The Past, Present, and Future of Genetically Engineered Mouse Models for Skeletal Biology
Abstract
1. Introduction
The Beginning of “Transgenic” Mice
2. Conditional Knockouts for Skeletal Biology
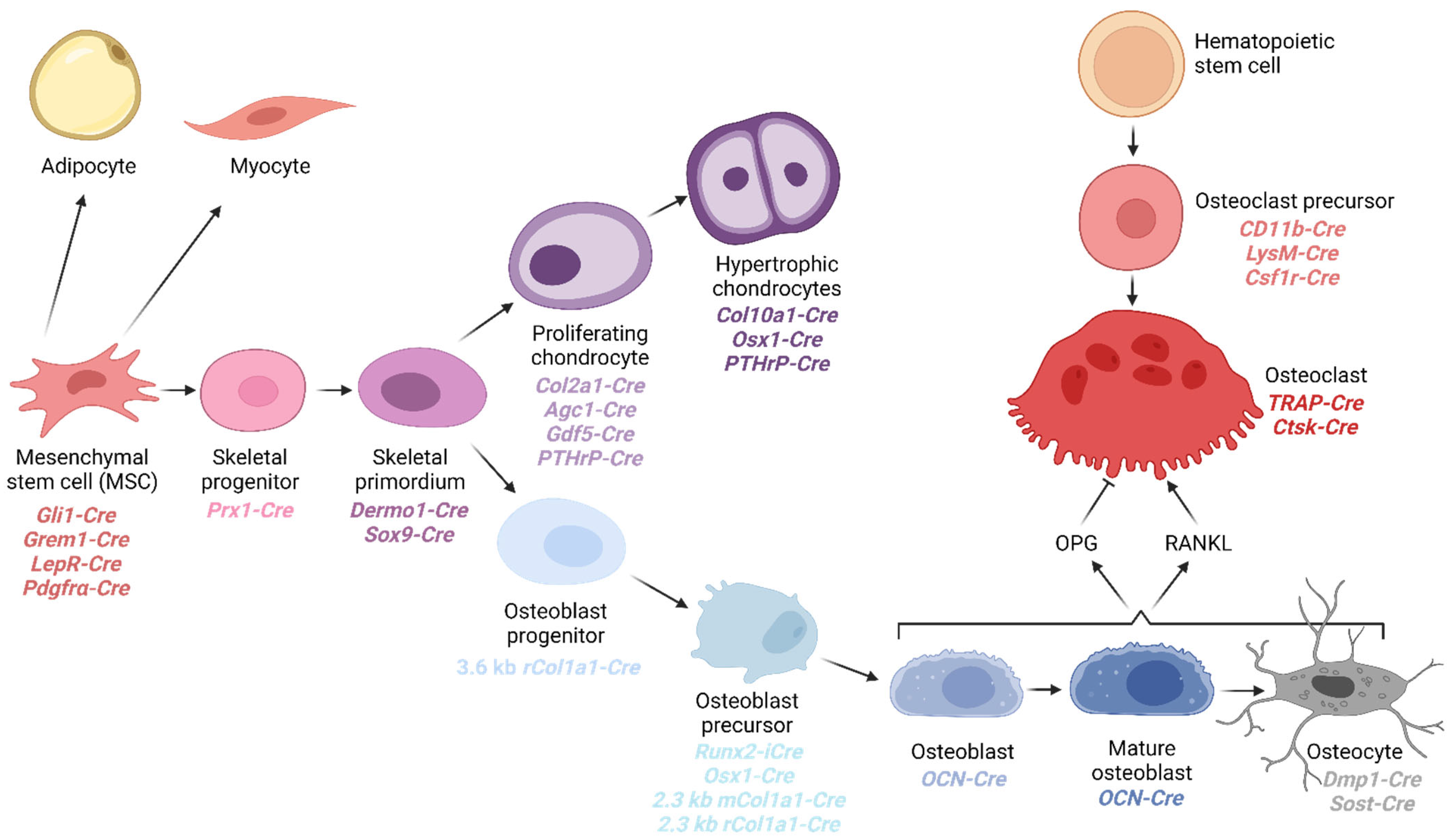
3. Cell Types of the Osteochondral Lineage
4. Mesenchymal Stem Cells within the Skeletal System
4.1. Gli1
4.2. Grem1
4.3. LepR
4.4. PDGFR-α
4.5. Additional BM-MSC Cre Drivers
5. Osteochondral Progenitor Cre Transgenic Strains
5.1. Prx1
5.2. Dermo1 (Twist2)
5.3. Sox9
6. Cre Transgenic Strains for Chondrocytes
6.1. Collagen II α1
6.2. Collagen 10α1
6.3. Agc1 (Aka Acan)
6.4. Gdf5
6.5. Prg4
6.6. PthrP
7. Osteoblast-Specific Transgenic Cre Strains
7.1. Runx2
7.2. Osterix1
7.3. Col1α1
7.4. Osteocalcin
8. Osteocyte Cres
8.1. DMP1
8.2. Sost
9. Osteoclasts
9.1. Myeloid Progenitors: Csf1r, LysM, and CD11b
9.2. TRAP and CtsK
10. The Future of Mouse Modeling
10.1. CRISPR/Cas9
10.2. Prime Editing and Base Editing
10.3. i-GONAD
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, K.; Fairfield, H.; Borgeia, S.; Curtain, M.; Hassan, M.G.; Dionne, L.; Karst, S.Y.; Coombs, H.; Bronson, R.T.; Reinholdt, L.G.; et al. Discovery and characterization of spontaneous mouse models of craniofacial dysmorphology. Dev. Biol. 2016, 415, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Fairfield, H.; Srivastava, A.; Ananda, G.; Liu, R.; Kircher, M.; Lakshminarayana, A.; Harris, B.S.; Karst, S.Y.; Dionne, L.A.; Kane, C.C.; et al. Exome sequencing reveals pathogenic mutations in 91 strains of mice with Mendelian disorders. Genome Res. 2015, 25, 948–957. [Google Scholar] [CrossRef]
- Brommage, R.; Ohlsson, C. High Fidelity of Mouse Models Mimicking Human Genetic Skeletal Disorders. Front. Endocrinol. 2019, 10, 934. [Google Scholar] [CrossRef] [PubMed]
- Rios, J.J.; Denton, K.; Russell, J.; Kozlitina, J.; Ferreira, C.R.; Lewanda, A.F.; Mayfield, J.E.; Moresco, E.; Ludwig, S.; Tang, M.; et al. Germline Saturation Mutagenesis Induces Skeletal Phenotypes in Mice. J. Bone Miner Res. 2021, 36, 1548–1565. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Mintz, B. Simian Virus 40 DNA Sequences in DNA of Healthy Adult Mice Derived from Preimplantation Blastocysts Injected with Viral DNA. Proc. Natl. Acad. Sci. USA 1974, 71, 1250–1254. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Sharing Laboratory Resources: Genetically Altered Mice: Summary of a Workshop Held at the National Academy of Sciences, 23–24 March 1993; National Research Council: Washington, DC, USA, 1994. [Google Scholar]
- Gordon, J.W.; Scangos, G.A.; Plotkin, D.J.; Barbosa, J.A.; Ruddle, F.H. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. USA 1980, 77, 7380–7384. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef]
- Bradley, A.; Evans, M.; Kaufman, M.H.; Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 1984, 309, 255–256. [Google Scholar] [CrossRef]
- Smithies, O. Forty years with homologous recombination. Nat. Med. 2001, 7, 1083–1086. [Google Scholar] [CrossRef]
- Soriano, P.; Montgomery, C.; Geske, R.; Bradley, A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 1991, 64, 693–702. [Google Scholar] [CrossRef]
- Copp, A.J. Death before birth: Clues from gene knockouts and mutations. Trends Genet. 1995, 11, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, D.L.; Abremski, K. Site-specific recombination by the bacteriophage P1 lox-Cre system: Cre-mediated synapsis of two lox sites. J. Mol. Biol. 1984, 178, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A. Cre recombinase: The universal reagent for genome tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Sauer, B.; Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 1988, 85, 5166–5170. [Google Scholar] [CrossRef] [PubMed]
- Sauer, B.; Henderson, N. Cre-stimulated recombination at loxP-containing DNA sequences placed into the mammalian genome. Nucleic Acids Res. 1989, 17, 147–161. [Google Scholar] [CrossRef]
- Sauer, B.; Henderson, N. Targeted insertion of exogenous DNA into the eukaryotic genome by the Cre recombinase. New Biol. 1990, 2, 441–449. [Google Scholar]
