Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators
Abstract
1. Introduction
2. An Overview of Proteasome Regulation
2.1. Unfolding-Dependent Protein Degradation
2.2. Degradation of Unstructured Proteins (Ubiquitin-Independent)
3. Proteasome Structure and Function
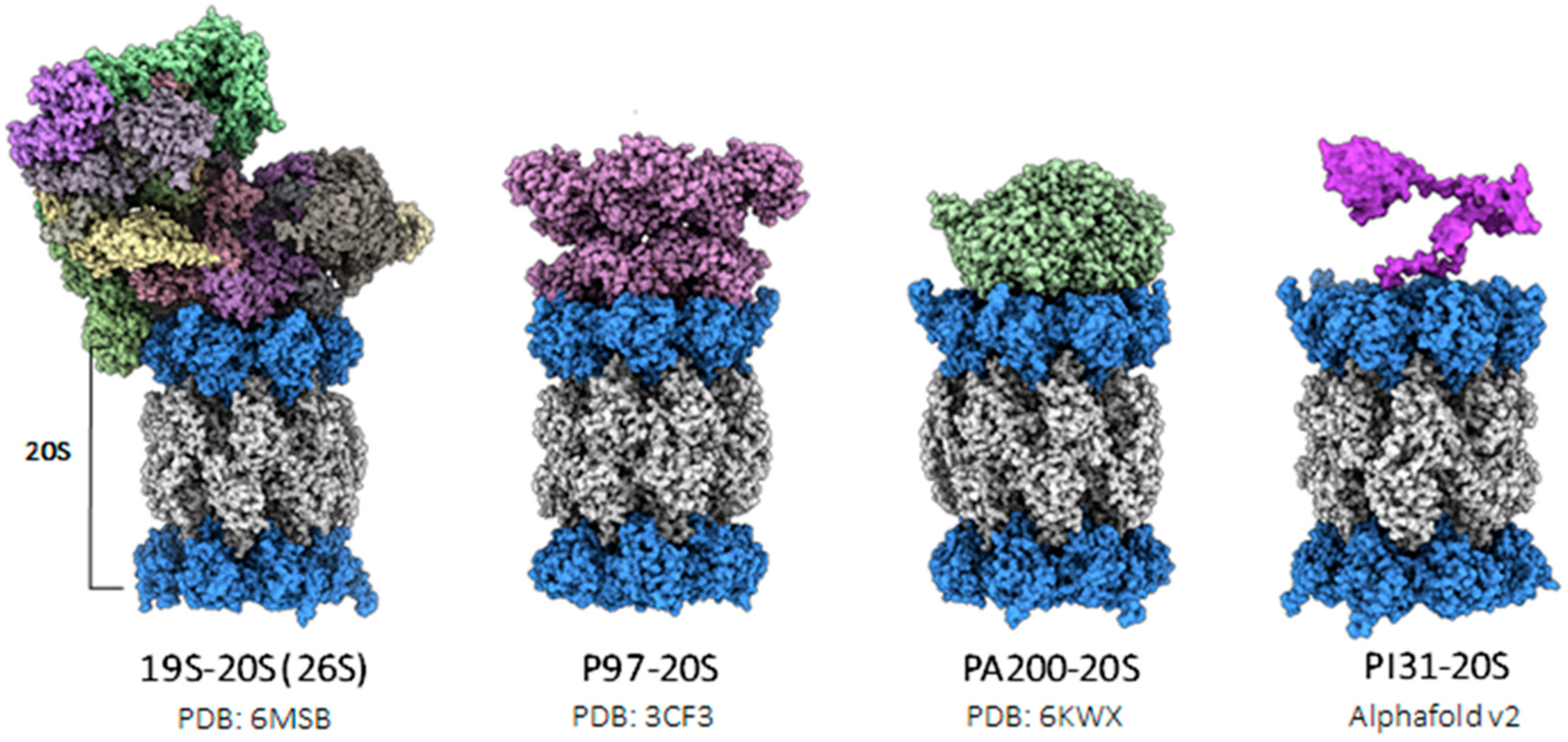
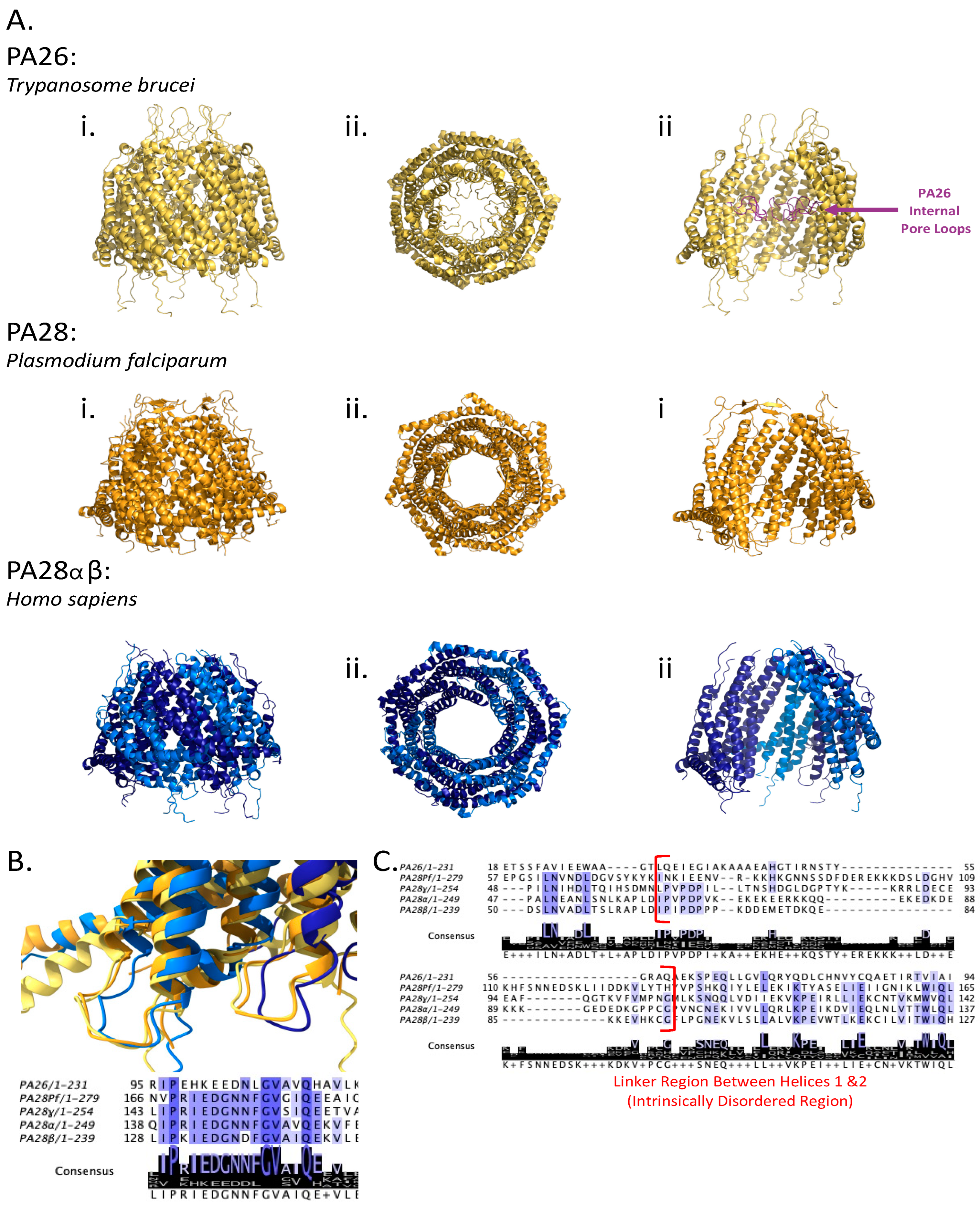
4. Diversity in the 11S Family of Proteasomal Regulators
5. PA28αβ
5.1. Structure-Function
5.2. Physiology
6. PA28γ
6.1. Structure-Function
6.2. Physiology
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Brodsky, J.L.; Werner, E.D.; Dubas, M.E.; Goeckeler, J.L.; Kruse, K.B.; McCracken, A.A. The requirement for molecular chaperones during endoplasmic reticulum- associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 1999, 274, 3453–3460. [Google Scholar] [CrossRef]
- Etlinger, J.D.; Goldberg, A.L. A soluble ATP-dependent proteolytic system responsible for the degradation of abnormal proteins in reticulocytes. Proc. Natl. Acad. Sci. USA 1977, 74, 54–58. [Google Scholar] [CrossRef]
- Konstantinova, I.M.; Tsimokha, A.S.; Mittenberg, A.G. Role of Proteasomes in Cellular Regulation. Int. Rev. Cell Mol. Biol. 2008, 267, 59–124. [Google Scholar] [CrossRef]
- Bassermann, F.; Eichner, R.; Pagano, M. The ubiquitin proteasome system—Implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 2003, 40, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M. The Anaphase-Promoting Complex: Proteolysis in Mitosis and Beyond. Mol. Cell 2002, 9, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Ben-Aroya, S.; Agmon, N.; Yuen, K.; Kwok, T.; McManus, K.; Kupiec, M.; Hieter, P. Proteasome Nuclear Activity Affects Chromosome Stability by Controlling the Turnover of Mms22, a Protein Important for DNA Repair. PLoS Genet. 2010, 6, e1000852. [Google Scholar] [CrossRef]
- Krogan, N.J.; Lam, M.H.; Fillingham, J.; Keogh, M.-C.; Gebbia, M.; Li, J.; Datta, N.; Cagney, G.; Buratowski, S.; Emili, A.; et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 2004, 16, 1027–1034. [Google Scholar] [CrossRef]
- Muratani, M.; Tansey, W.P. How the ubiquitin—Proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003, 4, 192–201. [Google Scholar] [CrossRef]
- Orlowski, R.Z. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999, 6, 303–313. [Google Scholar] [CrossRef]
- Niedermann, G. Immunological functions of the proteasome. Curr. Top. Microbiol. Immunol. 2002, 268, 91–136. [Google Scholar] [CrossRef] [PubMed]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M. Ubiquitin-dependent proteolysis: Its role in human diseases and the design of therapeutic strategies. Mol. Genet. Metab. 2002, 77, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Bajorek, M.; Köhler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Wolf, D.H.; Stolz, A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 117–124. [Google Scholar] [CrossRef]
- Gerega, A.; Rockel, B.; Peters, J.; Tamura, T.; Baumeister, W.; Zwickl, P. VAT, the Thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. J. Biol. Chem. 2005, 280, 42856–42862. [Google Scholar] [CrossRef]
- Coux, O.; Tanaka, K.; Goldberg, A.L. Structure and Functions of the 20S and 26S Proteasomes. Annu. Rev. Biochem. 2003, 65, 801–847. [Google Scholar] [CrossRef]
