Interaction of S100A6 Protein with the Four-Helical Cytokines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Surface Plasmon Resonance Studies
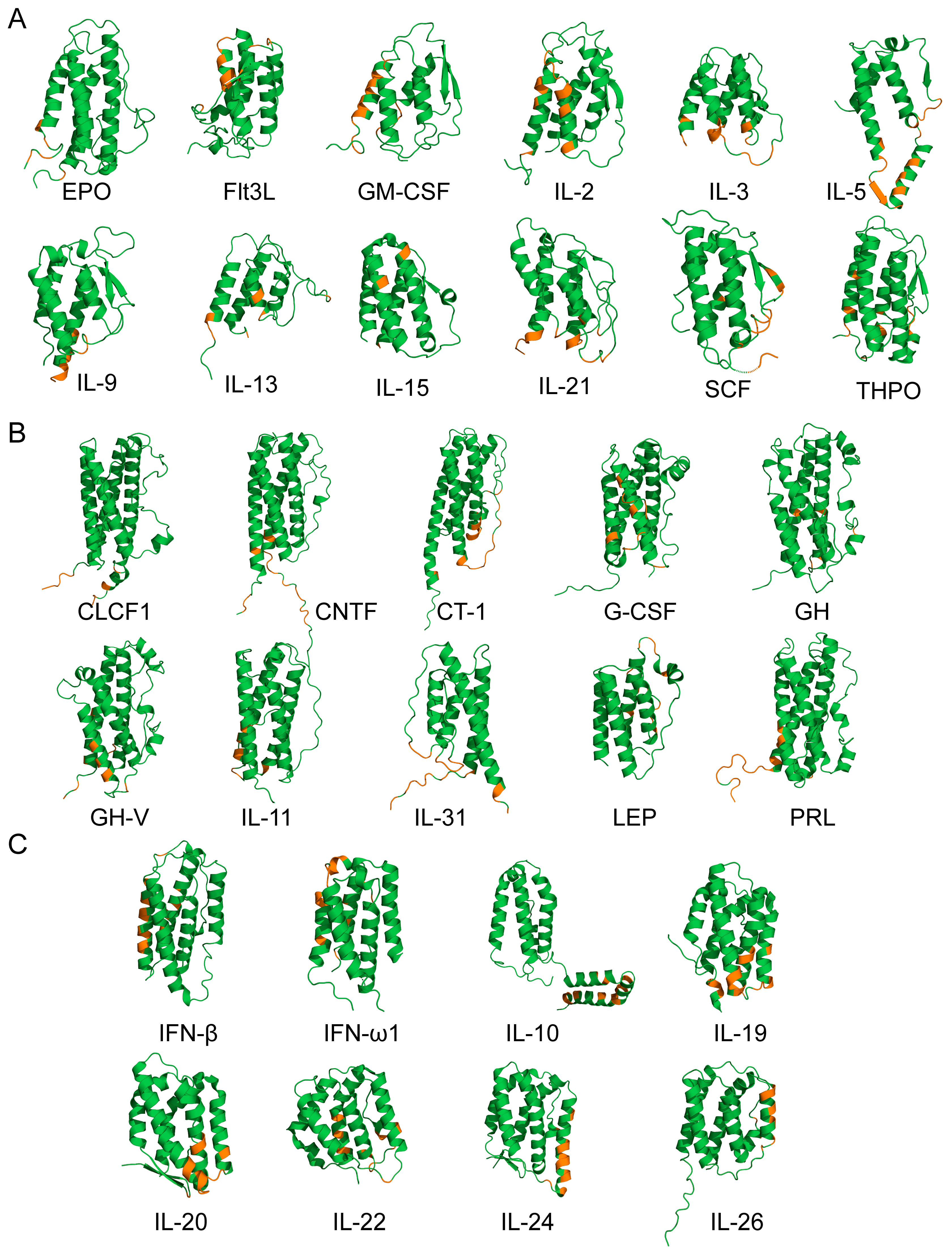
2.3. Structural Classification of Cytokines
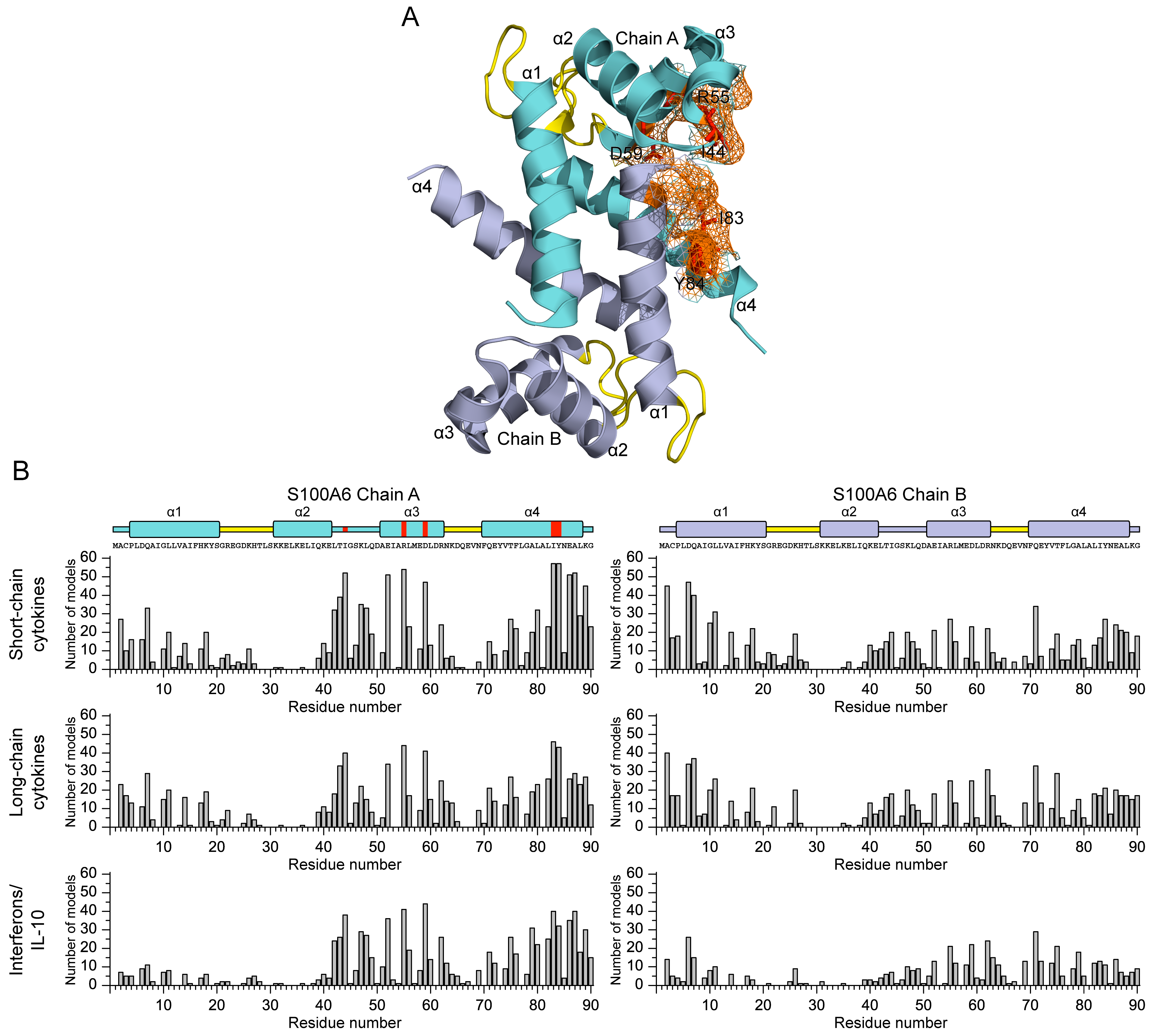
2.4. Structural Modeling of The S100A6–Cytokine Complexes
3. Results and Discussion
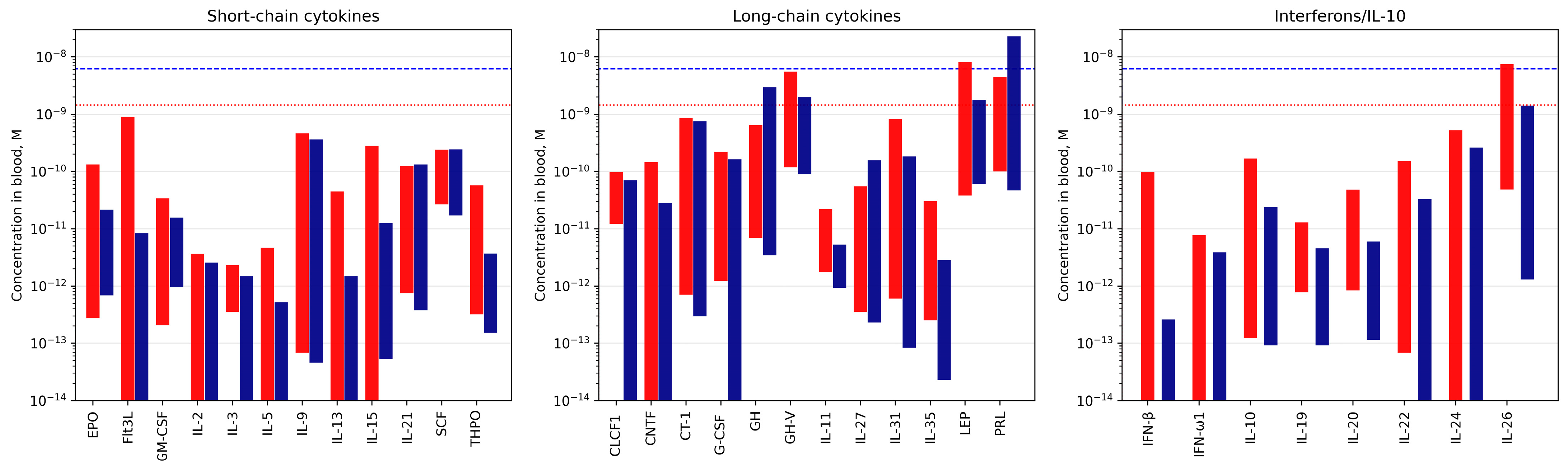
3.1. Selectivity of S100A6 Binding to The Four-Helical Cytokines
3.2. Structural Modeling of The S100A6–Cytokine Complexes
3.3. Potential Physiological Significance of the S100A6 Interactions with the Four-Helical Cytokines
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef] [PubMed]
- Sreejit, G.; Flynn, M.C.; Patil, M.; Krishnamurthy, P.; Murphy, A.J.; Nagareddy, P.R. S100 family proteins in inflammation and beyond. Adv. Clin. Chem. 2020, 98, 173–231. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, D.B.; Eubanks, J.O.; Ramakrishnan, D.; Criscitiello, M.F. Evolution of the S100 family of calcium sensor proteins. Cell Calcium 2013, 53, 170–179. [Google Scholar] [CrossRef]
- Sigrist, C.J.; de Castro, E.; Cerutti, L.; Cuche, B.A.; Hulo, N.; Bridge, A.; Bougueleret, L.; Xenarios, I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013, 41, D344–D347. [Google Scholar] [CrossRef]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc. Natl. Acad. Sci. USA 1972, 69, 581–584. [Google Scholar] [CrossRef]
- Gifford, J.L.; Walsh, M.P.; Vogel, H.J. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 2007, 405, 199–221. [Google Scholar] [CrossRef]
- Kawasaki, H.; Kretsinger, R.H. Structural and functional diversity of EF-hand proteins: Evolutionary perspectives. Protein Sci. 2017, 26, 1898–1920. [Google Scholar] [CrossRef]
- Fritz, G.; Heizmann, C.W. 3D Structures of the Calcium and Zinc Binding S100 Proteins. In Handbook of Metalloproteins; John Wiley & Sons: Hoboken, NJ, USA, 2004. [Google Scholar] [CrossRef]
- Gilston, B.A.; Skaar, E.P.; Chazin, W.J. Binding of transition metals to S100 proteins. Sci. China Life Sci. 2016, 59, 792–801. [Google Scholar] [CrossRef]
- Streicher, W.W.; Lopez, M.M.; Makhatadze, G.I. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys. Chem. 2010, 151, 181–186. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Bresnick, A.R.; Weber, D.J.; Zimmer, D.B. S100 proteins in cancer. Nat. Rev. Cancer 2015, 15, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Cristóvão, J.S.; Gomes, C.M. S100 Proteins in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 463. [Google Scholar] [CrossRef] [PubMed]
- Holzinger, D.; Tenbrock, K.; Roth, J. Alarmins of the S100-Family in Juvenile Autoimmune and Auto-Inflammatory Diseases. Front. Immunol. 2019, 10, 182. [Google Scholar] [CrossRef]
- Sattar, Z.; Lora, A.; Jundi, B.; Railwah, C.; Geraghty, P. The S100 Protein Family as Players and Therapeutic Targets in Pulmonary Diseases. Pulm. Med. 2021, 2021, 5488591. [Google Scholar] [CrossRef]
- Gonzalez, L.L.; Garrie, K.; Turner, M.D. Role of S100 proteins in health and disease. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118677. [Google Scholar] [CrossRef]
- Allgower, C.; Kretz, A.L.; von Karstedt, S.; Wittau, M.; Henne-Bruns, D.; Lemke, J. Friend or Foe: S100 Proteins in Cancer. Cancers 2020, 12, 2037. [Google Scholar] [CrossRef]
- Bresnick, A.R. S100 proteins as therapeutic targets. Biophys. Rev. 2018, 10, 1617–1629. [Google Scholar] [CrossRef]
- Yao, S.; Yang, X.; An, J.; Jin, H.; Wen, G.; Wang, H.; Tuo, B. Role of the S100 protein family in liver disease (Review). Int. J. Mol. Med. 2021, 48, 166. [Google Scholar] [CrossRef]
- Xiao, X.; Yang, C.; Qu, S.L.; Shao, Y.D.; Zhou, C.Y.; Chao, R.; Huang, L.; Zhang, C. S100 proteins in atherosclerosis. Clin. Chim. Acta 2020, 502, 293–304. [Google Scholar] [CrossRef]
- Engelkamp, D.; Schafer, B.W.; Mattei, M.G.; Erne, P.; Heizmann, C.W. Six S100 genes are clustered on human chromosome 1q21: Identification of two genes coding for the two previously unreported calcium-binding proteins S100D and S100E. Proc. Natl. Acad. Sci. USA 1993, 90, 6547–6551. [Google Scholar] [CrossRef] [PubMed]
- Schafer, B.W.; Wicki, R.; Engelkamp, D.; Mattei, M.G.; Heizmann, C.W. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: Rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics 1995, 25, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Lovering, R.C.; Heizmann, C.W. An update of the S100 nomenclature. Biochim. Biophys. Acta 2006, 1763, 1282–1283. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Mayorov, S.A.; Deryusheva, E.I.; Avkhacheva, N.V.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Permyakov, E.A.; Permyakov, S.E. Highly specific interaction of monomeric S100P protein with interferon beta. Int. J. Biol. Macromol. 2020, 143, 633–639. [Google Scholar] [CrossRef]
- Santamaria-Kisiel, L.; Rintala-Dempsey, A.C.; Shaw, G.S. Calcium-dependent and -independent interactions of the S100 protein family. Biochem. J. 2006, 396, 201–214. [Google Scholar] [CrossRef]
- Hsieh, H.L.; Schafer, B.W.; Cox, J.A.; Heizmann, C.W. S100A13 and S100A6 exhibit distinct translocation pathways in endothelial cells. J. Cell Sci. 2002, 115, 3149–3158. [Google Scholar] [CrossRef]
- Jurewicz, E.; Wyroba, E.; Filipek, A. Tubulin-dependent secretion of S100A6 and cellular signaling pathways activated by S100A6-integrin beta1 interaction. Cell. Signal. 2018, 42, 21–29. [Google Scholar] [CrossRef]
- Jurewicz, E.; Góral, A.; Filipek, A. S100A6 is secreted from Wharton’s jelly mesenchymal stem cells and interacts with integrin β1. Int. Journal. Biochem. Cell Biol. 2014, 55, 298–303. [Google Scholar] [CrossRef]
- Rumpret, M.; von Richthofen, H.J.; van der Linden, M.; Westerlaken, G.H.A.; Talavera Ormeno, C.; Low, T.Y.; Ovaa, H.; Meyaard, L. Recognition of S100 proteins by Signal Inhibitory Receptor on Leukocytes-1 negatively regulates human neutrophils. Eur. J. Immunol. 2021, 51, 2210–2217. [Google Scholar] [CrossRef]
- Moller, A.; Jauch-Speer, S.L.; Gandhi, S.; Vogl, T.; Roth, J.; Fehler, O. The roles of toll-like receptor 4, CD33, CD68, CD69, or CD147/EMMPRIN for monocyte activation by the DAMP S100A8/S100A9. Front. Immunol. 2023, 14, 1110185. [Google Scholar] [CrossRef]
- Ruma, I.M.; Putranto, E.W.; Kondo, E.; Murata, H.; Watanabe, M.; Huang, P.; Kinoshita, R.; Futami, J.; Inoue, Y.; Yamauchi, A.; et al. MCAM, as a novel receptor for S100A8/A9, mediates progression of malignant melanoma through prominent activation of NF-κB and ROS formation upon ligand binding. Clin. Exp. Metastasis 2016, 33, 609–627. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Yamamoto, M.; Miyai, M.; Maeda, T.; Hiruma, J.; Murata, H.; Kinoshita, R.; Winarsa Ruma, I.M.; Putranto, E.W.; Inoue, Y.; et al. Identification of an S100A8 Receptor Neuroplastin-beta and its Heterodimer Formation with EMMPRIN. J. Investig. Dermatol. 2016, 136, 2240–2250. [Google Scholar] [CrossRef]
- Tomonobu, N.; Kinoshita, R.; Sakaguchi, M. S100 Soil Sensor Receptors and Molecular Targeting Therapy Against Them in Cancer Metastasis. Transl. Oncol. 2020, 13, 100753. [Google Scholar] [CrossRef]
- Pankratova, S.; Klingelhofer, J.; Dmytriyeva, O.; Owczarek, S.; Renziehausen, A.; Syed, N.; Porter, A.E.; Dexter, D.T.; Kiryushko, D. The S100A4 Protein Signals through the ErbB4 Receptor to Promote Neuronal Survival. Theranostics 2018, 8, 3977–3990. [Google Scholar] [CrossRef]
- Warner-Schmidt, J.L.; Flajolet, M.; Maller, A.; Chen, E.Y.; Qi, H.; Svenningsson, P.; Greengard, P. Role of p11 in cellular and behavioral effects of 5-HT4 receptor stimulation. J. Neurosci. 2009, 29, 1937–1946. [Google Scholar] [CrossRef]