- Lakso, M.; Sauer, B.; Mosinger, B.; Lee, E.J.; Manning, R.W.; Yu, S.H.; Mulder, K.L.; Westphal, H. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc. Natl. Acad. Sci. USA 1992, 89, 6232–6236. [Google Scholar] [CrossRef]
- Orban, P.C.; Chui, D.; Marth, J.D. Tissue- and site-specific DNA recombination in transgenic mice. Proc. Natl. Acad. Sci. USA 1992, 89, 6861–6865. [Google Scholar] [CrossRef]
- Kim, H.; Kim, M.; Im, S.K.; Fang, S. Mouse Cre-LoxP system: General principles to determine tissue-specific roles of target genes. Lab. Anim. Res. 2018, 34, 147–159. [Google Scholar] [CrossRef]
- Kronenberg, H.M. Developmental regulation of the growth plate. Nature 2003, 423, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Galea, G.L.; Zein, M.R.; Allen, S.; Francis-West, P. Making and shaping endochondral and intramembranous bones. Dev. Dyn. 2020, 250, 414–449. [Google Scholar] [CrossRef] [PubMed]
- Kogianni, G.; Noble, B.S. The biology of osteocytes. Curr. Osteoporos. Rep. 2007, 5, 81–86. [Google Scholar] [CrossRef]
- Bar-Shavit, Z. The osteoclast: A multinucleated, hematopoietic-origin, bone-resorbing osteoimmune cell. J. Cell. Biochem. 2007, 102, 1130–1139. [Google Scholar] [CrossRef] [PubMed]
- Kendler, D.L.; Cosman, F.; Stad, R.K.; Ferrari, S. Denosumab in the Treatment of Osteoporosis: 10 Years Later: A Narrative Review. Adv. Ther. 2021, 39, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Lacey, D.L.; Boyle, W.J.; Simonet, W.S.; Kostenuik, P.J.; Dougall, W.C.; Sullivan, J.K.; Martin, J.S.; Dansey, R. Bench to bedside: Elucidation of the OPG–RANK–RANKL pathway and the development of denosumab. Nat. Rev. Drug Discov. 2012, 11, 401–419. [Google Scholar] [CrossRef]
- Simonet, W.S.; Lacey, D.L.; Dunstan, C.R.; Kelley, M.; Chang, M.S.; Luthy, R.; Nguyen, H.Q.; Wooden, S.; Bennett, L.; Boone, T.; et al. Osteoprotegerin: A Novel Secreted Protein Involved in the Regulation of Bone Density. Cell 1997, 89, 309–319. [Google Scholar] [CrossRef]
- VanKoevering, K.K.; Williams, B.O. Transgenic mouse strains for conditional gene deletion during skeletal development. IBMS BoneKEy 2008, 5, 151–170. [Google Scholar] [CrossRef]
- Ahn, S.; Joyner, A.L. Dynamic Changes in the Response of Cells to Positive Hedgehog Signaling during Mouse Limb Patterning. Cell 2004, 118, 505–516. [Google Scholar] [CrossRef]
- Shi, Y.; He, G.; Lee, W.-C.; McKenzie, J.A.; Silva, M.J.; Long, F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017, 8, 2043. [Google Scholar] [CrossRef]
- Worthley, D.L.; Churchill, M.; Compton, J.T.; Tailor, Y.; Rao, M.; Si, Y.; Levin, D.; Schwartz, M.G.; Uygur, A.; Hayakawa, Y.; et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell 2015, 160, 269–284. [Google Scholar] [CrossRef]
- DeFalco, J.; Tomishima, M.; Liu, H.; Zhao, C.; Cai, X.; Marth, J.D.; Enquist, L.; Friedman, J.M. Virus-Assisted Mapping of Neural Inputs to a Feeding Center in the Hypothalamus. Science 2001, 291, 2608–2613. [Google Scholar] [CrossRef]
- Zhou, B.O.; Yue, R.; Murphy, M.M.; Peyer, J.G.; Morrison, S.J. Faculty Opinions recommendation of Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell 2014, 15, 154–168. [Google Scholar] [CrossRef]
- Zhao, H.; Feng, J.; Ho, T.-V.; Grimes, W.; Urata, M.; Chai, Y. The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nature 2015, 17, 386–396. [Google Scholar] [CrossRef]
- Orr-Urtreger, A.; Lonai, P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development 1992, 115, 1045–1058. [Google Scholar] [CrossRef]
- Rivers, L.E.; Young, K.M.; Rizzi, M.; Jamen, F.; Psachoulia, K.; Wade, A.; Kessaris, N.; Richardson, W.D. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci. 2008, 11, 1392–1401. [Google Scholar] [CrossRef]
- Kang, S.H.; Fukaya, M.; Yang, J.K.; Rothstein, J.D.; Bergles, D.E. NG2+ CNS Glial Progenitors Remain Committed to the Oligodendrocyte Lineage in Postnatal Life and following Neurodegeneration. Neuron 2010, 68, 668–681. [Google Scholar] [CrossRef]