- Tanaka, K.; Ii, K.; Ichihara, A.; Waxman, L.; Goldberg, A.L. A high molecular weight protease in the cytosol of rat liver. I. Purification, enzymological properties, and tissue distribution. J. Biol. Chem. 1986, 261, 15197–15203. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 2003, 67, 425–479. [Google Scholar] [CrossRef]
- Bodnar, N.; Rapoport, T.; Meyer, H.; Ye, Y.; Freemont, P.; Hoppe, T.; Franz, A. Toward an understanding of the Cdc48/p97 ATPase. F1000Research 2017, 6, 1318. [Google Scholar] [CrossRef]
- Wu, X.; Rapoport, T.A. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018, 53, 22–28. [Google Scholar] [CrossRef]
- Erales, J.; Coffino, P. Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 216–221. [Google Scholar] [CrossRef]
- Hoyt, M.A.; Coffino, P. Ubiquitin-proteasome system. Cell. Mol. Life Sci. 2004, 61, 1596–1600. [Google Scholar] [CrossRef]
- Tanaka, K.; Waxman, L.; Goldberg, A.L. ATP serves two distinct roles in protein degradation in reticulocytes, one requiring and one independent of ubiquitin. J. Cell Biol. 1983, 96, 1580–1585. [Google Scholar] [CrossRef] [PubMed]
- Coskuner-Weber, O.; Mirzanli, O.; Uversky, V.N. Intrinsically disordered proteins and proteins with intrinsically disordered regions in neurodegenerative diseases. Biophys. Rev. 2022, 14, 679–707. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castaño, P.; Rizzuti, B.; Xia, Y.; Abian, O.; Peng, L.; Velázquez-Campoy, A.; Neira, J.; Iovanna, J. Targeting intrinsically disordered proteins involved in cancer. Cell. Mol. Life Sci. 2020, 77, 1695–1707. [Google Scholar] [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Midic, U.; Xie, H.; Xue, B.; Vucetic, S.; Iakoucheva, L.M.; Obradovic, Z.; Dunker, A.K. Unfoldomics of human diseases: Linking protein intrinsic disorder with diseases. BMC Genom. 2009, 10, S7. [Google Scholar] [CrossRef]
- Shang, F.; Taylor, A. Oxidative stress and recovery from oxidative stress are associated with altered ubiquitin conjugating and proteolytic activities in bovine lens epithelial cells. Biochemistry 1995, 307, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, F.K.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef] [PubMed]
- Pickering, A.M.; Davies, K.J.A. Differential roles of proteasome and immunoproteasome regulators Pa28αβ, Pa28γ and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef]
- Bader, N.; Grune, T. Protein oxidation and proteolysis. Biol. Chem. 2006, 387, 1351–1355. [Google Scholar] [CrossRef]
- Qian, M.-X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.-Y.; Ji, D.-Y.; Wang, G.-F.; Zhu, Q.-Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012. [Google Scholar] [CrossRef]
- Toste Rêgo, A.; da Fonseca, P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell 2019, 76, 138–147.e5. [Google Scholar] [CrossRef] [PubMed]
- Blickwedehl, J.; Agarwal, M.; Seong, C.; Pandita, R.K.; Melendy, T.; Sung, P.; Pandita, T.K.; Bangia, N. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc. Natl. Acad. Sci. USA 2008, 105, 16165–16170. [Google Scholar] [CrossRef] [PubMed]
- Chu-Ping, M.; Willy, P.J.; Slaughter, C.A.; DeMartino, G.N. PA28, an activator of the 20 S proteasome, is inactivated by proteolytic modification at its carboxyl terminus. J. Biol. Chem. 1993, 268, 22514–22519. [Google Scholar] [CrossRef]