- Klingelhofer, J.; Moller, H.D.; Sumer, E.U.; Berg, C.H.; Poulsen, M.; Kiryushko, D.; Soroka, V.; Ambartsumian, N.; Grigorian, M.; Lukanidin, E.M. Epidermal growth factor receptor ligands as new extracellular targets for the metastasis-promoting S100A4 protein. FEBS J. 2009, 276, 5936–5948. [Google Scholar] [CrossRef]
- Mohan, S.K.; Yu, C. The IL1alpha-S100A13 heterotetrameric complex structure: A component in the non-classical pathway for interleukin 1alpha secretion. J. Biol. Chem. 2011, 286, 14608–14617. [Google Scholar] [CrossRef]
- Carreira, C.M.; LaVallee, T.M.; Tarantini, F.; Jackson, A.; Lathrop, J.T.; Hampton, B.; Burgess, W.H.; Maciag, T. S100A13 is involved in the regulation of fibroblast growth factor-1 and p40 synaptotagmin-1 release in vitro. J. Biol. Chem. 1998, 273, 22224–22231. [Google Scholar] [CrossRef]
- Gupta, A.A.; Chou, R.H.; Li, H.C.; Yang, L.W.; Yu, C. Structural insights into the interaction of human S100B and basic fibroblast growth factor (FGF2): Effects on FGFR1 receptor signaling. Bba-Proteins Proteom. 2013, 1834, 2606–2619. [Google Scholar] [CrossRef]
- Riuzzi, F.; Sorci, G.; Donato, R. S100B protein regulates myoblast proliferation and differentiation by activating FGFR1 in a bFGF-dependent manner. J. Cell Sci. 2011, 124, 2389–2400. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Zemskova, M.Y.; Rystsov, G.K.; Vologzhannikova, A.A.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; et al. Specific S100 Proteins Bind Tumor Necrosis Factor and Inhibit Its Activity. Int. J. Mol. Sci. 2022, 23, 15956. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Deryusheva, E.I.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Erythropoietin Interacts with Specific S100 Proteins. Biomolecules 2022, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Sofin, A.D.; Avkhacheva, N.V.; Denesyuk, A.I.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; et al. Interferon Beta Activity Is Modulated via Binding of Specific S100 Proteins. Int. J. Mol. Sci. 2020, 21, 9473. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Sofin, A.D.; Avkhacheva, N.V.; Deryusheva, E.I.; Rastrygina, V.A.; Permyakova, M.E.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Interferon-β Activity Is Affected by S100B Protein. Int. J. Mol. Sci. 2022, 23, 1997. [Google Scholar] [CrossRef]
- Kazakov, A.S.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Specific cytokines of interleukin-6 family interact with S100 proteins. Cell Calcium 2022, 101, 102520. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Deryusheva, E.I.; Permyakova, M.E.; Sokolov, A.S.; Rastrygina, V.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Calcium-Bound S100P Protein Is a Promiscuous Binding Partner of the Four-Helical Cytokines. Int. J. Mol. Sci. 2022, 23, 12000. [Google Scholar] [CrossRef] [PubMed]
- Riuzzi, F.; Sorci, G.; Beccafico, S.; Donato, R. S100B engages RAGE or bFGF/FGFR1 in myoblasts depending on its own concentration and myoblast density. Implications for muscle regeneration. PLoS ONE 2012, 7, e28700. [Google Scholar] [CrossRef] [PubMed]
- Andreeva, A.; Kulesha, E.; Gough, J.; Murzin, A.G. The SCOP database in 2020: Expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020, 48, D376–D382. [Google Scholar] [CrossRef]
- Simon, M.A.; Ecsedi, P.; Kovacs, G.M.; Poti, A.L.; Remenyi, A.; Kardos, J.; Gogl, G.; Nyitray, L. High-throughput competitive fluorescence polarization assay reveals functional redundancy in the S100 protein family. FEBS J. 2020, 287, 2834–2846. [Google Scholar] [CrossRef]
- Simon, M.A.; Bartus, É.; Mag, B.; Boros, E.; Roszjár, L.; Gógl, G.; Travé, G.; Martinek, T.A.; Nyitray, L. Promiscuity mapping of the S100 protein family using a high-throughput holdup assay. Sci. Rep. 2022, 12, 5904. [Google Scholar] [CrossRef]
- Donato, R.; Sorci, G.; Giambanco, I. S100A6 protein: Functional roles. Cell. Mol. Life Sci. 2017, 74, 2749–2760. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, W.; Wilanowski, T.; Filipek, A. S100A6 - focus on recent developments. Biol. Chem. 2017, 398, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, W.; Filipek, A. S100A6 Protein-Expression and Function in Norm and Pathology. Int. J. Mol. Sci. 2023, 24, 1341. [Google Scholar] [CrossRef]
- Filipek, A.; Lesniak, W. S100A6 and Its Brain Ligands in Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 3979. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Ma, J.; Zhu, D.; Wang, Z.; Li, Y.; He, X.; Zhang, G.; Kang, X. The Role of S100A6 in Human Diseases: Molecular Mechanisms and Therapeutic Potential. Biomolecules 2023, 13, 1139. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Brocker, C.; Thompson, D.; Matsumoto, A.; Nebert, D.W.; Vasiliou, V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum. Genom. 2010, 5, 30–55. [Google Scholar] [CrossRef]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Otterbein, L.R.; Kordowska, J.; Witte-Hoffmann, C.; Wang, C.L.; Dominguez, R. Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: The calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure 2002, 10, 557–567. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Sokolov, A.S.; Vologzhannikova, A.A.; Permyakova, M.E.; Khorn, P.A.; Ismailov, R.G.; Denessiouk, K.A.; Denesyuk, A.I.; Rastrygina, V.A.; Baksheeva, V.E.; et al. Interleukin-11 binds specific EF-hand proteins via their conserved structural motifs. J. Biomol. Struct. Dyn. 2017, 35, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Kazakov, A.S.; Sokolov, A.S.; Rastrygina, V.A.; Solovyev, V.V.; Ismailov, R.G.; Mikhailov, R.V.; Ulitin, A.B.; Yakovenko, A.R.; Mirzabekov, T.A.; Permyakov, E.A.; et al. High-affinity interaction between interleukin-11 and S100P protein. Biochem. Biophys. Res. Commun. 2015, 468, 733–738. [Google Scholar] [CrossRef]
- Leclerc, E. Measuring binding of S100 proteins to RAGE by surface plasmon resonance. Methods Mol. Biol. 2013, 963, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, E.; Fritz, G.; Weibel, M.; Heizmann, C.W.; Galichet, A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J. Biol. Chem. 2007, 282, 31317–31331. [Google Scholar] [CrossRef]
- Mohan, S.K.; Gupta, A.A.; Yu, C. Interaction of the S100A6 mutant (C3S) with the V domain of the receptor for advanced glycation end products (RAGE). Biochem. Biophys. Res. Commun. 2013, 434, 328–333. [Google Scholar] [CrossRef]
- Loosen, S.H.; Benz, F.; Niedeggen, J.; Schmeding, M.; Schuller, F.; Koch, A.; Vucur, M.; Tacke, F.; Trautwein, C.; Roderburg, C.; et al. Serum levels of S100A6 are unaltered in patients with resectable cholangiocarcinoma. Clin. Transl. Med. 2016, 5, 39. [Google Scholar] [CrossRef]
- Permyakov, S.E.; Denesyuk, A.I.; Denessiouk, K.A.; Permyakova, M.E.; Kazakov, A.S.; Ismailov, R.G.; Rastrygina, V.A.; Sokolov, A.S.; Permyakov, E.A. Monomeric state of S100P protein: Experimental and molecular dynamics study. Cell Calcium 2019, 80, 152–159. [Google Scholar] [CrossRef]
- Guzel, C.; van den Berg, C.B.; Duvekot, J.J.; Stingl, C.; van den Bosch, T.P.P.; van der Weiden, M.; Steegers, E.A.P.; Steegers-Theunissen, R.P.M.; Luider, T.M. Quantification of Calcyclin and Heat Shock Protein 90 in Sera from Women with and without Preeclampsia by Mass Spectrometry. PROTEOMICS–Clin. Appl. 2019, 13, e1800181. [Google Scholar] [CrossRef]
- Wang, T.; Liang, Y.; Thakur, A.; Zhang, S.; Yang, T.; Chen, T.; Gao, L.; Chen, M.; Ren, H. Diagnostic significance of S100A2 and S100A6 levels in sera of patients with non-small cell lung cancer. Tumour Biol. 2016, 37, 2299–2304. [Google Scholar] [CrossRef]
- Cai, X.Y.; Lu, L.; Wang, Y.N.; Jin, C.; Zhang, R.Y.; Zhang, Q.; Chen, Q.J.; Shen, W.F. Association of increased S100B, S100A6 and S100P in serum levels with acute coronary syndrome and also with the severity of myocardial infarction in cardiac tissue of rat models with ischemia-reperfusion injury. Atherosclerosis 2011, 217, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Ismailov, R.G.; Xue, B.; Denesyuk, A.I.; Uversky, V.N.; Permyakov, E.A. Intrinsic disorder in S100 proteins. Mol. Biosyst. 2011, 7, 2164–2180. [Google Scholar] [CrossRef]