- Wattez, J.; Qiao, L.; Lee, S.; Natale, D.R.C.; Shao, J. The platelet-derived growth factor receptor alpha promoter-directed expression of cre recombinase in mouse placenta. Dev. Dyn. 2019, 248, 363–374. [Google Scholar] [CrossRef]
- O’Rourke, M.; Cullen, C.L.; Auderset, L.; Pitman, K.A.; Achatz, D.; Gasperini, R.; Young, K.M. Evaluating Tissue-Specific Recombination in a Pdgfralpha-CreERT2 Transgenic Mouse Line. PLoS ONE 2016, 11, e0162858. [Google Scholar] [CrossRef]
- Xu, J.; Wang, Y.; Li, Z.; Tian, Y.; Li, Z.; Lu, A.; Hsu, C.Y.; Negri, S.; Tang, C.; Tower, R.J.; et al. PDGFRalpha reporter activity identifies periosteal progenitor cells critical for bone formation and fracture repair. Bone Res. 2022, 10, 7. [Google Scholar] [CrossRef]
- Serowoky, M.A.; Arata, C.E.; Crump, J.G.; Mariani, F.V. Skeletal stem cells: Insights into maintaining and regenerating the skeleton. Development 2020, 147, dev179325. [Google Scholar] [CrossRef]
- Grcevic, D.; Pejda, S.; Matthews, B.G.; Repic, D.; Wang, L.; Li, H.; Kronenberg, M.S.; Jiang, X.; Maye, P.; Adams, D.J.; et al. In Vivo Fate Mapping Identifies Mesenchymal Progenitor Cells. Stem Cells 2012, 30, 187–196. [Google Scholar] [CrossRef]
- Matsushita, Y.; Nagata, M.; Kozloff, K.M.; Welch, J.D.; Mizuhashi, K.; Tokavanich, N.; Hallett, S.A.; Link, D.C.; Nagasawa, T.; Ono, W.; et al. A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat. Commun. 2020, 11, 332. [Google Scholar] [CrossRef]
- Kühn, R.; Schwenk, F.; Aguet, M.; Rajewsky, K. Inducible Gene Targeting in Mice. Science 1995, 269, 1427–1429. [Google Scholar] [CrossRef]
- Balordi, F.; Fishell, G. Mosaic Removal of Hedgehog Signaling in the Adult SVZ Reveals That the Residual Wild-Type Stem Cells Have a Limited Capacity for Self-Renewal. J. Neurosci. 2007, 27, 14248–14259. [Google Scholar] [CrossRef]
- Logan, M.; Martin, J.F.; Nagy, A.; Lobe, C.; Olson, E.N.; Tabin, C.J. Expression of Cre recombinase in the developing mouse limb bud driven by aPrxl enhancer. Genesis 2002, 33, 77–80. [Google Scholar] [CrossRef]
- Kawanami, A.; Matsushita, T.; Chan, Y.Y.; Murakami, S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem. Biophys. Res. Commun. 2009, 386, 477–482. [Google Scholar] [CrossRef]
- Li, L.; Cserjesi, P.; Olson, E.N. Dermo-1: A Novel Twist-Related bHLH Protein Expressed in the Developing Dermis. Dev. Biol. 1995, 172, 280–292. [Google Scholar] [CrossRef]
- Yu, K.; Xu, J.; Liu, Z.; Sosic, D.; Shao, J.; Olson, E.N.; Towler, D.A.; Ornitz, D.M. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development 2003, 130, 3063–3074. [Google Scholar] [CrossRef]
- Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999, 21, 70–71. [Google Scholar] [CrossRef]
- De Langhe, S.P.; Carraro, G.; Tefft, D.; Li, C.; Xu, X.; Chai, Y.; Minoo, P.; Hajihosseini, M.K.; Drouin, J.; Kaartinen, V.; et al. Formation and differentiation of multiple mesenchymal lineages during lung development is regulated by beta-catenin signaling. PLoS ONE 2008, 3, e1516. [Google Scholar] [CrossRef]
- Chen, H.; Zhuang, F.; Liu, Y.H.; Xu, B.; Del Moral, P.; Deng, W.; Chai, Y.; Kolb, M.; Gauldie, J.; Warburton, D.; et al. TGF-{beta} receptor II in Epithelia Versus Mesenchyme Plays Distinct Role in Developing Lung. Eur. Respir. J. 2008, 32, 285–295. [Google Scholar] [CrossRef]
- Sekiya, I.; Tsuji, K.; Koopman, P.; Watanabe, H.; Yamada, Y.; Shinomiya, K.; Nifuji, A.; Noda, M. SOX9 Enhances Aggrecan Gene Promoter/Enhancer Activity and Is Up-regulated by Retinoic Acid in a Cartilage-derived Cell Line, TC6. J. Biol. Chem. 2000, 275, 10738–10744. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; Huang, W.; Harley, V.R.; Goodfellow, P.N.; de Crombrugghe, B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol. Cell Biol. 1997, 17, 2336–2346. [Google Scholar] [CrossRef] [PubMed]
- Bridgewater, L.C.; Lefebvre, V.; de Crombrugghe, B. Chondrocyte-specific Enhancer Elements in the Col11a2 Gene Resemble the Col2a1 Tissue-specific Enhancer. J. Biol. Chem. 1998, 273, 14998–15006. [Google Scholar] [CrossRef] [PubMed]
