Abstract
S100A10 (p11, annexin II light chain, calpactin light chain) is a multifunctional protein with a wide range of physiological activity. S100A10 is unique among the S100 family members of proteins since it does not bind to Ca2+, despite its sequence and structural similarity. This review focuses on studies highlighting the structure, regulation, and binding partners of S100A10. The binding partners of S100A10 were collated and summarized.
1. Introduction
S100A10 (p11) is one of twenty-five different S100 family members of proteins, of which most are genes located on chromosome 1q21, which is amplified in several neoplastic diseases. Although the S100 proteins demonstrate high-level sequence and structural similarity, they are markedly different in their functions and the proteins they interact with. S100A10 is unique among the twenty-five S100 proteins as it does not bind to Ca2+ and is present in a conformation that resembles the Ca2+-bound form of other S100 proteins, suggesting it perpetually exists in the active state. S100A10 interacts with several proteins but predominantly exists in a heterotetrameric complex with Annexin A2 (p36), which plays an important role in the cellular localization, stability, and functionality of S100A10. Over the past decade, there have been several excellent reviews that have been published on S100A10. Our focus in this review is to highlight the recent advances in the structure, regulation, binding partners, interactions, and functions of S100A10.
2. Structure
The first demonstration of the importance of calcium (Ca2+) as a signaling molecule was the evidence that Ca2+ played a key role in muscle contraction. This discovery was made by Sidney Ringer, who reported that “phosphate of calcium added to saturation to saline can sustain contractility [1]. Subsequently, Heildebrun reported that “calcium, even in rather high dilution, causes immediate and pronounced shortening of frog muscle”, and that “this effect is not shared by any one of the other cations normally present in any quantity in muscle, i.e., K+, Na+, and Mg2+” [2]. Bailey also envisioned the importance of Ca2+ ions in muscle contraction and reported that actomyosin ATPase was stimulated by Ca2+ and, to a much lesser extent, Mn2+ [3]. This work was corroborated by Bozler, who demonstrated that the removal of Ca2+ by the chelator, EDTA, relaxed muscle fibers [4]. Subsequently, Ebashi reported that Ca2+, at a micromolar concentration, caused the contraction of actomyosin preparations [5]. Ebashi later revealed that the receptor that mediated Ca2+-dependent regulation of muscle contraction was a protein he named troponin. Therefore, troponin was the first intracellular Ca2+-binding protein identified [6]. Later, it was shown that a subunit of troponin, troponin-C, was the Ca2+-binding protein and that the binding of Ca2+ to troponin-C induced a conformational change in troponin that resulted in the activation of actomyosin ATPase. This established that Ca2+ and a receptor of Ca2+, namely a Ca2+-binding protein, regulated muscle contraction.
An understanding of how proteins bound Ca2+ was provided by Kretsinger, who first reported the X-ray crystallographic structure of the carp muscle Ca2+-binding protein, parvalbumin [7]. He demonstrated that parvalbumin possessed a Ca2+-binding domain that he referred to as an EF-hand. The designation “EF-hand” was derived from the structural orientation of the two α-helices (E and F) that form together with the Ca2+-binding loop of 12 amino acids. The EF-hand motif is the most frequently occurring Ca2+-binding motif in eukaryotic systems [8].
There are more than 66 subfamilies of EF–hand proteins [9], which include well-known families such as the calmodulin (CaM) and S100 families. The conformational change induced by the binding of Ca2+ to each of these families is fundamentally different. The Ca2+-induced conformational change in all S100 proteins results from a shift in the orientation of Helix III [10], whereas the conformational change in calmodulin involves a dramatic opening of both EF-hands, which is not observed in the S100 proteins. Furthermore, calmodulin utilizes both EF-hand domains to bind to a target molecule, whereas an S100 dimer binds two target molecules on opposite faces of the structure. CaM has two independent globular domains tethered by a flexible linker, which enables the two binding sites to bind to the same target. In contrast, the S100 dimer forms a single, highly interdigitated globular domain, with the two symmetrically disposed binding sites on opposite faces of the structure. Like other EF-hand proteins, the conformational change results in the exposure of a hydrophobic patch that serves as the key factor driving the binding of targets [9,10,11]. In contrast, a unique dimeric structural organization has been observed for the S100A6/RAGE receptor complex, where the two symmetrical binding sites for the RAGE receptor are localized to the same face of the S100A6 dimer [12].
The S100 protein family is the largest subfamily of EF-hand proteins. They were initially identified for their solubility in 100% ammonium sulfate [13]. This was true, however, for the first two proteins discovered, namely S100B and S100A1, but it is no longer a shared feature among the twenty-three S100 members identified so far in humans, which include S100A1, S100A2, S100A3, S100A4, S100A5, S100A6, S100A7, S100A7A, S100A7B, S100A8, S100A9, S100A10, S100A11, S100A11P, S100A12, S100A13, S100A14, S100A15, S100A16, S100A17, S100A18, S100B, S100G, S100P, and S100Z. The S100 proteins are abundant, low molecular weight (10–12 kDa), acidic proteins exhibiting distinct tissue-specific expression. All, except S100G, exist as dimers, particularly but not exclusively homodimers, under physiological conditions. The dimer is formed by noncovalent interactions between helices I and IV. NMR, mass spectrometry, and Green Fluorescent Protein (GFP) trap experiments consistently show that S100A1:S100B, S100A1:S100P, and S100A11:S100B heterodimers are the predominant species formed compared to their corresponding homodimers [14,15,16].