- Yatime, L.; Betzer, C.; Jensen, R.K.; Mortensen, S.; Jensen, P.H.; Andersen, G.R. The Structure of the RAGE:S100A6 Complex Reveals a Unique Mode of Homodimerization for S100 Proteins. Structure 2016, 24, 2043–2052. [Google Scholar] [CrossRef]
- Rousseau, F.; Gauchat, J.F.; McLeod, J.G.; Chevalier, S.; Guillet, C.; Guilhot, F.; Cognet, I.; Froger, J.; Hahn, A.F.; Knappskog, P.M.; et al. Inactivation of cardiotrophin-like cytokine, a second ligand for ciliary neurotrophic factor receptor, leads to cold-induced sweating syndrome in a patient. Proc. Natl. Acad. Sci. USA 2006, 103, 10068–10073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, K.; Jiang, X. S100A6 as a potential serum prognostic biomarker and therapeutic target in gastric cancer. Dig. Dis. Sci. 2014, 59, 2136–2144. [Google Scholar] [CrossRef]
- Grote Beverborg, N.; Verweij, N.; Klip, I.T.; van der Wal, H.H.; Voors, A.A.; van Veldhuisen, D.J.; Gansevoort, R.T.; Bakker, S.J.; van der Harst, P.; van der Meer, P. Erythropoietin in the general population: Reference ranges and clinical, biochemical and genetic correlates. PLoS ONE 2015, 10, e0125215. [Google Scholar] [CrossRef]
- Szuber, N.; Lavu, S.; Mudireddy, M.; Nicolosi, M.; Penna, D.; Vallapureddy, R.R.; Lasho, T.L.; Finke, C.; Hanson, C.A.; Ketterling, R.P.; et al. Serum erythropoietin levels in essential thrombocythemia: Phenotypic and prognostic correlates. Blood Cancer J. 2018, 8, 118. [Google Scholar] [CrossRef]
- Davidovic, S.; Babic, N.; Jovanovic, S.; Barisic, S.; Grkovic, D.; Miljkovic, A. Serum erythropoietin concentration and its correlation with stage of diabetic retinopathy. BMC Ophthalmol. 2019, 19, 227. [Google Scholar] [CrossRef]
- Mannello, F.; Fabbri, L.; Ciandrini, E.; Tonti, G.A. Increased levels of erythropoietin in nipple aspirate fluid and in ductal cells from breast cancer patients. Anal. Cell. Oncol. 2008, 30, 51–61. [Google Scholar] [CrossRef]
- Varda, M.M.; Tomassetti, S. Erythropoietin Levels in Patients with Polycythemia Vera and Secondary Erythrocytosis. Blood 2022, 140, 12181–12182. [Google Scholar] [CrossRef]
- Tas, F.; Oguz, H.; Argon, A.; Duranyildiz, D.; Camlica, H.; Yasasever, V.; Topuz, E. The value of serum levels of IL-6, TNF-alpha, and erythropoietin in metastatic malignant melanoma: Serum IL-6 level is a valuable prognostic factor at least as serum LDH in advanced melanoma. Med. Oncol. 2005, 22, 241–246. [Google Scholar] [CrossRef]
- Lyman, S.D.; Seaberg, M.; Hanna, R.; Zappone, J.; Brasel, K.; Abkowitz, J.L.; Prchal, J.T.; Schultz, J.C.; Shahidi, N.T. Plasma/serum levels of flt3 ligand are low in normal individuals and highly elevated in patients with Fanconi anemia and acquired aplastic anemia. Blood 1995, 86, 4091–4096. [Google Scholar] [CrossRef]
- Zwierzina, H.; Anderson, J.E.; Rollinger-Holzinger, I.; Torok-Storb, B.; Nuessler, V.; Lyman, S.D. Endogenous FLT-3 ligand serum levels are associated with disease stage in patients with myelodysplastic syndromes. Leukemia 1999, 13, 553–557. [Google Scholar] [CrossRef]
- Peterlin, P.; Gaschet, J.; Guillaume, T.; Garnier, A.; Eveillard, M.; Le Bourgeois, A.; Cherel, M.; Debord, C.; Le Bris, Y.; Theisen, O.; et al. FLT3 ligand plasma levels in acute myeloid leukemia. Haematologica 2019, 104, e240–e243. [Google Scholar] [CrossRef]
- Tobon, G.J.; Saraux, A.; Gottenberg, J.E.; Quartuccio, L.; Fabris, M.; Seror, R.; Devauchelle-Pensec, V.; Morel, J.; Rist, S.; Mariette, X.; et al. Role of Fms-like tyrosine kinase 3 ligand as a potential biologic marker of lymphoma in primary Sjogren’s syndrome. Arthritis Rheum. 2013, 65, 3218–3227. [Google Scholar] [CrossRef]
- Papagoras, C.; Tsiami, S.; Chrysanthopoulou, A.; Mitroulis, I.; Baraliakos, X. Serum granulocyte-macrophage colony-stimulating factor (GM-CSF) is increased in patients with active radiographic axial spondyloarthritis and persists despite anti-TNF treatment. Arthritis Res. Ther. 2022, 24, 195. [Google Scholar] [CrossRef]
- Riccio, A.; De Caterina, M.; Natale, D.; Grimaldi, E.; Pronesti, G.; Montagnani, S.; Postiglione, L. Serum Levels of Granulocyte Macrophage Colony Stimulating Factor (GM-CSF) in a Group of Patients with Systemic Sclerosis. Int. J. Immunopathol. Pharmacol. 1996, 9, 9–12. [Google Scholar] [CrossRef]
- Balleari, E.; Bason, C.; Visani, G.; Gobbi, M.; Ottaviani, E.; Ghio, R. Serum levels of granulocyte-macrophage colony-stimulating factor and granulocyte colony-stimulating factor in treated patients with chronic myelogenous leukemia in chronic phase. Haematologica 1994, 79, 7–12. [Google Scholar]
- Fiehn, C.; Wermann, M.; Pezzutto, A.; Hufner, M.; Heilig, B. Plasma GM-CSF concentrations in rheumatoid arthritis, systemic lupus erythematosus and spondyloarthropathy. Z. Rheumatol. 1992, 51, 121–126. [Google Scholar]
- Carrieri, P.B.; Provitera, V.; De Rosa, T.; Tartaglia, G.; Gorga, F.; Perrella, O. Profile of cerebrospinal fluid and serum cytokines in patients with relapsing-remitting multiple sclerosis: A correlation with clinical activity. Immunopharmacol. Immunotoxicol. 1998, 20, 373–382. [Google Scholar] [CrossRef]
- Kleiner, G.; Marcuzzi, A.; Zanin, V.; Monasta, L.; Zauli, G. Cytokine levels in the serum of healthy subjects. Mediators Inflamm. 2013, 2013, 434010. [Google Scholar] [CrossRef] [PubMed]
- Huan, X.; Zhao, R.; Song, J.; Zhong, H.; Su, M.; Yan, C.; Wang, Y.; Chen, S.; Zhou, Z.; Lu, J.; et al. Increased serum IL-2, IL-4, IL-5 and IL-12p70 levels in AChR subtype generalized myasthenia gravis. BMC Immunol. 2022, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Guo, Q.; Wang, Y.; Su, R.; Gao, C.; Zhao, J.; Li, X.; Wang, C. Increased Serum Interleukin-2 Levels Are Associated with Abnormal Peripheral Blood Natural Killer Cell Levels in Patients with Active Rheumatoid Arthritis. Mediat. Inflamm. 2020, 2020, 6108342. [Google Scholar] [CrossRef] [PubMed]
- Kasumagic-Halilovic, E.; Cavaljuga, S.; Ovcina-Kurtovic, N.; Zecevic, L. Serum Levels of Interleukin-2 in Patients with Alopecia Areata: Relationship with Clinical Type and Duration of the Disease. Skin. Appendage Disord. 2018, 4, 286–290. [Google Scholar] [CrossRef]
- Ceyhan, B.B.; Enc, F.Y.; Sahin, S. IL-2 and IL-10 levels in induced sputum and serum samples of asthmatics. J. Investig. Allergol. Clin. Immunol. 2004, 14, 80–85. [Google Scholar]
- Kutukculer, N.; Vergin, C.; Cetingul, N.; Kavakli, K.; Caglayan, S.; Oztop, S.; Nisli, G. Plasma interleukin-3 (IL-3) and IL-7 concentrations in children with homozygous beta-thalassemia. J. Trop. Pediatr. 1997, 43, 366–367. [Google Scholar] [CrossRef]
- Biancotto, A.; Wank, A.; Perl, S.; Cook, W.; Olnes, M.J.; Dagur, P.K.; Fuchs, J.C.; Langweiler, M.; Wang, E.; McCoy, J.P. Baseline levels and temporal stability of 27 multiplexed serum cytokine concentrations in healthy subjects. PLoS ONE 2013, 8, e76091. [Google Scholar] [CrossRef]
- Xiu, M.H.; Lin, C.G.; Tian, L.; Tan, Y.L.; Chen, J.; Chen, S.; Tan, S.P.; Wang, Z.R.; Yang, F.D.; Chen, D.C.; et al. Increased IL-3 serum levels in chronic patients with schizophrenia: Associated with psychopathology. Psychiatry Res. 2015, 229, 225–229. [Google Scholar] [CrossRef]
- Vasiliades, G.; Kopanakis, N.; Vasiloglou, M.; Zografos, G.; Margaris, H.; Masselou, K.; Kokosi, E.; Liakakos, T. Role of the hematopoietic cytokines SCF, IL-3, GM-CSF and M-CSF in the diagnosis of pancreatic and ampullary cancer. Int. J. Biol. Markers 2012, 27, e186–e194. [Google Scholar] [CrossRef]