- Ng, L.-J.; Wheatley, S.; Muscat, G.E.; Conway-Campbell, J.; Bowles, J.; Wright, E.; Bell, D.M.; Tam, P.P.; Cheah, K.S.; Koopman, P. SOX9 Binds DNA, Activates Transcription, and Coexpresses with Type II Collagen during Chondrogenesis in the Mouse. Dev. Biol. 1997, 183, 108–121. [Google Scholar] [CrossRef]
- Akiyama, H.; Kim, J.-E.; Nakashima, K.; Balmes, G.; Iwai, N.; Deng, J.M.; Zhang, Z.; Martin, J.F.; Behringer, R.R.; Nakamura, T.; et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. USA 2005, 102, 14665–14670. [Google Scholar] [CrossRef]
- Ovchinnikov, D.A.; Deng, J.M.; Ogunrinu, G.; Behringer, R.R. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis 2000, 26, 145–146. [Google Scholar] [CrossRef]
- Long, F.; Zhang, X.M.; Karp, S.; Yang, Y.; McMahon, A.P. Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 2001, 128, 5099–5108. [Google Scholar] [CrossRef]
- Schipani, E.; Ryan, H.E.; Didrickson, S.; Kobayashi, T.; Knight, M.; Johnson, R.S. Hypoxia in cartilage: HIF-1α is essential for chondrocyte growth arrest and survival. Genes Dev. 2001, 15, 2865–2876. [Google Scholar] [CrossRef]
- Sakai, K.; Hiripi, L.; Glumoff, V.; Brandau, O.; Eerola, R.; Vuorio, E.; Bösze, Z.; Fässler, R.; Aszódi, A. Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 2001, 19, 761–767. [Google Scholar] [CrossRef]
- Terpstra, L.; Prud’Homme, J.; Arabian, A.; Takeda, S.; Karsenty, G.; Dedhar, S.; St-Arnaud, R. Reduced chondrocyte proliferation and chondrodysplasia in mice lacking the integrin-linked kinase in chondrocytes. J. Cell Biol. 2003, 162, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Haigh, J.J.; Gerber, H.-P.; Ferrara, N.; Wagner, E.F. Conditional inactivation of VEGF-A in areas of collagen2a1 expression results in embryonic lethality in the heterozygous state. Development 2000, 127, 1445–1453. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, E.; Nguyen, M.T.; Mackem, S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 2006, 235, 2603–2612. [Google Scholar] [CrossRef]
- Grover, J.; Roughley, P.J. Generation of a transgenic mouse in which Cre recombinase is expressed under control of the type II collagen promoter and doxycycline administration. Matrix Biol. 2006, 25, 158–165. [Google Scholar] [CrossRef]
- Chen, M.; Lichtler, A.C.; Sheu, T.-J.; Xie, C.; Zhang, X.; O’Keefe, R.J.; Chen, D. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis 2007, 45, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Hilton, M.J.; Tu, X.; Long, F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev. Biol. 2007, 308, 93–105. [Google Scholar] [CrossRef]
- Couasnay, G.; Madel, M.; Lim, J.; Lee, B.; Elefteriou, F. Sites of Cre-recombinase activity in mouse lines targeting skeletal cells. J. Bone Miner. Res. 2021, 36, 1661–1679. [Google Scholar] [CrossRef]
- Yang, G.; Cui, F.; Hou, N.; Cheng, X.; Zhang, J.; Wang, Y.; Jiang, N.; Gao, X.; Yang, X. Transgenic mice that express Cre recombinase in hypertrophic chondrocytes. Genesis 2005, 42, 33–36. [Google Scholar] [CrossRef]
- Kim, Y.; Murao, H.; Yamamoto, K.; Deng, J.M.; Behringer, R.R.; Nakamura, T.; Akiyama, H. Generation of transgenic mice for conditional overexpression of Sox9. J. Bone Miner. Metab. 2010, 29, 123–129. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Chen, F.; Bian, H.; Wang, Q.; Zhang, X.; Sun, L.; Gu, J.; Lu, Y.; Zheng, Q. Hypertrophic chondrocyte-specific Col10a1 controlling elements in Cre recombinase transgenic studies. Am. J. Transl. Res. 2019, 11, 6672–6679. [Google Scholar] [PubMed]
- Gebhard, S.; Hattori, T.; Bauer, E.; Schlund, B.; Bösl, M.R.; de Crombrugghe, B.; von der Mark, K. Specific expression of Cre recombinase in hypertrophic cartilage under the control of a BAC-Col10a1 promoter. Matrix Biol. 2008, 27, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tsang, K.Y.; Tang, H.C.; Chan, D.; Cheah, K.S.E. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc. Natl. Acad. Sci. USA 2014, 111, 12097–12102. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.P.; Jang, C.W.; Deng, J.M.; Zhang, Z.; Behringer, R.R.; de Crombrugghe, B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 2009, 47, 805–814. [Google Scholar] [CrossRef]