- Minis, A.; Rodriguez, J.A.; Levin, A.; Liu, K.; Govek, E.E.; Hatten, M.E.; Steller, H. The proteasome regulator PI31 is required for protein homeostasis, synapse maintenance, and neuronal survival in mice. Proc. Natl. Acad. Sci. USA 2019, 116, 24639–24650. [Google Scholar] [CrossRef]
- Hsu, H.C.; Wang, J.; Kjellgren, A.; Li, H.; DeMartino, G.N. High-resolution structure of mammalian PI31—20S proteasome complex reveals mechanism of proteasome inhibition. J. Biol. Chem. 2023, 299, 104862. [Google Scholar] [CrossRef]
- Huber, E.M.; Groll, M. The Mammalian Proteasome Activator PA28 Forms an Asymmetric α4β3 Complex. Structure 2017, 25, 1473–1480.e3. [Google Scholar] [CrossRef]
- Realini, C.; Jensen, C.C.; Zhang, Z.G.; Johnston, S.C.; Knowlton, J.R.; Hill, C.P.; Rechsteiner, M. Characterization of recombinant REGα, REGβ, and REGγ proteasome activators. J. Biol. Chem. 1997, 272, 25483–25492. [Google Scholar] [CrossRef]
- Förster, A.; Masters, E.I.; Whitby, F.G.; Robinson, H.; Hill, C.P. The 1.9 Å structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol. Cell 2005, 18, 589–599. [Google Scholar] [CrossRef]
- Whitby, F.G.; Masters, E.I.; Kramer, L.; Knowlton, J.R.; Yao, Y.; Wang, C.C.; Hill, C.P. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 2000, 408, 115–120. [Google Scholar] [CrossRef]
- Stadtmueller, B.M.; Hill, C.P. Proteasome Activators. Mol. Cell 2011, 41, 8–19. [Google Scholar] [CrossRef]
- Cascio, P. PA28αβ: The Enigmatic Magic Ring of the Proteasome? Biomolecules 2014, 4, 566–584. [Google Scholar] [CrossRef]
- Li, J.; Rechsteiner, M. Molecular dissection of the 11S REG (PA28) proteasome activators. Biochimie 2001, 83, 373–383. [Google Scholar] [CrossRef]
- Thomas, T.A.; Smith, D.M. Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. J. Biol. Chem. 2022, 298, 102140. [Google Scholar] [CrossRef] [PubMed]
- Jonik-Nowak, B.; Menneteau, T.; Fesquet, D.; Baldin, V.; Bonne-Andrea, C.; Méchali, F.; Fabre, B.; Boisguerin, P.; de Rossi, S.; Henriquet, C.; et al. PIP30/FAM192A is a novel regulator of the nuclear proteasome activator PA28γ. Proc. Natl. Acad. Sci. USA 2018, 115, E6477–E6486. [Google Scholar] [CrossRef] [PubMed]
- Wilk, S.; Chen, W.E.; Magnusson, R.P. Properties of the Beta Subunit of the Proteasome Activator PA28 (11S REG). Arch. Biochem. Biophys. 2000, 384, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Frayssinhes, J.-Y.A.; Cerruti, F.; Laulin, J.; Cattaneo, A.; Bachi, A.; Apcher, S.; Coux, O.; Cascio, P. PA28γ-20S proteasome is a proteolytic complex committed to degrade unfolded proteins. Cell. Mol. Life Sci. CMLS 2021, 79, 45. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Löwe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4Å resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Groll, M.; Huber, R. Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 2003, 35, 606–616. [Google Scholar] [CrossRef]