- Lee, H.L.; Jang, J.W.; Lee, S.W.; Yoo, S.H.; Kwon, J.H.; Nam, S.W.; Bae, S.H.; Choi, J.Y.; Han, N.I.; Yoon, S.K. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci. Rep. 2019, 9, 3260. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.C.; Lee, K.H.; Lee, H.B.; Rhee, Y.K. Serum levels of interleukins (IL)-4, IL-5, IL-13, and interferon-gamma in acute asthma. J. Asthma 2001, 38, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Benedict, S.; Safa, W.; Joseph, M. Serum interleukin-5 levels are elevated in mild and moderate persistent asthma irrespective of regular inhaled glucocorticoid therapy. BMC Pulm. Med. 2004, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, D.; Youroukova, V.; Ivanova-Todorova, E.; Tumangelova-Yuzeir, K.; Velikova, T. Serum levels of IL-5, IL-6, IL-8, IL-13 and IL-17A in pre-defined groups of adult patients with moderate and severe bronchial asthma. Respir. Med. 2019, 154, 144–154. [Google Scholar] [CrossRef]
- Perret, J.; McDonald, C.; Apostolopoulos, V. Elevated serum interleukin-5 levels in severe chronic obstructive pulmonary disease. Acta Biochim. Biophys. Sin. 2017, 49, 560–563. [Google Scholar] [CrossRef]
- Fischer, M.; Bijman, M.; Molin, D.; Cormont, F.; Uyttenhove, C.; van Snick, J.; Sundstrom, C.; Enblad, G.; Nilsson, G. Increased serum levels of interleukin-9 correlate to negative prognostic factors in Hodgkin’s lymphoma. Leukemia 2003, 17, 2513–2516. [Google Scholar] [CrossRef]
- Dantas, A.T.; Marques, C.D.L.; da Rocha Junior, L.F.; Cavalcanti, M.B.; Gonçalves, S.M.C.; Cardoso, P.R.G.; Mariz, H.d.A.; Rego, M.J.B.d.M.; Duarte, A.L.B.P.; Pitta, I.d.R.; et al. Increased Serum Interleukin-9 Levels in Rheumatoid Arthritis and Systemic Lupus Erythematosus: Pathogenic Role or Just an Epiphenomenon? Dis. Markers 2015, 2015, 519638. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, Y.; Zhang, S.; Gao, F. Association between low expression levels of interleukin-9 and colon cancer progression. Exp. Ther. Med. 2015, 10, 942–946. [Google Scholar] [CrossRef]
- Erpenbeck, V.J.; Hohlfeld, J.M.; Volkmann, B.; Hagenberg, A.; Geldmacher, H.; Braun, A.; Krug, N. Segmental allergen challenge in patients with atopic asthma leads to increased IL-9 expression in bronchoalveolar lavage fluid lymphocytes. J. Allergy Clin. Immunol. 2003, 111, 1319–1327. [Google Scholar] [CrossRef]
- Mahdaviani, S.A.; Eskian, M.; Khorasanizadeh, M.; Bashardoost, B.; Tashayoie Nejad, S.; Jamaati, H.R.; Rezaei, A.; Sadr, M.; Aryan, Z.; Rezaei, N. Interleukin 9 serum level and single nucleotide polymorphism in patients with asthma. Acta Biomed. 2021, 92, e2021206. [Google Scholar] [CrossRef] [PubMed]
- Defendenti, C.; Sarzi-Puttini, P.; Saibeni, S.; Bollani, S.; Bruno, S.; Almasio, P.L.; Declich, P.; Atzeni, F. Significance of serum Il-9 levels in inflammatory bowel disease. Int. J. Immunopathol. Pharmacol. 2015, 28, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Feng, L.; Ge, X.; Lu, K.; Wang, X. Interleukin-9 promotes cell survival and drug resistance in diffuse large B-cell lymphoma. J. Exp. Clin. Cancer Res. 2016, 35, 106. [Google Scholar] [CrossRef]
- Ciprandi, G.; De Amici, M.; Castellazzi, A.M.; Tosca, M.A.; Marseglia, G. Serum IL-9 levels depend on allergen exposure: Preliminary study. Int. Arch. Allergy Immunol. 2011, 154, 246–248. [Google Scholar] [CrossRef]
- Yanaba, K.; Yoshizaki, A.; Asano, Y.; Kadono, T.; Sato, S. Serum interleukin 9 levels are increased in patients with systemic sclerosis: Association with lower frequency and severity of pulmonary fibrosis. J. Rheumatol. 2011, 38, 2193–2197. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, C.P.; Gómez-Arauz, A.Y.; Torres-Castro, I.; Manjarrez-Reyna, A.N.; Palomera, L.F.; Olivos-García, A.; Mendoza-Tenorio, E.; Sánchez-Medina, G.A.; Islas-Andrade, S.; Melendez-Mier, G.; et al. Serum Levels of Interleukin-13 Increase in Subjects with Insulin Resistance but Do Not Correlate with Markers of Low-Grade Systemic Inflammation. J. Diabetes Res. 2018, 2018, 7209872. [Google Scholar] [CrossRef]
- Nabavi, M.; Arshi, S.; Bahrami, A.; Aryan, Z.; Bemanian, M.H.; Esmaeilzadeh, H.; Jalali, F.; Pousti, S.B.; Rezaei, N. Increased level of interleukin-13, but not interleukin-4 and interferon-γ in chronic rhinosinusitis with nasal polyps. Allergol. Immunopathol. 2014, 42, 465–471. [Google Scholar] [CrossRef]
- Cingu, A.K.; Turkcu, F.M.; Aktas, S.; Sahin, A.; Ayyildiz, O. Serum IL-4, IL-12, IL-13, IL-27, and IL-33 levels in active and inactive ocular Behcet’s disease. Int. Ophthalmol. 2020, 40, 3441–3451. [Google Scholar] [CrossRef]
- Spadaro, A.; Rinaldi, T.; Riccieri, V.; Valesini, G.; Taccari, E. Interleukin 13 in synovial fluid and serum of patients with psoriatic arthritis. Ann. Rheum. Dis. 2002, 61, 174–176. [Google Scholar] [CrossRef]
- Versace, A.G.; Bitto, A.; Ioppolo, C.; Aragona, C.O.; La Rosa, D.; Roberts, W.N.; D’Angelo, T.; Cinquegrani, A.; Cirmi, S.; Irrera, N.; et al. IL-13 and IL-33 Serum Levels Are Increased in Systemic Sclerosis Patients With Interstitial Lung Disease. Front. Med. 2022, 9, 825567. [Google Scholar] [CrossRef]
- Mielnik, P.; Chwalinska-Sadowska, H.; Wiesik-Szewczyk, E.; Maslinski, W.; Olesinska, M. Serum concentration of interleukin 15, interleukin 2 receptor and TNF receptor in patients with polymyositis and dermatomyositis: Correlation to disease activity. Rheumatol. Int. 2012, 32, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Kaibe, M.; Ohishi, M.; Ito, N.; Yuan, M.; Takagi, T.; Terai, M.; Tatara, Y.; Komai, N.; Rakugi, H.; Ogihara, T. Serum Interleukin-15 Concentration in Patients With Essential Hypertension*. Am. J. Hypertens. 2005, 18, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.-C.; Kim, H.-Y.; Ahn, S.-Y.; Kim, D.-S. Raised serum interleukin 15 levels in Kawasaki disease. Ann. Rheum. Dis. 2003, 62, 264–266. [Google Scholar] [CrossRef]
- Mengus, C.; Le Magnen, C.; Trella, E.; Yousef, K.; Bubendorf, L.; Provenzano, M.; Bachmann, A.; Heberer, M.; Spagnoli, G.C.; Wyler, S. Elevated levels of circulating IL-7 and IL-15 in patients with early stage prostate cancer. J. Transl. Med. 2011, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Rentzos, M.; Cambouri, C.; Rombos, A.; Nikolaou, C.; Anagnostouli, M.; Tsoutsou, A.; Dimitrakopoulos, A.; Triantafyllou, N.; Vassilopoulos, D. IL-15 is elevated in serum and cerebrospinal fluid of patients with multiple sclerosis. J. Neurol. Sci. 2006, 241, 25–29. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Caligiuri, M.A. Interleukin 15: Biology and relevance to human disease. Blood 2001, 97, 14–32. [Google Scholar] [CrossRef]
- Lauw, F.N.; Simpson, A.J.H.; Prins, J.M.; Smith, M.D.; Kurimoto, M.; van Deventer, S.J.H.; Speelman, P.; Chaowagul, W.; White, N.J.; van der Poll, T. Elevated Plasma Concentrations of Interferon (IFN)-γ and the IFN-γ—Inducing Cytokines Interleukin (IL)-18, IL-12, and IL-15 in Severe Melioidosis. J. Infect. Dis. 1999, 180, 1878–1885. [Google Scholar] [CrossRef]
- Vivanco-Cid, H.; Maldonado-Renteria, M.J.; Sanchez-Vargas, L.A.; Izaguirre-Hernandez, I.Y.; Hernandez-Flores, K.G.; Remes-Ruiz, R. Dynamics of interleukin-21 production during the clinical course of primary and secondary dengue virus infections. Immunol. Lett. 2014, 161, 89–95. [Google Scholar] [CrossRef]
- de Oliveira, P.S.; Cardoso, P.R.; Lima, E.V.; Pereira, M.C.; Duarte, A.L.; Pitta Ida, R.; Rego, M.J.; Pitta, M.G. IL-17A, IL-22, IL-6, and IL-21 Serum Levels in Plaque-Type Psoriasis in Brazilian Patients. Mediat. Inflamm. 2015, 2015, 819149. [Google Scholar] [CrossRef]
- Bae, Y.J.; Kim, M.H.; Lee, H.Y.; Uh, Y.; Namgoong, M.K.; Cha, B.H.; Chun, J.K. Elevated Serum Levels of IL-21 in Kawasaki Disease. Allergy Asthma Immunol. Res. 2012, 4, 351–356. [Google Scholar] [CrossRef]