- Roelofs, A.J.; Zupan, J.; Riemen, A.H.K.; Kania, K.; Ansboro, S.; White, N.; Clark, S.M.; De Bari, C. Joint morphogenetic cells in the adult mammalian synovium. Nat. Commun. 2017, 8, 15040. [Google Scholar] [CrossRef]
- Rountree, R.B.; Schoor, M.; Chen, H.; Marks, M.E.; Harley, V.; Mishina, Y.; Kingsley, D.M. BMP Receptor Signaling Is Required for Postnatal Maintenance of Articular Cartilage. PLoS Biol. 2004, 2, e355. [Google Scholar] [CrossRef]
- Shwartz, Y.; Viukov, S.; Krief, S.; Zelzer, E. Faculty Opinions recommendation of Joint Development Involves a Continuous Influx of Gdf5-Positive Cells. Cell Rep. 2016, 15, 2577–2587. [Google Scholar] [CrossRef]
- Decker, R.S.; Um, H.-B.; Dyment, N.A.; Cottingham, N.; Usami, Y.; Enomoto-Iwamoto, M.; Kronenberg, M.S.; Maye, P.; Rowe, D.W.; Koyama, E.; et al. Cell origin, volume and arrangement are drivers of articular cartilage formation, morphogenesis and response to injury in mouse limbs. Dev. Biol. 2017, 426, 56–68. [Google Scholar] [CrossRef]
- Kozhemyakina, E.; Zhang, M.; Ionescu, A.; Ayturk, U.M.; Ono, N.; Kobayashi, A.; Kronenberg, H.; Warman, M.L.; Lassar, A.B. Identification of a Prg4-Expressing Articular Cartilage Progenitor Cell Population in Mice. Arthritis Rheumatol. 2015, 67, 1261–1273. [Google Scholar] [CrossRef]
- Mizuhashi, K.; Ono, W.; Matsushita, Y.; Sakagami, N.; Takahashi, A.; Saunders, T.L.; Nagasawa, T.; Kronenberg, H.M.; Ono, N. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature 2018, 563, 254–258. [Google Scholar] [CrossRef]
- Karsenty, G. Minireview: Transcriptional control of osteoblast differentiation. Endocrinology 2001, 142, 2731–2733. [Google Scholar] [CrossRef]
- Rauch, A.; Seitz, S.; Baschant, U.; Schilling, A.F.; Illing, A.; Stride, B.; Kirilov, M.; Mandic, V.; Takacz, A.; Schmidt-Ullrich, R.; et al. Glucocorticoids Suppress Bone Formation by Attenuating Osteoblast Differentiation via the Monomeric Glucocorticoid Receptor. Cell Metab. 2010, 11, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Rodda, S.J.; McMahon, A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 2006, 133, 3231–3244. [Google Scholar] [CrossRef] [PubMed]
- Maes, C.; Kobayashi, T.; Selig, M.K.; Torrekens, S.; Roth, S.I.; Mackem, S.; Carmeliet, G.; Kronenberg, H.M. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell 2010, 19, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Rossert, J.A.; Chen, S.S.; Eberspaecher, H.; Smith, C.N.; de Crombrugghe, B. Identification of a minimal sequence of the mouse pro-alpha 1(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc. Natl. Acad. Sci. USA 1996, 93, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Rossert, J.; Eberspaecher, H.; De Crombrugghe, B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 1995, 129, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Dacquin, R.; Starbuck, M.; Schinke, T.; Karsenty, G. Mouse α1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev. Dyn. 2002, 224, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Woitge, H.W.; Braut, A.; Kronenberg, M.S.; Lichtler, A.C.; Mina, M.; Kream, B.E. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int. J. Dev. Biol. 2004, 48, 645–653. [Google Scholar] [CrossRef]
- Zha, L.; Hou, N.; Wang, J.; Yang, G.; Gao, Y.; Chen, L.; Yang, X. Collagen1α1 promoter drives the expression of Cre recombinase in osteoblasts of transgenic mice. J. Genet. Genom. 2008, 35, 525–530. [Google Scholar] [CrossRef]
- Kim, J.-E.; Nakashima, K.; de Crombrugghe, B. Transgenic Mice Expressing a Ligand-Inducible Cre Recombinase in Osteoblasts and Odontoblasts: A New Tool to Examine Physiology and Disease of Postnatal Bone and Tooth. Am. J. Pathol. 2004, 165, 1875–1882. [Google Scholar] [CrossRef]