- Baumeister, W.; Cejka, Z.; Kania, M.; Seemüller, E. The Proteasome: A Macromolecular Assembly Designed to Confine Proteolysis to a Nanocompartment. Biol. Chem. 1997, 378, 121–206. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, A.F.; Akopian, T.N.; Castillo, V.; Goldberg, A.L. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol. Cell 1999, 4, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Realini, C.; Rogers, S.W.; Rechsteiner, M. KEKE motifs. FEBS Lett. 1994, 348, 109–113. [Google Scholar] [CrossRef]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the Proteasomal ATPases’ Carboxyl Termini in the 20S Proteasome’s α Ring Opens the Gate for Substrate Entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Früh, K.; Yang, Y. Antigen presentation by MHC class I and its regulation by interferon gamma. Curr. Opin. Immunol. 1999, 11, 76–81. [Google Scholar] [CrossRef]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28αβ Reduces Size and Increases Hydrophilicity of 20S Immunoproteasome Peptide Products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef]
- Stohwasser, R.; Soza, A.; Eggers, M.; Koszinowski, U.H.; Kloetzel, P.M. PA28αβ double and PA28β single transfectant mouse B8 cell lines reveal enhanced presentation of a mouse cytomegalovirus (MCMV) pp89 MHC class I epitope. Mol. Immunol. 2000, 37, 13–19. [Google Scholar] [CrossRef]
- Driscoll, J.; Brown, M.G.; Finley, D.; Monaco, J.J. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature 1993, 365, 262–264. [Google Scholar] [CrossRef]
- Gaczynska, M.; Rock, K.L.; Goldberg, A.L. γ-Interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 1993, 365, 264–267. [Google Scholar] [CrossRef]
- Hisamatsu, H.; Shimbara, N.; Saito, Y.; Kristensen, P.; Hendil, K.B.; Fujiwara, T.; Takahashi, E.; Tanahashi, N.; Tamura, T.; Ichihara, A.; et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J. Exp. Med. 1996, 183, 1807–1816. [Google Scholar] [CrossRef]
- Rock, K.L.; Gramm, C.; Rothstein, L.; Clark, K.; Stein, R.; Dick, L.; Hwang, D.; Goldberg, A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994, 78, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Huang, L.; Krutchinsky, A.; Wong, M.L.; Standing, K.G.; Burlingame, A.L.; Wang, C.C. Structural and functional characterizations of the proteasome-activating protein PA26 from Trypanosoma brucei. J. Biol. Chem. 1999, 274, 33921–33930. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.C.; Metcalfe, R.D.; Hanssen, E.; Yang, T.; Gillett, D.L.; Leis, A.P.; Morton, C.J.; Kuiper, M.J.; Parker, M.W.; Spillman, N.J.; et al. The structure of the PA28—20S proteasome complex from Plasmodium falciparum and implications for proteostasis. Nat. Microbiol. 2019, 4, 1990–2000. [Google Scholar] [CrossRef]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of Proteasome Regulators in Eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef] [PubMed]
- Masson, P.; Andersson, O.; Petersen, U.M.; Young, P. Identification and Characterization of a DrosophilaNuclear Proteasome Regulator: A Homolog of Human 11 S REGγ (PA28γ). J. Biol. Chem. 2001, 276, 1383–1390. [Google Scholar] [CrossRef]
- Murray, B.W.; Sültmann, H.; Klein, J. Identification and Linkage of the Proteasome Activator Complex PA28 Subunit Genes in Zebrafish. Scand. J. Immunol. 2000, 51, 571–576. [Google Scholar] [CrossRef]
- Zhang, Z.; Clawson, A.; Rechsteiner, M. The Proteasome Activator 11 S Regulator or PA28. J. Biol. Chem. 1998, 273, 30660–30668. [Google Scholar] [CrossRef]