- Bono, P.; Krause, A.; von Mehren, M.; Heinrich, M.C.; Blanke, C.D.; Dimitrijevic, S.; Demetri, G.D.; Joensuu, H. Serum KIT and KIT ligand levels in patients with gastrointestinal stromal tumors treated with imatinib. Blood 2004, 103, 2929–2935. [Google Scholar] [CrossRef] [PubMed]
- Makowska, J.S.; Cieslak, M.; Kowalski, M.L. Stem cell factor and its soluble receptor (c-kit) in serum of asthmatic patients- correlation with disease severity. BMC Pulm. Med. 2009, 9, 27. [Google Scholar] [CrossRef] [PubMed]
- Kalmarzi, R.N.; Foroutan, A.; Abdi, M.; Ataee, P.; Jalili, A.; Babaei, E.; Kashefi, H.; Mohamadi, S.; Sigari, N.; Kooti, W. Serum level of stem cell factor and its soluble receptor in aspirin-exacerbated respiratory disease. Immunotherapy 2019, 11, 1283–1291. [Google Scholar] [CrossRef]
- Mashayekhi, F.; Shabani, S.; Sasani, S.T.; Salehi, Z. The association of stem cell factor and soluble c-Kit (s-cKit) receptor serum concentrations with the severity and risk prediction of autism spectrum disorders. Metab. Brain Dis. 2022, 37, 619–624. [Google Scholar] [CrossRef]
- Leyhe, T.; Hoffmann, N.; Stransky, E.; Laske, C. Increase of SCF plasma concentration during donepezil treatment of patients with early Alzheimer’s disease. Int. J. Neuropsychopharmacol. 2009, 12, 1319–1326. [Google Scholar] [CrossRef]
- Temel, T.; Cansu, D.U.; Temel, H.E.; Ozakyol, A.H. Serum thrombopoietin levels and its relationship with thrombocytopenia in patients with cirrhosis. Hepat. Mon. 2014, 14, e18556. [Google Scholar] [CrossRef] [PubMed]
- Usuki, K.; Tahara, T.; Iki, S.; Endo, M.; Osawa, M.; Kitazume, K.; Kato, T.; Miyazaki, H.; Urabe, A. Serum thrombopoietin level in various hematological diseases. Stem Cells 1996, 14, 558–565. [Google Scholar] [CrossRef]
- Ballmaier, M.; Schulze, H.; Strauβ, G.; Cherkaoui, K.; Wittner, N.; Lynen, S.; Wolters, S.; Bogenberger, J.; Welte, K. Thrombopoietin in Patients With Congenital Thrombocytopenia and Absent Radii: Elevated Serum Levels, Normal Receptor Expression, But Defective Reactivity to Thrombopoietin. Blood 1997, 90, 612–619. [Google Scholar] [CrossRef]
- Tsukishiro, S.; Suzumori, N.; Nishikawa, H.; Arakawa, A.; Suzumori, K. Preoperative serum thrombopoietin levels are higher in patients with ovarian cancer than with benign cysts. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 140, 67–70. [Google Scholar] [CrossRef]
- Makar, R.S.; Zhukov, O.S.; Sahud, M.A.; Kuter, D.J. Thrombopoietin levels in patients with disorders of platelet production: Diagnostic potential and utility in predicting response to TPO receptor agonists. Am. J. Hematol. 2013, 88, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Chebotareva, N.; Vinogradov, A.; Cao, V.; Gindis, A.; Berns, A.; Alentov, I.; Sergeeva, N. Serum levels of plasminogen activator urokinase receptor and cardiotrophin-like cytokine factor 1 in patients with nephrotic syndrome. Clin. Nephrol. 2022, 97, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Muller-Deile, J.; Sarau, G.; Kotb, A.M.; Jaremenko, C.; Rolle-Kampczyk, U.E.; Daniel, C.; Kalkhof, S.; Christiansen, S.H.; Schiffer, M. Novel diagnostic and therapeutic techniques reveal changed metabolic profiles in recurrent focal segmental glomerulosclerosis. Sci. Rep. 2021, 11, 4577. [Google Scholar] [CrossRef] [PubMed]
- Robak, T.; Gladalska, A.; Stepien, H.; Robak, E. Serum levels of interleukin-6 type cytokines and soluble interleukin-6 receptor in patients with rheumatoid arthritis. Mediat. Inflamm. 1998, 7, 347–353. [Google Scholar] [CrossRef]
- Laaksovirta, H.; Soinila, S.; Hukkanen, V.; Roytta, M.; Soilu-Hanninen, M. Serum level of CNTF is elevated in patients with amyotrophic lateral sclerosis and correlates with site of disease onset. Eur. J. Neurol. 2008, 15, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Druzhkova, T.A.; Yakovlev, A.A.; Rider, F.K.; Zinchuk, M.S.; Guekht, A.B.; Gulyaeva, N.V. Elevated Serum Cortisol Levels in Patients with Focal Epilepsy, Depression, and Comorbid Epilepsy and Depression. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef]
- Shpak, A.A.; Guekht, A.B.; Druzhkova, T.A.; Kozlova, K.I.; Gulyaeva, N.V. Ciliary neurotrophic factor in patients with primary open-angle glaucoma and age-related cataract. Mol. Vis. 2017, 23, 799–809. [Google Scholar]
- Brondino, N.; Rocchetti, M.; Fusar-Poli, L.; Damiani, S.; Goggi, A.; Chiodelli, G.; Corti, S.; Visai, L.; Politi, P. Increased CNTF levels in adults with autism spectrum disorders. World J. Biol. Psychiatry 2019, 20, 742–746. [Google Scholar] [CrossRef]
- Perugini, J.; Di Mercurio, E.; Giuliani, A.; Sabbatinelli, J.; Bonfigli, A.R.; Tortato, E.; Severi, I.; Cinti, S.; Olivieri, F.; le Roux, C.W.; et al. Ciliary neurotrophic factor is increased in the plasma of patients with obesity and its levels correlate with diabetes and inflammation indices. Sci. Rep. 2022, 12, 8331. [Google Scholar] [CrossRef]
- Wang, K.; Chu, C.; Hu, J.; Wang, Y.; Zheng, W.; Lv, Y.; Yan, Y.; Ma, Q.; Mu, J. Effect of Salt Intake on the Serum Cardiotrophin-1 Levels in Chinese Adults. Ann. Nutr. Metab. 2018, 73, 302–309. [Google Scholar] [CrossRef]
- Cakir, I.; Uluhan, M. Cardiotrophin-1 and leptin as cardiovascular risk markers in male patients with obstructive sleep apnea syndrome. Arch. Med. Sci. Atheroscler. Dis. 2018, 3, e123–e128. [Google Scholar] [CrossRef]
- Calabro, P.; Limongelli, G.; Riegler, L.; Maddaloni, V.; Palmieri, R.; Golia, E.; Roselli, T.; Masarone, D.; Pacileo, G.; Golino, P.; et al. Novel insights into the role of cardiotrophin-1 in cardiovascular diseases. J. Mol. Cell. Cardiol. 2009, 46, 142–148. [Google Scholar] [CrossRef]
- Monserrat, L.; Lopez, B.; Gonzalez, A.; Hermida, M.; Fernandez, X.; Ortiz, M.; Barriales-Villa, R.; Castro-Beiras, A.; Diez, J. Cardiotrophin-1 plasma levels are associated with the severity of hypertrophy in hypertrophic cardiomyopathy. Eur. Heart J. 2011, 32, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, M.; Tsutsumi, H.; Kumakawa, T.; Abe, H.; Hirai, M.; Kurosawa, S.; Mori, M.; Fukushima, M. Levels of serum granulocyte colony-stimulating factor in patients with infections. Blood 1990, 76, 1962–1964. [Google Scholar] [CrossRef]
- Bordbar, E.; Malekzadeh, M.; Ardekani, M.T.; Doroudchi, M.; Ghaderi, A. Serum levels of G-CSF and IL-7 in Iranian breast cancer patients. Asian Pac. J. Cancer Prev. 2012, 13, 5307–5312. [Google Scholar] [CrossRef] [PubMed]
- Pauksen, K.; Elfman, L.; Ulfgren, A.K.; Venge, P. Serum levels of granulocyte-colony stimulating factor (G-CSF) in bacterial and viral infections, and in atypical pneumonia. Br. J. Haematol. 1994, 88, 256–260. [Google Scholar] [CrossRef]
- Morozumi, K.; Namiki, S.; Kudo, T.; Aizawa, M.; Ioritani, N. Serum G-CSF May Be a More Valuable Biomarker than Image Evaluation in G-CSF-Producing Urothelial Carcinoma: A Case Report. Case Rep. Oncol. 2017, 10, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Galvan, S.T.; Flores-Lopez, M.; Romero-Sanchiz, P.; Requena-Ocana, N.; Porras-Perales, O.; Nogueira-Arjona, R.; Mayoral, F.; Araos, P.; Serrano, A.; Muga, R.; et al. Plasma concentrations of granulocyte colony-stimulating factor (G-CSF) in patients with substance use disorders and comorbid major depressive disorder. Sci. Rep. 2021, 11, 13629. [Google Scholar] [CrossRef] [PubMed]
- Watari, K.; Asano, S.; Shirafuji, N.; Kodo, H.; Ozawa, K.; Takaku, F.; Kamachi, S. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood 1989, 73, 117–122. [Google Scholar] [CrossRef]
- Gross-Weege, W.; Dumon, K.; Dahmen, A.; Schneider, E.M.; Röher, H.D. Granulocyte colony-stimulating factor (G-CSF) serum levels in surgical intensive care patients. Infection 1997, 25, 213–216. [Google Scholar] [CrossRef]