- Zhang, M.; Xuan, S.; Bouxsein, M.L.; von Stechow, D.; Akeno, N.; Faugere, M.C.; Malluche, H.; Zhao, G.; Rosen, C.J.; Efstratiadis, A.; et al. Osteoblast-specific Knockout of the Insulin-like Growth Factor (IGF) Receptor Gene Reveals an Essential Role of IGF Signaling in Bone Matrix Mineralization. J. Biol. Chem. 2002, 277, 44005–44012. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, Y.; Kode, A.; Xu, L.; Mosialou, I.; Silva, B.C.; Ferron, M.; Clemens, T.L.; Economides, A.N.; Kousteni, S. Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. J. Bone Miner. Res. 2011, 26, 2012–2025. [Google Scholar] [CrossRef]
- Delgado-Calle, J.M.; Bellido, T. The osteocyte as a signaling cell. Physiol. Rev. 2022, 102, 379–410. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.B.; Robling, A.G. The Wnt pathway: An important control mechanism in bone’s response to mechanical loading. Bone 2021, 153, 116087. [Google Scholar] [CrossRef] [PubMed]
- Robling, A.G.; Turner, C.H. Mechanical Signaling for Bone Modeling and Remodeling. Crit. Rev. Eukaryot. Gene Expr. 2009, 19, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Hayashi, M.; Fukunaga, T.; Kurata, K.; Oh-Hora, M.; Feng, J.Q.; Bonewald, L.F.; Kodama, T.; Wutz, A.; Wagner, E.F.; et al. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 2011, 17, 1231–1234. [Google Scholar] [CrossRef]
- Xiong, J.; Onal, M.; Jilka, R.L.; Weinstein, R.S.; Manolagas, S.C.; O’Brien, C.A. Matrix-embedded cells control osteoclast formation. Nat. Med. 2011, 17, 1235–1241. [Google Scholar] [CrossRef]
- Sawakami, K.; Robling, A.G.; Ai, M.; Pitner, N.D.; Liu, D.; Warden, S.J.; Li, J.; Maye, P.; Rowe, D.W.; Duncan, R.L.; et al. The Wnt Co-receptor LRP5 Is Essential for Skeletal Mechanotransduction but Not for the Anabolic Bone Response to Parathyroid Hormone Treatment. J. Biol. Chem. 2006, 281, 23698–23711. [Google Scholar] [CrossRef]
- Lu, Y.; Xie, Y.; Zhang, S.; Dusevich, V.; Bonewald, L.; Feng, J. DMP1-targeted Cre Expression in Odontoblasts and Osteocytes. J. Dent. Res. 2007, 86, 320–325. [Google Scholar] [CrossRef]
- Kalajzic, I.; Braut, A.; Guo, D.; Jiang, X.; Kronenberg, M.; Mina, M.; Harris, M.; Harris, S.; Rowe, D. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone 2004, 35, 74–82. [Google Scholar] [CrossRef]
- Lim, J.; Burclaff, J.; He, G.; Mills, J.C.; Long, F. Unintended targeting of Dmp1-Cre reveals a critical role for Bmpr1a signaling in the gastrointestinal mesenchyme of adult mice. Bone Res. 2017, 5, 16049. [Google Scholar] [CrossRef] [PubMed]
- Bivi, N.; Condon, K.W.; Allen, M.R.; Farlow, N.; Passeri, G.; Brun, L.R.; Rhee, Y.; Bellido, T.; Plotkin, L.I. Cell autonomous requirement of connexin 43 for osteocyte survival: Consequences for endocortical resorption and periosteal bone formation. J. Bone Miner. Res. 2011, 27, 374–389. [Google Scholar] [CrossRef]
- Powell, W.F.; Barry, K.J.; Tulum, I.; Kobayashi, T.; Harris, S.E.; Bringhurst, F.R.; Pajevic, P.D. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J. Endocrinol. 2011, 209, 21–32. [Google Scholar] [CrossRef]
- van Bezooijen, R.L.; Roelen, B.A.; Visser, A.; van der Wee-Pals, L.; de Wilt, E.; Karperien, M.; Hamersma, H.; Papapoulos, S.E.; Dijke, P.T.; Löwik, C.W. Sclerostin Is an Osteocyte-expressed Negative Regulator of Bone Formation, But Not a Classical BMP Antagonist. J. Exp. Med. 2004, 199, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Piemontese, M.; Onal, M.; Campbell, J.; Goellner, J.J.; Dusevich, V.; Bonewald, L.; Manolagas, S.C.; O’brien, C.A. Osteocytes, not Osteoblasts or Lining Cells, are the Main Source of the RANKL Required for Osteoclast Formation in Remodeling Bone. PLoS ONE 2015, 10, e0138189. [Google Scholar] [CrossRef]