- Zhang, Z.; Realini, C.; Clawson, A.; Endicott, S.; Rechsteiner, M. Proteasome activation by REG molecules lacking homolog-specific inserts. J. Biol. Chem. 1998, 273, 9501–9509. [Google Scholar] [CrossRef]
- Li, T.; Naqvi, N.I.; Yang, H.; Teo, T.S. Identification of a 26S Proteasome-Associated UCH in Fission Yeast. Biochem. Biophys. Res. Commun. 2000, 272, 270–275. [Google Scholar] [CrossRef]
- Song, X.; Von Kampen, J.; Slaughter, C.A.; DeMartino, G.N. Relative functions of the α and β subunits of the proteasome activator, PA28. J. Biol. Chem. 1997, 272, 27994–28000. [Google Scholar] [CrossRef]
- Zhao, J.; Makhija, S.; Zhou, C.; Zhang, H.; Wang, Y.; Muralidharan, M.; Huang, B.; Cheng, Y. Structural insights into the human PA28—20S proteasome enabled by efficient tagging and purification of endogenous proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2207200119. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Y.; Xu, C.; Chen, K.; Zhao, Q.; Wang, S.; Yin, Y.; Peng, C.; Ding, Z.; Cong, Y. Cryo-EM of mammalian PA28αβ-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28αβ. Nat. Commun. 2021, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P.; Goldberg, A.L. Preparation of Hybrid (19S-20S-PA28) Proteasome Complexes and Analysis of Peptides Generated during Protein Degradation. Methods Enzymol. 2005, 398, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P.; Call, M.; Petre, B.M.; Walz, T.; Goldberg, A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002, 21, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Borissenko, L.; Groll, M. 20S proteasome and its inhibitors: Crystallographic knowledge for drug development. Chem. Rev. 2007, 107, 687–717. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Goldberg, A.L. Degradation of Cell Proteins and the Generation of MHC Class I-Presented Peptides. Annu. Rev. Immunol. 1999, 17, 739–779. [Google Scholar] [CrossRef]
- Schwarz, K.; van Den Broek, M.; Kostka, S.; Kraft, R.; Soza, A.; Schmidtke, G.; Kloetzel, P.-M.; Groettrup, M. Overexpression of the Proteasome Subunits LMP2, LMP7, and MECL-1, But Not PA28α/β, Enhances the Presentation of an Immunodominant Lymphocytic Choriomeningitis Virus T Cell Epitope. J. Immunol. 2000, 165, 768–778. [Google Scholar] [CrossRef]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J.A. The immunoproteasome, the 20S proteasome and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–595. [Google Scholar] [CrossRef]
- Pickering, A.M.; Linder, R.A.; Zhang, H.; Forman, H.J.; Davies, K.J.A. Nrf2-dependent Induction of Proteasome and Pa28αβ Regulator Are Required for Adaptation to Oxidative Stress. J. Biol. Chem. 2012, 287, 10021–10031. [Google Scholar] [CrossRef]
- Wu, D.G.; Wang, Y.N.; Zhou, Y.; Gao, H.; Zhao, B. Inhibition of the Proteasome Regulator PA28 Aggravates Oxidized Protein Overload in the Diabetic Rat Brain. Cell. Mol. Neurobiol. 2023, 43, 2857–2869. [Google Scholar] [CrossRef]
- Nikaido, T.; Shimada, K.; Shibata, M.; Hata, M.; Sakamoto, M.; Takasaki, Y.; Sato, C.; Takahashi, T.; Nishida, Y. Cloning and nucleotide sequence of cDNA for Ki antigen, a highly conserved nuclear protein detected with sera from patients with systemic lupus erythematosus. Clin. Exp. Immunol. 1990, 79, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, J.; Pratt, G.; Wilk, S.; Rechsteiner, M. Purification procedures determine the proteasome activation properties of REGγ (PA28γ). Arch. Biochem. Biophys. 2004, 425, 158–164. [Google Scholar] [CrossRef] [PubMed]
