycle 2020, 19, 268–289. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, N.D.; Chen, M.; Cryns, V.L.; Anderson, R.A. Lipid transfer proteins initiate nuclear phosphoinositide signaling. BioRxiv 2023. [Google Scholar] [CrossRef]
- Chen, M.; Choi, S.; Wen, T.; Chen, C.; Thapa, N.; Lee, J.H.; Cryns, V.L.; Anderson, R.A. A p53-phosphoinositide signalosome regulates nuclear AKT activation. Nat. Cell Biol. 2022, 24, 1099–1113. [Google Scholar] [CrossRef]
- Chen, M.; Horn, H.T.; Wen, T.; Cryns, V.L.; Anderson, R.A. Assessing In Situ Phosphoinositide-Protein Interactions Through Fluorescence Proximity Ligation Assay in Cultured Cells. Methods Mol. Biol. 2021, 2251, 133–142. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, T.; Thapa, N.; Cryns, V.L.; Anderson, R.A. Regulation of Phosphoinositide Signaling by Scaffolds at Cytoplasmic Membranes. Biomolecules 2023, 13, 1297. https://doi.org/10.3390/biom13091297
Wen T, Thapa N, Cryns VL, Anderson RA. Regulation of Phosphoinositide Signaling by Scaffolds at Cytoplasmic Membranes. Biomolecules. 2023; 13(9):1297. https://doi.org/10.3390/biom13091297
Chicago/Turabian StyleWen, Tianmu, Narendra Thapa, Vincent L. Cryns, and Richard A. Anderson. 2023. "Regulation of Phosphoinositide Signaling by Scaffolds at Cytoplasmic Membranes" Biomolecules 13, no. 9: 1297. https://doi.org/10.3390/biom13091297
APA StyleWen, T., Thapa, N., Cryns, V. L., & Anderson, R. A. (2023). Regulation of Phosphoinositide Signaling by Scaffolds at Cytoplasmic Membranes. Biomolecules, 13(9), 1297. https://doi.org/10.3390/biom13091297