- Triantafillidis, J.K.; Merikas, E.; Govosdis, V.; Konstandellou, E.; Cheracakis, P.; Barbatzas, C.; Tzourmakliotis, D.; Peros, G. Increased fasting serum levels of growth hormone and gastrin in patients with gastric and large bowel cancer. Hepatogastroenterology 2003, 50 (Suppl. S2), cclvi–cclx. [Google Scholar]
- Elkarow, M.H.; Hamdy, A. A Suggested Role of Human Growth Hormone in Control of the COVID-19 Pandemic. Front. Endocrinol. 2020, 11, 569633. [Google Scholar] [CrossRef] [PubMed]
- Akirov, A.; Masri-Iraqi, H.; Dotan, I.; Shimon, I. The Biochemical Diagnosis of Acromegaly. J. Clin. Med. 2021, 10. [Google Scholar] [CrossRef]
- Ishikawa, M.; Yokoya, S.; Tachibana, K.; Hasegawa, Y.; Yasuda, T.; Tokuhiro, E.; Hashimoto, Y.; Tanaka, T. Serum levels of 20-kilodalton human growth hormone (GH) are parallel those of 22-kilodalton human GH in normal and short children. J. Clin. Endocrinol. Metab. 1999, 84, 98–104. [Google Scholar] [CrossRef]
- Christiansen, M. Placental growth hormone and growth hormone binding protein are first trimester maternal serum markers of Down syndrome. Prenat. Diagn. 2009, 29, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Mittal, P.; Espinoza, J.; Hassan, S.; Kusanovic, J.P.; Edwin, S.S.; Nien, J.K.; Gotsch, F.; Than, N.G.; Erez, O.; Mazaki-Tovi, S.; et al. Placental growth hormone is increased in the maternal and fetal serum of patients with preeclampsia. J. Matern. Fetal Neonatal Med. 2007, 20, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, J.; Lv, X.; Yang, Q.; Yao, S.; Zhang, D. Clinical value of serum and exhaled breath condensate inflammatory factor IL-11 levels in non-small cell lung cancer: Clinical value of IL-11 in non-small cell lung cancer. Int. J. Biol. Markers 2021, 36, 64–76. [Google Scholar] [CrossRef]
- Ye, J.; Wang, Z.; Ye, D.; Wang, Y.; Wang, M.; Ji, Q.; Huang, Y.; Liu, L.; Shi, Y.; Shi, L.; et al. Increased Interleukin-11 Levels Are Correlated with Cardiac Events in Patients with Chronic Heart Failure. Mediat. Inflamm. 2019, 2019, 1575410. [Google Scholar] [CrossRef]
- Trontzas, P.; Kamper, E.F.; Potamianou, A.; Kyriazis, N.C.; Kritikos, H.; Stavridis, J. Comparative study of serum and synovial fluid interleukin-11 levels in patients with various arthritides. Clin. Biochem. 1998, 31, 673–679. [Google Scholar] [CrossRef]
- Ren, C.; Chen, Y.; Han, C.; Fu, D.; Chen, H. Plasma interleukin-11 (IL-11) levels have diagnostic and prognostic roles in patients with pancreatic cancer. Tumour Biol. 2014, 35, 11467–11472. [Google Scholar] [CrossRef]
- Wu, P.; Lin, B.; Huang, S.; Meng, J.; Zhang, F.; Zhou, M.; Hei, X.; Ke, Y.; Yang, H.; Huang, D. IL-11 Is Elevated and Drives the Profibrotic Phenotype Transition of Orbital Fibroblasts in Thyroid-Associated Ophthalmopathy. Front. Endocrinol. 2022, 13, 846106. [Google Scholar] [CrossRef]
- Hassan, W.A.; Hamaad, G.A.; Sayed, E.A.; El Behisy, M.M.; Gomaa, M.K. Clinical significance of interleukin 27 serum concentration in patients with systemic sclerosis: Relation to clinical, laboratory and radiological parameters. Egypt. Rheumatol. Rehabil. 2019, 46, 101–107. [Google Scholar] [CrossRef]
- Swaminathan, S.; Hu, Z.; Rupert, A.W.; Higgins, J.M.; Dewar, R.L.; Stevens, R.; Chen, Q.; Rehm, C.A.; Metcalf, J.A.; Baseler, M.W.; et al. Plasma Interleukin-27 (IL-27) Levels Are Not Modulated in Patients with Chronic HIV-1 Infection. PLoS ONE 2014, 9, e98989. [Google Scholar] [CrossRef] [PubMed]
- Hassan, T.; Abdel Rahman, D.; Raafat, N.; Fathy, M.; Shehab, M.; Hosny, A.; Fawzy, R.; Zakaria, M. Contribution of interleukin 27 serum level to pathogenesis and prognosis in children with immune thrombocytopenia. Medicine 2022, 101, e29504. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Nemati, M.; Rezayati, M.T. Serum levels of interleukin (IL)-27 in patients with ischemic heart disease. Cytokine 2011, 56, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; You, H.; Zhang, X. Levels of Interleukin 27 and Interleukin 35 in the Serum and Vitreous of Patients with Proliferative Diabetic Retinopathy. Ocul. Immunol. Inflamm. 2018, 26, 273–279. [Google Scholar] [CrossRef]
- Lukawska-Tatarczuk, M.; Franek, E.; Czupryniak, L.; Joniec-Maciejak, I.; Pawlak, A.; Wojnar, E.; Zielinski, J.; Mirowska-Guzel, D.; Mrozikiewicz-Rakowska, B. Sirtuin 1, Visfatin and IL-27 Serum Levels of Type 1 Diabetic Females in Relation to Cardiovascular Parameters and Autoimmune Thyroid Disease. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Forrester, M.A.; Robertson, L.; Bayoumi, N.; Keavney, B.D.; Barker, R.N.; Vickers, M.A. Human interleukin-27: Wide individual variation in plasma levels and complex inter-relationships with interleukin-17A. Clin. Exp. Immunol. 2014, 178, 373–383. [Google Scholar] [CrossRef]
- Ezzat, M.H.; Hasan, Z.E.; Shaheen, K.Y. Serum measurement of interleukin-31 (IL-31) in paediatric atopic dermatitis: Elevated levels correlate with severity scoring. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 334–339. [Google Scholar] [CrossRef]
- Ginaldi, L.; De Martinis, M.; Ciccarelli, F.; Saitta, S.; Imbesi, S.; Mannucci, C.; Gangemi, S. Increased levels of interleukin 31 (IL-31) in osteoporosis. BMC Immunol. 2015, 16, 60. [Google Scholar] [CrossRef]
- Mohd Ashari, N.S.; Syuhada Mohd Amin, S.N.; Wan Abdul Hamid, W.Z.; Musa, M.; Rahman, A.A.; Mohamad, I. Determination of interleukin 31 (IL-31) serum levels in allergic rhinitis patients. Int. J. Pediatr. Adolesc. Med. 2014, 1, 69–72. [Google Scholar] [CrossRef]
- Rosine, N.; Etcheto, A.; Hendel-Chavez, H.; Seror, R.; Briot, K.; Molto, A.; Chanson, P.; Taoufik, Y.; Wendling, D.; Lories, R.; et al. Increase In Il-31 Serum Levels Is Associated With Reduced Structural Damage In Early Axial Spondyloarthritis. Sci. Rep. 2018, 8, 7731. [Google Scholar] [CrossRef] [PubMed]
- Swierczynska, K.; Krajewski, P.K.; Nowicka-Suszko, D.; Bialynicki-Birula, R.; Krajewska, M.; Szepietowski, J.C. The Serum Level of IL-31 in Patients with Chronic Kidney Disease-Associated Pruritus: What Can We Expect? Toxins 2022, 14. [Google Scholar] [CrossRef] [PubMed]
- Raap, U.; Wichmann, K.; Bruder, M.; Stander, S.; Wedi, B.; Kapp, A.; Werfel, T. Correlation of IL-31 serum levels with severity of atopic dermatitis. J. Allergy Clin. Immunol. 2008, 122, 421–423. [Google Scholar] [CrossRef]
- Zeng, X.; Zhang, Z.; Gao, Q.Q.; Wang, Y.Y.; Yu, X.Z.; Zhou, B.; Xi, M.R. Clinical Significance of Serum Interleukin-31 and Interleukin-33 Levels in Patients of Endometrial Cancer: A Case Control Study. Dis. Markers 2016, 2016, 9262919. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yuan, Y. Serum level of interleukin-17 and interleukin-35 as a biomarker for diagnosis of thyroid cancer. J. Cancer Res. Ther. 2015, 11 Suppl. 2, C209–C211. [Google Scholar] [CrossRef]
- Ahmed, H.A.; Maklad, A.M.; Khaled, S.A.; Elyamany, A. Interleukin-27 and interleukin-35 in de novo acute myeloid leukemia: Expression and significance as biological markers. J. Blood Med. 2019, 10, 341–349. [Google Scholar] [CrossRef]
- Su, Y.; Feng, S.; Luo, L.; Liu, R.; Yi, Q. Association between IL-35 and coronary arterial lesions in children with Kawasaki disease. Clin. Exp. Med. 2019, 19, 87–92. [Google Scholar] [CrossRef]
- Mann, H.; Krystufkova, O.; Zamecnik, J.; Hacek, J.; Hulejova, H.; Filkova, M.; Vencovsky, J.; Senolt, L. Interleukin-35 in idiopathic inflammatory myopathies. Cytokine 2021, 137, 155350. [Google Scholar] [CrossRef]
- Li, W.; Gao, R.; Xin, T.; Gao, P. Different expression levels of interleukin-35 in asthma phenotypes. Respir. Res. 2020, 21, 89. [Google Scholar] [CrossRef]
- Zhang, N.; Dai, H.; Dong, X.; Liu, W.; Jiang, H.; Zhao, Q.; Gao, Y.; Feng, Z.; Dong, Z.; Hu, Y.; et al. Level of interleukin-35 in patients with idiopathic membranous nephropathy and its predictive value for remission time. Front. Immunol. 2022, 13, 926368. [Google Scholar] [CrossRef]