- De la Peña, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362, eaav0725. [Google Scholar] [CrossRef]
- Dong, Y.; Zhang, S.; Wu, Z.; Li, X.; Wang, W.L.; Zhu, Y.; Stoilova-McPhie, S.; Lu, Y.; Finley, D.; Mao, Y. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 2018, 565, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.D.; Hao, J.; Shen, C.H.; Deng, X.M.; Yun, C.H. Atomic resolution Cryo-EM structure of human proteasome activator PA28γ. Int. J. Biol. Macromol. 2022, 219, 500–507. [Google Scholar] [CrossRef]
- Liu, Y.; Qiao, F.; Leiferman, P.C.; Ross, A.; Schlenker, E.H.; Wang, H. FOXOs modulate proteasome activity in human-induced pluripotent stem cells of Huntington’s disease and their derived neural cells. Hum. Mol. Genet. 2017, 26, 4416–4428. [Google Scholar] [CrossRef]
- Moncsek, A.; Gruner, M.; Meyer, H.; Lehmann, A.; Kloetzel, P.M.; Stohwasser, R. Evidence for anti-apoptotic roles of proteasome activator 28γ via inhibiting caspase activity. Apoptosis 2015, 20, 1211–1228. [Google Scholar] [CrossRef]
- Murata, S.; Kawahara, H.; Tohma, S.; Yamamoto, K.; Kasahara, M.; Nabeshima, Y.-I.; Tanaka, K.; Chiba, T. Growth retardation in mice lacking the proteasome activator PA28γ. J. Biol. Chem. 1999, 274, 38211–38215. [Google Scholar] [CrossRef]
- Moriishi, K.; Mochizuki, R.; Moriya, K.; Miyamoto, H.; Mori, Y.; Abe, T.; Murata, S.; Tanaka, K.; Miyamura, T.; Suzuki, T.; et al. Critical role of PA28γ in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 1661–1666. [Google Scholar] [CrossRef]
- Levy-Barda, A.; Lerenthal, Y.; Davis, A.J.; Chung, Y.M.; Essers, J.; Shao, Z.; van Vliet, N.; Chen, D.J.; Hu, M.C.-T.; Kanaar, R.; et al. Involvement of the nuclear proteasome activator PA28γ in the cellular response to DNA double-strand breaks. Cell Cycle 2011, 10, 4300–4310. [Google Scholar] [CrossRef]
- Uchimura, Y.; Barton, L.F.; Rada, C.; Neuberger, M.S. REG-γ associates with and modulates the abundance of nuclear activation-induced deaminase. J. Exp. Med. 2011, 208, 2385–2391. [Google Scholar] [CrossRef]
- Huang, L.; Haratake, K.; Miyahara, H.; Chiba, T. Proteasome activators, PA28γ and PA200, play indispensable roles in male fertility. Sci. Rep. 2016, 6, 23171. [Google Scholar] [CrossRef]
- Fesquet, D.; Llères, D.; Grimaud, C.; Viganò, C.; Méchali, F.; Boulon, S.; Coux, O.; Bonne-Andrea, C.; Baldin, V. The 20S proteasome activator PA28γ controls the compaction of chromatin. J. Cell Sci. 2021, 134, jcs257717. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, S.; Tan, J.; Tian, T.; Ran, L.; Rodier, J.F.; Ren, G. REG gamma: A potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med. Oncol. 2011, 28, 31–41. [Google Scholar] [CrossRef]