- Maurel, D.B.; Matsumoto, T.; Vallejo, J.A.; Johnson, M.L.; Dallas, S.L.; Kitase, Y.; Brotto, M.; Wacker, M.J.; Harris, M.A.; Harris, S.E.; et al. Characterization of a novel murine Sost ER(T2) Cre model targeting osteocytes. Bone Res. 2019, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Zhou, J.-F.; Sellers, R.S.; Li, J.-F.; Nguyen, A.V.; Wang, Y.; Orlofsky, A.; Liu, Q.; Hume, D.A.; Pollard, J.W.; et al. A Novel Mouse Model of Inflammatory Bowel Disease Links Mammalian Target of Rapamycin-Dependent Hyperproliferation of Colonic Epithelium to Inflammation-Associated Tumorigenesis. Am. J. Pathol. 2010, 176, 952–967. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Clausen, B.; Burkhardt, C.; Reith, W.; Renkawitz, R.; Förster, I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999, 8, 265–277. [Google Scholar] [CrossRef]
- Ferron, M.; Vacher, J. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis 2005, 41, 138–145. [Google Scholar] [CrossRef]
- Chiu, W.; McManus, J.; Notini, A.; Cassady, A.; Zajac, J.; Davey, R. Transgenic mice that express Cre recombinase in osteoclasts. Genesis 2004, 39, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Hayman, A.R. Tartrate-resistant acid phosphatase (TRAP) and the osteoclast/immune cell dichotomy. Autoimmunity 2008, 41, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Lecaille, F.; Brömme, D.; Lalmanach, G. Biochemical properties and regulation of cathepsin K activity. Biochimie 2008, 90, 208–226. [Google Scholar] [CrossRef]
- Dossa, T.; Arabian, A.; Windle, J.J.; Dedhar, S.; Teitelbaum, S.L.; Ross, F.P.; Roodman, G.D.; St-Arnaud, R. Osteoclast-specific inactivation of the integrin-linked kinase (ILK) inhibits bone resorption. J. Cell. Biochem. 2010, 110, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Copeland, N.G.; Jenkins, N.A.; Court, D.L. Recombineering: A powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001, 2, 769–779. [Google Scholar] [CrossRef]
- Nakamura, T.; Imai, Y.; Matsumoto, T.; Sato, S.; Takeuchi, K.; Igarashi, K.; Harada, Y.; Azuma, Y.; Krust, A.; Yamamoto, Y.; et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell 2007, 130, 811–823. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, M.A.; Sbacchi, S.; Correa-Tapia, M.; Naumann, R.; Klemm, J.; Chambon, P.; Al-Robaiy, S.; Blessing, M.; Hoflack, B. Transgenic Mice for a Tamoxifen-Induced, Conditional Expression of the Cre Recombinase in Osteoclasts. PLoS ONE 2012, 7, e37592. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 as a versatile tool for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.-W.W.; le Sage, C.; Larrieu, D.; Demir, M.; Jackson, S.P. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 2016, 6, 24356. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Kurt, I.C.; Zhou, R.; Iyer, S.; Garcia, S.P.; Miller, B.R.; Langner, L.M.; Grünewald, J.; Joung, J.K. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. 2020, 39, 41–46. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Gurumurthy, C.B.; Sato, M.; Nakamura, A.; Inui, M.; Kawano, N.; Islam, A.; Ogiwara, S.; Takabayashi, S.; Matsuyama, M.; Nakagawa, S.; et al. Creation of CRISPR-based germline-genome-engineered mice without ex vivo handling of zygotes by i-GONAD. Nat. Protoc. 2019, 14, 2452–2482. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M.; Sato, M.; Miura, H.; Takabayashi, S.; Matsuyama, M.; Koyano, T.; Arifin, N.; Nakamura, S.; Wada, K.; Gurumurthy, C.B. i-GONAD: A robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 2018, 19, 1–15. [Google Scholar] [CrossRef]
- Iwata, S.; Sasaki, T.; Nagahara, M.; Iwamoto, T. An efficient i-GONAD method for creating and maintaining lethal mutant mice using an inversion balancer identified from the C3H/HeJJcl strain. G3 Genes Genomes Genet. 2021, 11, jkab194. [Google Scholar] [CrossRef] [PubMed]
- Namba, M.; Kobayashi, T.; Koyano, T.; Kohno, M.; Ohtsuka, M.; Matsuyama, M. GONAD: A new method for germline genome editing in mice and rats. Dev. Growth Differ. 2021, 63, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Kawai, S.; Ooki, Y.; Chiba, T.; Ishii, C.; Nozawa, T.; Utsuki, H.; Umemura, M.; Takahashi, S.; Takahashi, Y. Functional validation of epitope-tagged ATF5 knock-in mice generated by improved genome editing of oviductal nucleic acid delivery (i-GONAD). Cell Tissue Res. 2021, 385, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Shang, R.; Zhang, H.; Bi, P. Generation of mouse conditional knockout alleles in one step using the i-GONAD method. Genome Res. 2020, 31, 121–130. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Aoshima, T.; Ito, R.; Shinmura, R.; Ohtsuka, M.; Akasaka, E.; Sato, M.; Takabayashi, S. Modification of i-GONAD Suitable for Production of Genome-Edited C57BL/6 Inbred Mouse Strain. Cells 2020, 9, 957. [Google Scholar] [CrossRef]