- Chen, B.; Liu, Y.N.; Ji, L.; Liu, P.L.; He, J.; Gan, Y.Y.; Ji, G.J.; Zhu, S.Y.; Zhang, W.H. Elevated levels of interleukin-35 and interleukin-37 in adult patients with obstructive sleep apnea. J. Clin. Lab. Anal. 2021, 35, e23790. [Google Scholar] [CrossRef] [PubMed]
- Szczepankiewicz, D.; Sobkowiak, P.; Narozna, B.; Wojsyk-Banaszak, I.; Breborowicz, A.; Szczepankiewicz, A. Leptin gene polymorphism affects leptin level in childhood asthma. World J. Pediatr. 2018, 14, 601–606. [Google Scholar] [CrossRef]
- Kvistad, S.S.; Myhr, K.M.; Holmoy, T.; Benth, J.S.; Wergeland, S.; Beiske, A.G.; Bjerve, K.S.; Hovdal, H.; Midgard, R.; Sagen, J.V.; et al. Serum levels of leptin and adiponectin are not associated with disease activity or treatment response in multiple sclerosis. J. Neuroimmunol. 2018, 323, 73–77. [Google Scholar] [CrossRef]
- Fahmi, R.M.; Kamel, A.E.; Elsayed, D.A.; Zidan, A.A.; Sarhan, N.T. Serum levels of leptin and adiponectin in patients with multiple sclerosis. Egypt. J. Neurol. Psychiatry Neurosurg. 2021, 57, 114. [Google Scholar] [CrossRef]
- Demiray, G.; Degirmencioglu, S.; Ugurlu, E.; Yaren, A. Effects of Serum Leptin and Resistin Levels on Cancer Cachexia in Patients With Advanced-Stage Non-Small Cell Lung Cancer. Clin. Med. Insights Oncol. 2017, 11, 1179554917690144. [Google Scholar] [CrossRef]
- Duan, D.M.; Jhang, J.Y.; Wu, S.; Teng, M.S.; Hsu, L.A.; Ko, Y.L. Modification effect of sex and obesity on the correlation of LEP polymorphisms with leptin levels in Taiwanese obese women. Mol. Genet. Genom. Med. 2020, 8, e1113. [Google Scholar] [CrossRef] [PubMed]
- Ko, B.J.; Lee, M.; Park, H.S.; Han, K.; Cho, G.J.; Hwang, T.G.; Kim, J.H.; Lee, S.H.; Lee, H.Y.; Kim, S.M. Elevated vaspin and leptin levels are associated with obesity in prepubertal Korean children. Endocr. J. 2013, 60, 609–616. [Google Scholar] [CrossRef]
- Juhasz, E.; Kiss, E.; Simonova, E.; Patocs, A.; Reismann, P. Serum prolactin as a biomarker for the study of intracerebral dopamine effect in adult patients with phenylketonuria: A cross-sectional monocentric study. Eur. J. Med. Res. 2016, 21, 22. [Google Scholar] [CrossRef]
- Jacobi, A.M.; Rohde, W.; Ventz, M.; Riemekasten, G.; Burmester, G.R.; Hiepe, F. Enhanced serum prolactin (PRL) in patients with systemic lupus erythematosus: PRL levels are related to the disease activity. Lupus 2001, 10, 554–561. [Google Scholar] [CrossRef]
- Keen, M.A.; Hassan, I. Serum prolactin levels in psoriasis and its association with disease activity: A case-control study. Indian J. Dermatol. 2014, 59, 562–566. [Google Scholar] [CrossRef]
- Cohen, A.D.; Cohen, Y.; Maislos, M.; Buskila, D. Prolactin serum level in patients with breast cancer. Isr. Med. Assoc. J. 2000, 2, 287–289. [Google Scholar] [PubMed]
- Al-Nami, M.S.; Al-Kuraishy, H.M.; Al-Gareeb, A.I.; Al-Mamoori, F. Metabolic profile and prolactin serum levels in men with type 2 diabetes mellitus: Old-new rubric. Int. J. Crit. Illn. Inj. Sci. 2019, 9, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Pech Torres, R.E.; Cedillo Rivera, R.M.; Lorono Pino, M.A.; Sanchez Burgos, G.G. Serum levels of IFN-beta are associated with days of evolution but not with severity of dengue. J. Med. Virol. 2016, 88, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.W.; Tang, K.T.; Fang, W.F.; Lee, T.I.; Lin, J.D. Differential serum interferon-beta levels in autoimmune thyroid diseases. Arch. Med. Sci. 2022, 18, 1231–1240. [Google Scholar] [CrossRef]
- Liao, A.P.; Salajegheh, M.; Nazareno, R.; Kagan, J.C.; Jubin, R.G.; Greenberg, S.A. Interferon beta is associated with type 1 interferon-inducible gene expression in dermatomyositis. Ann. Rheum. Dis. 2011, 70, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Garza, M.T.; Elva Cruz-Vega, D.; Maldonado-Bernal, C. IL10 as Cancer Biomarker. Transl. Res. Cancer 2021. [Google Scholar] [CrossRef]
- Sobhan, M.R.; Farshchian, M.; Hoseinzadeh, A.; Ghasemibasir, H.R.; Solgi, G. Serum Levels of IL-10 and IL-22 Cytokines in Patients with Psoriasis. Iran. J. Immunol. 2016, 13, 317–323. [Google Scholar]
- Della Bella, C.; Antico, A.; Panozzo, M.P.; Capitani, N.; Benagiano, M.; Petrone, L.; Azzurri, A.; Pratesi, S.; D’Elios, S.; Cianchi, F.; et al. Elevated IL-19 Serum Levels in Patients With Pernicious Anemia and Autoimmune Gastritis. Front. Immunol. 2022, 13, 887256. [Google Scholar] [CrossRef]
- Saleh, H.M.; Deif, M.A.; El-Husseiny, R.M. Assessment of serum interleukin-19 in acne vulgaris patients of different clinical severities. J. Cosmet. Dermatol. 2021, 20, 3034–3040. [Google Scholar] [CrossRef]
- Konrad, R.J.; Higgs, R.E.; Rodgers, G.H.; Ming, W.; Qian, Y.W.; Bivi, N.; Mack, J.K.; Siegel, R.W.; Nickoloff, B.J. Assessment and Clinical Relevance of Serum IL-19 Levels in Psoriasis and Atopic Dermatitis Using a Sensitive and Specific Novel Immunoassay. Sci. Rep. 2019, 9, 5211. [Google Scholar] [CrossRef]
- Rong, B.; Liu, Y.; Li, M.; Fu, T.; Gao, W.; Liu, H. Correlation of serum levels of HIF-1alpha and IL-19 with the disease progression of COPD: A retrospective study. Int. J. Chron. Obstruct. Pulmon. Dis. 2018, 13, 3791–3803. [Google Scholar] [CrossRef] [PubMed]
- Dumycz, K.; Kunkiel, K.; Stelmaszczyk-Emmel, A.; Józefczuk, P.; Ambrozej, D.; Feleszko, W. The role and association of plasma level of IL-19 and pro-inflammatory cytokines (IL-17A, IL-4 and IL-1β) with severity of atopic dermatitis in children. World Allergy Organ. J. 2020, 13, 100217. [Google Scholar] [CrossRef]
- Saheb Sharif-Askari, F.; Saheb Sharif-Askari, N.; Hafezi, S.; Goel, S.; Ali Hussain Alsayed, H.; Ansari, A.W.; Mahboub, B.; Al-Muhsen, S.; Temsah, M.H.; Hamid, Q.; et al. Upregulation of interleukin-19 in saliva of patients with COVID-19. Sci. Rep. 2022, 12, 16019. [Google Scholar] [CrossRef] [PubMed]
- Hussien, D.T.; Shabana, A.A.; Hassan, A.S.; Elmarghany, E.B. Assessment of serum interleukin-20 level in rheumatoid arthritis patients: Relation to disease activity and ultrasound measures. Egypt. Rheumatol. 2022, 44, 181–186. [Google Scholar] [CrossRef]
- Michalak-Stoma, A.; Bartosinska, J.; Kowal, M.; Juszkiewicz-Borowiec, M.; Gerkowicz, A.; Chodorowska, G. Serum levels of selected Th17 and Th22 cytokines in psoriatic patients. Dis. Markers 2013, 35, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.H.; Li, H.H.; Hsieh, M.Y.; Liu, M.F.; Huang, K.Y.; Chin, L.S.; Chen, P.C.; Cheng, H.H.; Chang, M.S. Function of interleukin-20 as a proinflammatory molecule in rheumatoid and experimental arthritis. Arthritis Rheum. 2006, 54, 2722–2733. [Google Scholar] [CrossRef]
- Naumnik, W.; Naumnik, B.; Niklińska, W.; Ossolińska, M.; Chyczewska, E. Clinical Implications of Hepatocyte Growth Factor, Interleukin-20, and Interleukin-22 in Serum and Bronchoalveolar Fluid of Patients with Non-Small Cell Lung Cancer. In Advancements in Clinical Research; Pokorski, M., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 41–49. [Google Scholar]
- Kragstrup, T.W.; Otkjaer, K.; Holm, C.; Jorgensen, A.; Hokland, M.; Iversen, L.; Deleuran, B. The expression of IL-20 and IL-24 and their shared receptors are increased in rheumatoid arthritis and spondyloarthropathy. Cytokine 2008, 41, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Valentina, M.; Jan, F.; Peder, N.L.; Bo, Z.; Hongjie, D.; Pernille, K. Cytokine detection and simultaneous assessment of rheumatoid factor interference in human serum and synovial fluid using high-sensitivity protein arrays on plasmonic gold chips. BMC Biotechnol. 2015, 15, 73. [Google Scholar] [CrossRef]
- Aydogdu, E.; Pamuk, O.N.; Donmez, S.; Pamuk, G.E. Decreased interleukin-20 level in patients with systemic sclerosis: Are they related with angiogenesis? Clin. Rheumatol. 2013, 32, 1599–1603. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, Z.; Xing, H.; Wang, L.; Zhang, G.; Yu, N.; Wang, J.; Guo, W.; Jiang, J. Elevated Th22 cells and related cytokines in patients with epithelial ovarian cancer. Medicine 2017, 96, e8359. [Google Scholar] [CrossRef]