- Yi, Z.; Yang, D.; Liao, X.; Guo, F.; Wang, Y.; Wang, X. PSME3 induces epithelial—Mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp. Cell Res. 2017, 358, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Okamura, T.; Taniguchi, S.-I.; Ohkura, T.; Yoshida, A.; Shimizu, H.; Sakai, M.; Maeta, H.; Fukui, H.; Ueta, Y.; Hisatome, I.; et al. Abnormally High Expression of Proteasome Activator-γ in Thyroid Neoplasm. J. Clin. Endocrinol. Metab. 2003, 88, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dang, Y.; Zhang, J.; Yan, W.; Zhai, W.; Chen, H.; Li, K.; Tong, L.; Gao, X.; Amjad, A.; et al. REGγ is critical for skin carcinogenesis by modulating the Wnt/β-catenin pathway. Nat. Commun. 2015, 6, 6875. [Google Scholar] [CrossRef]
- Roessler, M.; Rollinger, W.; Mantovani-Endl, L.; Hagmann, M.-L.; Palme, S.; Berndt, P.; Engel, A.M.; Pfeffer, M.; Karl, J.; Bodenmüller, H.; et al. Identification of PSME3 as a Novel Serum Tumor Marker for Colorectal Cancer by Combining Two-dimensional Polyacrylamide Gel Electrophoresis with a Strictly Mass Spectrometry-based Approach for Data Analysis. Mol. Cell. Proteom. 2006, 5, 2092–2101. [Google Scholar] [CrossRef]
- Xiong, S.; Zheng, Y.; Jiang, P.; Liu, R.; Liu, X.; Qian, J.; Gu, J.; Chang, L.; Ge, D.; Chu, Y. PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br. J. Cancer 2013, 110, 353–362. [Google Scholar] [CrossRef]
- Xu, X.; Liu, D.; Ji, N.; Li, T.; Li, L.; Jiang, L.; Li, J.; Zhang, P.; Zeng, X.; Chen, Q. A novel transcript variant of proteasome activator 28γ: Identification and function in oral cancer cells. Int. J. Oncol. 2015, 47, 188–194. [Google Scholar] [CrossRef]
- Jeon, J.; Kim, W.; Jang, J.; Isacson, O.; Seo, H. Gene therapy by proteasome activator, PA28γ, improves motor coordination and proteasome function in Huntington’s disease YAC128 mice. Neuroscience 2016, 324, 20–28. [Google Scholar] [CrossRef]
- Seo, H.; Sonntag, K.C.; Kim, W.; Cattaneo, E.; Isacson, O. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS ONE 2007, 2, e238. [Google Scholar] [CrossRef]
- Yersak, J.M.; Montie, H.L.; Chevalier-Larsen, E.S.; Liu, Y.; Huang, L.; Rechsteiner, M.; Merry, D.E. The 11S Proteasomal Activator REGγ Impacts Polyglutamine-Expanded Androgen Receptor Aggregation and Motor Neuron Viability through Distinct Mechanisms. Front. Mol. Neurosci. 2017, 10, 159. [Google Scholar] [CrossRef]
- Xie, Y.; Gao, Y.; Gao, R.; Yang, W.; Dong, Z.; Moses, R.E.; Sun, A.; Li, X.; Ge, J. The proteasome activator REGγ accelerates cardiac hypertrophy by declining PP2Acα—SOD2 pathway. Cell Death Differ. 2020, 27, 2952–2972. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Moriishi, K.; Fukuda, K.; Shirakura, M.; Ishii, K.; Shoji, I.; Wakita, T.; Miyamura, T.; Matsuura, Y.; Suzuki, T. Proteasomal Turnover of Hepatitis C Virus Core Protein Is Regulated by Two Distinct Mechanisms: A Ubiquitin-Dependent Mechanism and a Ubiquitin-Independent but PA28γ-Dependent Mechanism. J. Virol. 2009, 83, 2389–2392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tu, J.; Cao, C.; Yang, T.; Gao, L. Proteasome activator PA28γ-dependent degradation of coronavirus disease (COVID-19) nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 529, 251–256. [Google Scholar] [CrossRef]
- Cascio, P. PA28γ: New Insights on an Ancient Proteasome Activator. Biomolecules 2021, 11, 228. [Google Scholar] [CrossRef]
- Ahadi, S.; Zhou, W.; Rose, S.M.S.-F.; Sailani, M.R.; Contrepois, K.; Avina, M.; Ashland, M.; Brunet, A.; Snyder, M. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 2020, 26, 83–90. [Google Scholar] [CrossRef]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.M.; Ping, L.; Zhou, M.; Yin, L.; Higginbotham, L.A.; Guajardo, A.; White, B.; Troncoso, J.C.; et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020, 26, 769–780. [Google Scholar] [CrossRef]