- Sato, M.; Takabayashi, S.; Akasaka, E.; Nakamura, S. Recent Advances and Future Perspectives of In Vivo Targeted Delivery of Genome-Editing Reagents to Germ cells, Embryos, and Fetuses in Mice. Cells 2020, 9, 799. [Google Scholar] [CrossRef]
- Sato, M.; Miyagasako, R.; Takabayashi, S.; Ohtsuka, M.; Hatada, I.; Horii, T. Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice. Cells 2020, 9, 546. [Google Scholar] [CrossRef]
- Koyano, T.; Namba, M.; Kobayashi, T.; Nakakuni, K.; Nakano, D.; Fukushima, M.; Nishiyama, A.; Matsuyama, M. The p21 dependent G2 arrest of the cell cycle in epithelial tubular cells links to the early stage of renal fibrosis. Sci. Rep. 2019, 9, 12059. [Google Scholar] [CrossRef]
- Lambert, L.J.; Challa, A.K.; Niu, A.; Zhou, L.; Tucholski, J.; Johnson, M.S.; Nagy, T.R.; Eberhardt, A.W.; Estep, P.N.; Kesterson, R.A.; et al. Increased trabecular bone and improved biomechanics in an osteocalcin null rat model created by CRISPR/Cas9 technology. Dis. Model. Mech. 2016, 9, 1169–1179. [Google Scholar] [CrossRef]
- Ubels, J.L.; Diegel, C.R.; Foxa, G.E.; Ethen, N.J.; Lensing, J.N.; Madaj, Z.B.; Williams, B.O. Low-Density Lipoprotein Receptor–Related Protein 5–Deficient Rats Have Reduced Bone Mass and Abnormal Development of the Retinal Vasculature. CRISPR J. 2020, 3, 284–298. [Google Scholar] [CrossRef]
- Williams, D.K.; Pinzón, C.; Huggins, S.; Pryor, J.H.; Falck, A.; Herman, F.; Oldeschulte, J.; Chavez, M.B.; Foster, B.L.; White, S.H.; et al. Genetic engineering a large animal model of human hypophosphatasia in sheep. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, C.; Zhang, Y.; Jiang, Y.; Qin, Y.; Pang, D.; Zhang, G.; Liu, H.; Xie, Z.; Yuan, H.; et al. A CRISPR-engineered swine model of COL2A1 deficiency recapitulates altered early skeletal developmental defects in humans. Bone 2020, 137, 115450. [Google Scholar] [CrossRef] [PubMed]
- dos Santos-Neto, P.; Cuadro, F.; Souza-Neves, M.; Crispo, M.; Menchaca, A. Refinements in embryo manipulation applied to CRISPR technology in livestock. Theriogenology 2023, 208, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Jabbar, A.; Zulfiqar, F.; Mahnoor, M.; Mushtaq, N.; Zaman, M.H.; Din, A.S.U.; Khan, M.A.; Ahmad, H.I. Advances and Perspectives in the Application of CRISPR-Cas9 in Livestock. Mol. Biotechnol. 2021, 63, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Menchaca, A.; dos Santos-Neto, P.C.; Mulet, A.P.; Crispo, M. CRISPR in livestock: From editing to printing. Theriogenology 2020, 150, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Lamas-Toranzo, I.; Ramos-Ibeas, P.; Pericuesta, E.; Bermejo-Álvarez, P. Directions and applications of CRISPR technology in livestock research. Anim. Reprod. 2018, 15, 292–300. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalski, M.N.; Williams, B.O. The Past, Present, and Future of Genetically Engineered Mouse Models for Skeletal Biology. Biomolecules 2023, 13, 1311. https://doi.org/10.3390/biom13091311
Michalski MN, Williams BO. The Past, Present, and Future of Genetically Engineered Mouse Models for Skeletal Biology. Biomolecules. 2023; 13(9):1311. https://doi.org/10.3390/biom13091311
Chicago/Turabian StyleMichalski, Megan N., and Bart O. Williams. 2023. "The Past, Present, and Future of Genetically Engineered Mouse Models for Skeletal Biology" Biomolecules 13, no. 9: 1311. https://doi.org/10.3390/biom13091311
APA StyleMichalski, M. N., & Williams, B. O. (2023). The Past, Present, and Future of Genetically Engineered Mouse Models for Skeletal Biology. Biomolecules, 13(9), 1311. https://doi.org/10.3390/biom13091311