- Tsirakis, G.; Pappa, C.A.; Kolovou, A.; Kokonozaki, M.; Neonakis, I.; Alexandrakis, M.G. Clinical significance of interleukin-22 in multiple myeloma. Hematology 2015, 20, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.M.; Matias, B.F.; Rodrigues, C.M.; Murta, E.F.; Michelin, M.A. IL-17 and IL-22 serum cytokine levels in patients with squamous intraepithelial lesion and invasive cervical carcinoma. Eur. J. Gynaecol. Oncol. 2013, 34, 466–468. [Google Scholar] [PubMed]
- Abdullah, H.N.; Abdulwahid, A.G. Expression of Serum IL-22, IL-23, and TLR9 as Tumor Markers in Untreated Breast Cancer Patients. J. Drug Deliv. 2020, 10, 472–476. [Google Scholar] [CrossRef]
- Thomas-Dupont, P.; Remes-Troche, J.M.; Izaguirre-Hernández, I.Y.; Sánchez-Vargas, L.A.; Maldonado-Rentería, M.d.J.; Hernández-Flores, K.G.; Torre, A.; Bravo-Sarmiento, E.; Vivanco-Cid, H. Elevated circulating levels of IL-21 and IL-22 define a cytokine signature profile in type 2 autoimmune hepatitis patients. Ann. Hepatol. 2016, 15, 550–558. [Google Scholar]
- Khoshroo, M.; Yazdanpanah, M.J.; Yasrebi, S. Serum Interleukin-24 Levels in Gastric and Breast Cancers and Non-cancerous Inflammations. Middle East J. of Cancer 2021, 12, 183–189. [Google Scholar] [CrossRef]
- Li, R.C.; Guo, J.; Su, L.C.; Huang, A.F. Elevated levels of IL-24 in systemic lupus erythematosus patients. Lupus 2019, 28, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Brilland, B.; Bach-Bunner, M.; Gomes, C.N.; Larochette, V.; Foucher, E.; Plaisance, M.; Saulnier, P.; Costedoat-Chalumeau, N.; Ghillani, P.; Belizna, C.; et al. Serum Interleukin-26 Is a New Biomarker for Disease Activity Assessment in Systemic Lupus Erythematosus. Front. Immunol. 2021, 12, 663192. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Dong, X.; Liu, J.; Lv, H.; Ge, Y. Serum Interleukin-26 is a Potential Biomarker for the Differential Diagnosis of Neurosyphilis and Syphilis at Other Stages. Infect. Drug Resist. 2022, 15, 3693–3702. [Google Scholar] [CrossRef]
- Louhaichi, S.; Mlika, M.; Hamdi, B.; Hamzaoui, K.; Hamzaoui, A. Sputum IL-26 Is Overexpressed in Severe Asthma and Induces Proinflammatory Cytokine Production and Th17 Cell Generation: A Case-Control Study of Women. J. Asthma Allergy 2020, 13, 95–107. [Google Scholar] [CrossRef]
- Kaabachi, W.; Bouali, E.; Berraïes, A.; Dhifallh, I.B.; Hamdi, B.; Hamzaoui, K.; Hamzaoui, A. Interleukin-26 is overexpressed in Behçet’s disease and enhances Th17 related −cytokines. Immunol. Lett. 2017, 190, 177–184. [Google Scholar] [CrossRef]
- Pinero, P.; Juanola, O.; Gutierrez, A.; Zapater, P.; Gimenez, P.; Steinert, A.; Sempere, L.; Gonzalez-Navajas, J.M.; Niess, J.H.; Frances, R. IL26 modulates cytokine response and anti-TNF consumption in Crohn’s disease patients with bacterial DNA. J. Mol. Med. 2017, 95, 1227–1236. [Google Scholar] [CrossRef] [PubMed]
- Corvaisier, M.; Delneste, Y.; Jeanvoine, H.; Preisser, L.; Blanchard, S.; Garo, E.; Hoppe, E.; Barre, B.; Audran, M.; Bouvard, B.; et al. IL-26 is overexpressed in rheumatoid arthritis and induces proinflammatory cytokine production and Th17 cell generation. PLoS Biol. 2012, 10, e1001395. [Google Scholar] [CrossRef]

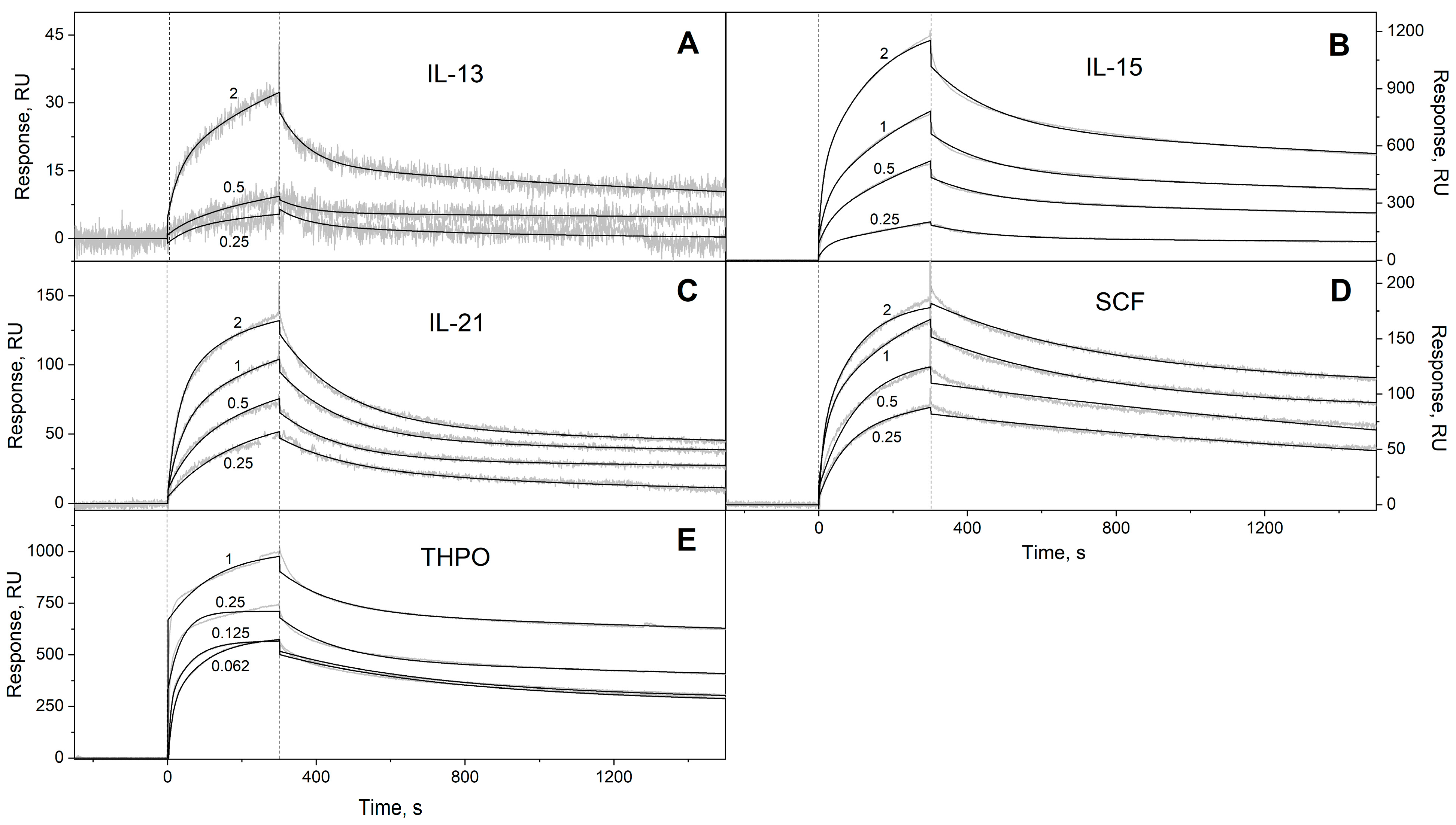
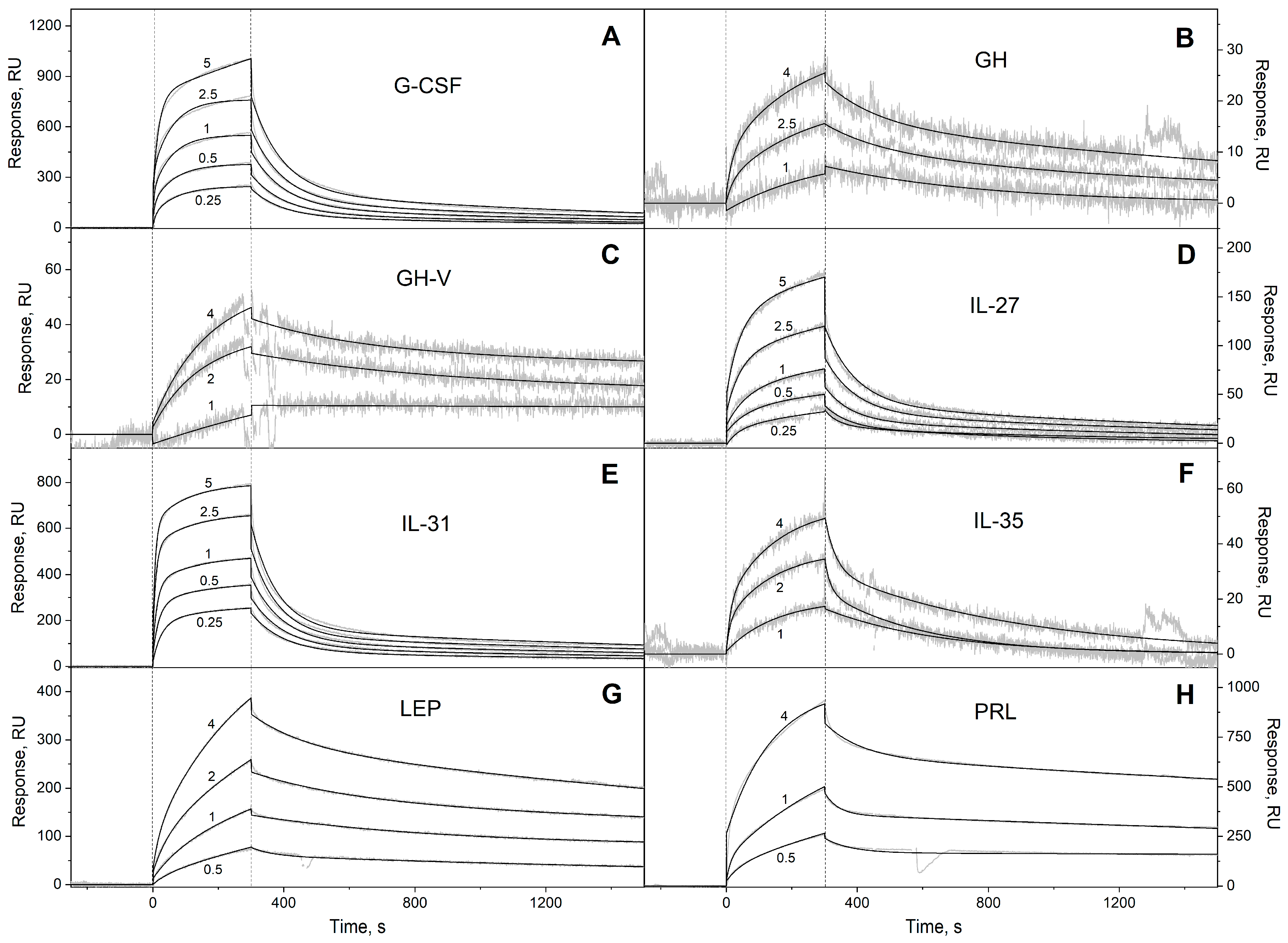
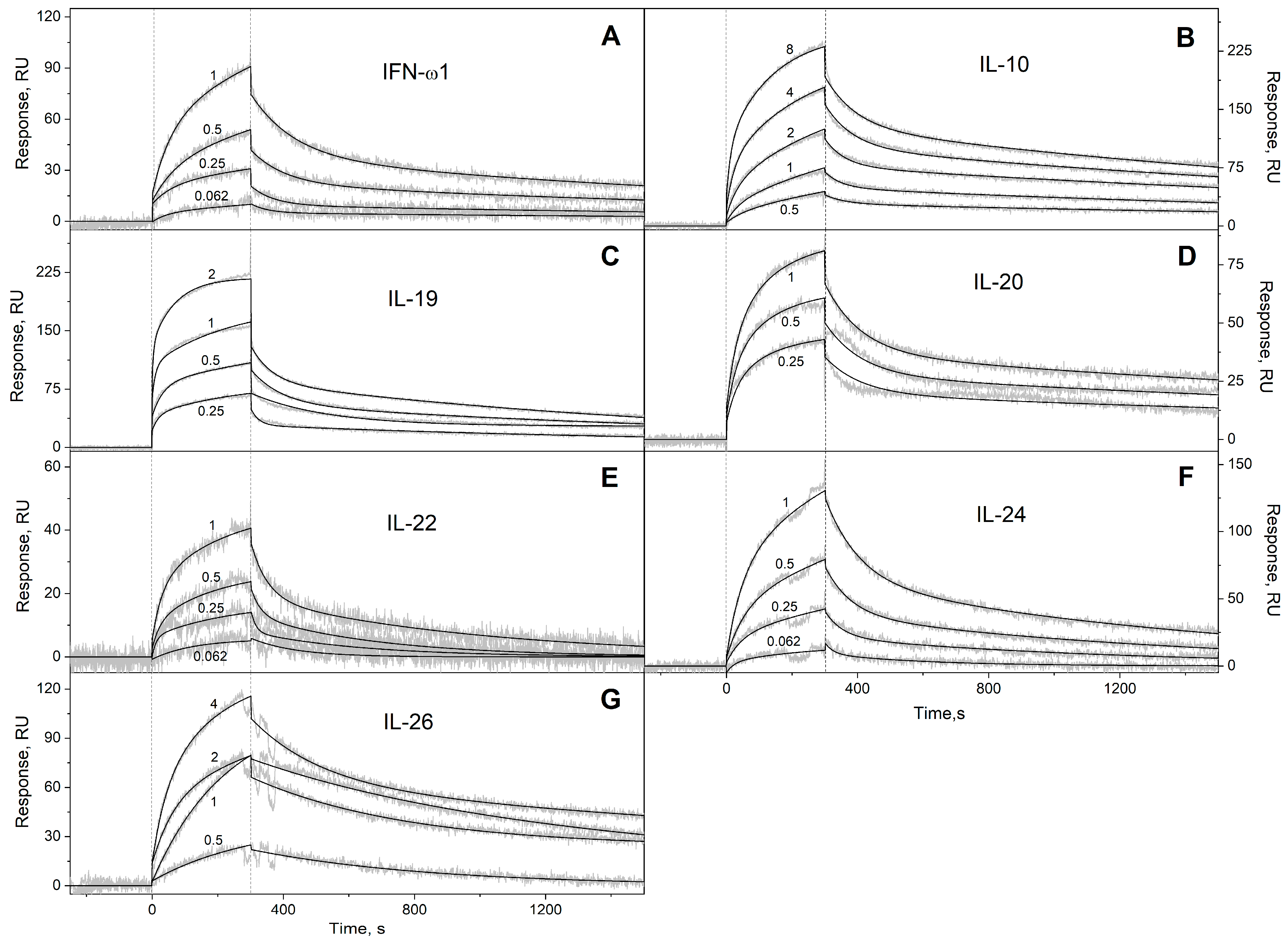

| Cytokine | Kd1, M | Kd2, M | Reference |
|---|---|---|---|
| Short-chain cytokines | |||
| EPO | (1.5 ± 0.3) × 10−7 | (6.5 ± 2.6) × 10−7 | [43] |
| Long-chain cytokines | |||
| CLCF1 | (1.1 ± 0.8) × 10−6 | (3.7 ± 0.8) × 10−6 | [46] |
| CNTF | (1.1 ± 1.0) × 10−7 | (2.07 ± 0.08) × 10−6 | [46] |
| CT-1 | (1.6 ± 0.5) × 10−6 | (1.2 ± 0.4) × 10−5 | [46] |
| IL-11 | (8.3 ± 1.9) × 10−6 | (7.9 ± 2.1) × 10−6 | [46] |
| Interferons/IL-10 | |||
| IFN-β | (8.2 ± 2.4) × 10−8 (0.70 ± 0.03) × 10−9 * | (2.67 ± 0.57) × 10−7 (2.81 ± 0.49) × 10−7 * | [44] |
| Cytokine | kd1, s−1 | Kd1, M | kd2, s−1 | Kd2, M |
|---|---|---|---|---|
| Short-chain cytokines | ||||
| Flt3L | (8.83 ± 2.54) × 10−4 | (1.22 ± 0.27) × 10−7 | (8.32 ± 4.05) × 10−4 | (1.25 ± 0.46) × 10−7 |
| GM-CSF | (7.92 ± 4.02) × 10−4 | (2.26 ± 1.18) × 10−6 | (1.76 ± 0.49) × 10−2 | (3.63 ± 1.14) × 10−6 |
| IL-2 * | (4.97 ± 0.78) × 10−3 | (1.20 ± 0.04) × 10−5 | n/a | n/a |
| IL-3 | (2.71 ± 0.78) × 10−4 | (6.11 ± 2.19) × 10−9 | (7.28 ± 0.65) × 10−3 | (4.02 ± 2.03) × 10−7 |
| IL-5 | (2.47 ± 0.45) × 10−4 | (1.16 ± 0.52) × 10−8 | (5.07 ± 0.20) × 10−3 | (3.23 ± 0.73) × 10−8 |
| IL-9 | (5.97 ± 2.79) × 10−3 | (1.69 ± 0.90) × 10−7 | (5.85 ± 2.72) × 10−3 | (2.71 ± 2.42) × 10−7 |
| IL-13 | (2.64 ± 0.98) × 10−4 | (1.97 ± 1.50) × 10−7 | (1.36 ± 0.06) × 10−2 | (1.91 ± 1.07) × 10−6 |
| IL-15 | (1.62 ± 0.24) × 10−4 | (5.70 ± 0.46) × 10−8 | (6.29 ± 0.26) × 10−3 | (2.15 ± 1.32) × 10−7 |
| IL-21 | (1.88 ± 1.33) × 10−4 | (4.57 ± 1.30) × 10−8 | (5.68 ± 0.34) × 10−3 | (3.87 ± 0.86) × 10−7 |
| SCF | (2.57 ± 0.92) × 10−4 | (8.72 ± 4.95) × 10−9 | (4.51 ± 1.52) × 10−3 | (1.90 ± 1.45) × 10−6 |
| THPO | (6.00 ± 3.29) × 10−5 | (2.89 ± 1.83) × 10−10 | (4.47 ± 2.46) × 10−3 | (3.96 ± 2.65) × 10−9 |
| Long-chain cytokines | ||||
| G-CSF | (7.72 ± 0.31) × 10−4 | (3.91 ± 2.06) × 10−9 | (1.04 ± 0.04) × 10−2 | (1.68 ± 1.29) × 10−6 |
| GH | (3.73 ± 2.36) × 10−4 | (3.42 ± 2.85) × 10−7 | (5.87 ± 1.16) × 10−3 | (4.16 ± 2.65) × 10−7 |
| GH-V | (7.92 ± 3.35) × 10−5 | (4.62 ± 2.41) × 10−8 | (3.06 ± 1.27) × 10−3 | (1.78 ± 0.92) × 10−6 |
| IL-27 # | (9.61 ± 4.18) × 10−4 | (1.16 ± 0.63) × 10−6 | (1.47 ± 0.62) × 10−2 | (2.80 ± 0.85) × 10−6 |
| IL-31 | (4.64 ± 0.13) × 10−4 | (1.06 ± 0.96) × 10−7 | (1.11 ± 0.12) × 10−2 | (2.56 ± 1.95) × 10−7 |
| IL-35 # | (2.59 ± 0.81) × 10−3 | (2.34 ± 1.26) × 10−6 | (3.91 ± 1.55) × 10−2 | (4.65 ± 2.42) × 10−6 |
| LEP | (3.18 ± 2.17) × 10−3 | (3.25 ± 1.92) × 10−7 | (2.87 ± 1.00) × 10−4 | (3.85 ± 1.06) × 10−7 |
| PRL | (1.59 ± 0.23) × 10−4 | (8.40 ± 0.58) × 10−8 | (1.55 ± 0.82) × 10−2 | (6.35 ± 0.96) × 10−7 |
| Interferons/IL-10 | ||||
| IFN-ω1 | (4.71 ± 0.22) × 10−4 | (2.28 ± 0.64) × 10−7 | (1.29 ± 0.55) × 10−2 | (1.11 ± 0.63) × 10−6 |
| IL-10 | (3.98 ± 0.28) × 10−4 | (3.61 ± 1.09) × 10−7 | (1.50 ± 0.37) × 10−2 | (2.13 ± 0.67) × 10−6 |
| IL-19 | (6.28 ± 0.86) × 10−4 | (1.47 ± 1.08) × 10−7 | (2.77 ± 1.69) × 10−2 | (4.96 ± 4.04) × 10−7 |
| IL-20 | (3.08 ± 0.86) × 10−4 | (4.51 ± 1.12) × 10−8 | (9.14 ± 2.51) × 10−3 | (3.85 ± 0.59) × 10−7 |
| IL-22 | (3.57 ± 2.56) × 10−3 | (5.93 ± 0.84) × 10−7 | (3.26 ± 2.67) × 10−2 | (1.31 ± 0.49) × 10−6 |
| IL-24 | (5.73 ± 0.85) × 10−4 | (2.39 ± 0.94) × 10−7 | (7.79 ± 1.72) × 10−3 | (6.01 ± 1.57) × 10−7 |
| IL-26 | (5.98 ± 2.92) × 10−4 | (2.78 ± 2.31) × 10−7 | (2.56 ± 1.39) × 10−3 | (6.00 ± 2.63) × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazakov, A.S.; Deryusheva, E.I.; Rastrygina, V.A.; Sokolov, A.S.; Permyakova, M.E.; Litus, E.A.; Uversky, V.N.; Permyakov, E.A.; Permyakov, S.E. Interaction of S100A6 Protein with the Four-Helical Cytokines. Biomolecules 2023, 13, 1345. https://doi.org/10.3390/biom13091345
Kazakov AS, Deryusheva EI, Rastrygina VA, Sokolov AS, Permyakova ME, Litus EA, Uversky VN, Permyakov EA, Permyakov SE. Interaction of S100A6 Protein with the Four-Helical Cytokines. Biomolecules. 2023; 13(9):1345. https://doi.org/10.3390/biom13091345
Chicago/Turabian StyleKazakov, Alexey S., Evgenia I. Deryusheva, Victoria A. Rastrygina, Andrey S. Sokolov, Maria E. Permyakova, Ekaterina A. Litus, Vladimir N. Uversky, Eugene A. Permyakov, and Sergei E. Permyakov. 2023. "Interaction of S100A6 Protein with the Four-Helical Cytokines" Biomolecules 13, no. 9: 1345. https://doi.org/10.3390/biom13091345