- Nativio, R.; Lan, Y.; Donahue, G.; Sidoli, S.; Berson, A.; Srinivasan, A.R.; Shcherbakova, O.; Amlie-Wolf, A.; Nie, J.; Cui, X.; et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 2020, 52, 1024–1035. [Google Scholar] [CrossRef]
- Tu, J.; Zhang, H.; Yang, T.; Liu, Y.; Kibreab, S.; Zhang, Y.; Gao, L.; Moses, R.E.; O’Malley, B.W.; Xiao, J.; et al. Aging-associated REGγ proteasome decline predisposes to tauopathy. J. Biol. Chem. 2022, 298, 102571. [Google Scholar] [CrossRef]
- Lee, H.J.; Alirzayeva, H.; Koyuncu, S.; Rueber, A.; Noormohammadi, A.; Vilchez, D. Cold temperature extends longevity and prevents disease-related protein aggregation through PA28γ-induced proteasomes. Nat. Aging 2023, 3, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, R. Proteasome activator PA28γ regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008, 27, 852–864. [Google Scholar] [CrossRef]
- Li, X.; Amazit, L.; Long, W.; Lonard, D.M.; Monaco, J.J.; O’Malley, B.W. Ubiquitin- and ATP-Independent Proteolytic Turnover of p21 by the REGγ-Proteasome Pathway. Mol. Cell 2007, 26, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lonard, D.M.; Jung, S.Y.; Malovannaya, A.; Feng, Q.; Qin, J.; Tsai, S.Y.; Tsai, M.-J.; O’Malley, B.W. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGγ proteasome. Cell 2006, 124, 381–392. [Google Scholar] [CrossRef]
- Nie, J.; Wu, M.; Wang, J.; Xing, G.; He, F.; Zhang, L. REGγ proteasome mediates degradation of the ubiquitin ligase Smurf1. FEBS Lett. 2010, 584, 3021–3027. [Google Scholar] [CrossRef]
- Fan, J.; Liu, L.; Liu, Q.; Cui, Y.; Yao, B.; Zhang, M.; Gao, Y.; Fu, Y.; Dai, H.; Pan, J.; et al. CKIP-1 limits foam cell formation and inhibits atherosclerosis by promoting degradation of Oct-1 by REGγ. Nat. Commun. 2019, 10, 425. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, P.; Wetzel, R.; Tanaka, M.; Nukina, N.; Goldberg, A.L. Eukaryotic Proteasomes Cannot Digest Polyglutamine Sequences and Release Them during Degradation of Polyglutamine-Containing Proteins. Mol. Cell 2004, 14, 95–104. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Garcia-Calvo, M.; Overkleeft, H.S.; Peterson, E.; Pennington, M.W.; Ploegh, H.L.; Thornberry, N.A.; Goldberg, A.L. The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 2003, 278, 35869–35877. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, T.; Salcedo-Tacuma, D.; Smith, D.M. Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules 2023, 13, 1326. https://doi.org/10.3390/biom13091326
Thomas T, Salcedo-Tacuma D, Smith DM. Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules. 2023; 13(9):1326. https://doi.org/10.3390/biom13091326
Chicago/Turabian StyleThomas, Taylor, David Salcedo-Tacuma, and David M. Smith. 2023. "Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators" Biomolecules 13, no. 9: 1326. https://doi.org/10.3390/biom13091326
APA StyleThomas, T., Salcedo-Tacuma, D., & Smith, D. M. (2023). Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules, 13(9), 1326. https://doi.org/10.3390/biom13091326