The S100 monomers are typically composed of an EF-hand motif at the amino-terminal (EF1) and one at the carboxyl-terminal (EF2) connected by a region referred to as a “hinge”. The EF1 motif contains a long helix (HI), a loop (L), and a short helix (HII). The EF1 Ca2+-binding site is called a ‘pseudo’ EF-hand because it possesses two extra residues in the loop, and the coordination of Ca2+ is accomplished mainly by backbone carbonyls. In contrast, the canonical EF2 Ca2+-binding site contains a short helix (HIII), a loop (L), and a long helix (HIV) and binds Ca2+ through acidic side chains (Figure 1).
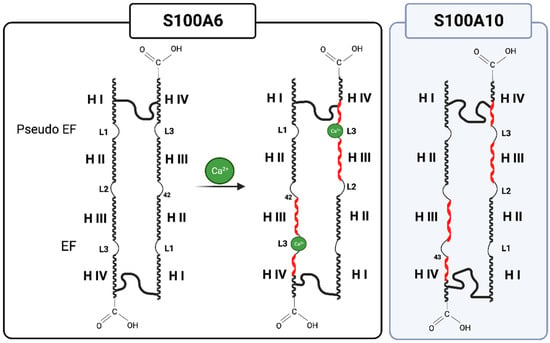
Figure 1.
S100 (ex. S100A6) monomers, typically containing two EF-hand motifs consisting of a short helix, loop, and long helix, undergo a conformation change upon Ca2+ binding, exposing a hydrophobic patch in helix III and IV to bind target proteins (red helix). S100A10 resembles the Ca2+-loaded S100 protein, suggesting that it exists in an active, Ca2+-independent conformation. Created with “Biorender.com (accessed on 5 September 2023)”.
As discussed, Ca2+-binding to the S100 proteins exposes a hydrophobic region comprising residues from both helix III and IV, which serves to bind target proteins. Structural studies have established that Ca2+-binding to the S100 protein, except S100A10, is required to facilitate most S100-annexin interactions [17]. Of the twenty-five members of the S100 protein family, seven (S100A1, S100A4, S100A6, S100A10, S100A11, S100A12, and S100B) interact with at least one of the 12 human annexin proteins. S100 proteins, such as S100A6, for example, appear to form complexes with several annexin proteins (A2, A5, A6, and A11) [18,19] (Figure 2).
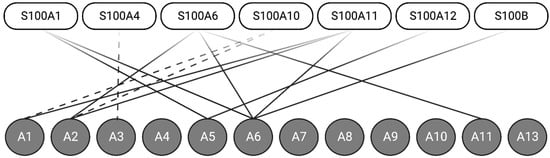
Figure 2.
Seven S100 proteins can form Ca2+-dependent (solid-line) or Ca2+-independent (dashed-line) interactions with one of the twelve annexin proteins. Annexins A1, A2, A4, and A6 can form interactions with multiple S100 proteins. Created with “Biorender.com (accessed on 8 September 2023)”.
Annexins are a family of Ca2+-binding proteins that interact with biological membranes. There are twelve human annexins. All annexins have four repeating, highly homogenous amino acid sequences (eight repeats in the case of annexin VI), known as the annexin repeat [20] (Figure 3). The annexins typically have three different Ca2+-binding sites distinct from the EF-hand sites. The type II Ca2+-binding sites have the canonical sequence of the endonexin fold, KGXGT-(38X)-D/E, a Ca2+-binding site characteristic of the annexin protein family. The annexins are noted for a single type II site in the second, third, and fourth domains, two type III Ca2+-binding sites in the first domain of the protein, and a type AB’ site [21]. Annexin A2 has been extensively studied and shown to bind to multiple proteins, as well as phospholipids [22], polysaccharides such as heparan [23], fucoidan [24], and RNA [25] (reviewed in [26,27]). Specific and well-established binding sites on annexin A2 include residues K279 and K281, which form a phosphoinositide-specific binding site that contributes significantly to the specificity of annexin A2 binding to phosphoinositide-containing membranes [28], and the last nine residues of the carboxyl-terminal, LLYLCGGDD, that form the F-actin binding site [29].
S100A10 differs from the others as it cannot bind Ca2+ ions because of a three-residue deletion in EF-1 and the mutation of acidic Ca2+-coordinating residues in EF-2. However, S100A10 is virtually identical to Ca2+-loaded S100 proteins, suggesting S100A10 has adapted an active conformation [17]. Multiple laboratories have studied the AIIt complex in great detail (reviewed in [26]). AIIt is a highly symmetric complex with the S100A10 antiparallel dimer in the center and an annexin A2 monomer on either side of S100A10. The N-terminus of each annexin A2 monomer is associated with a region formed by EF1-Helix I from one S100A10 monomer and the EF2-Helix IV and hinge from the other S100A10 monomer. Recently, the Ca2+-free and seven Ca2+-loaded AIIt complexes have been analyzed by molecular dynamics [30]. The advantage of this approach was that the modeler added the last five residues of S100A10, Q93, K94, G95, K96, and K97, which were absent from the crystal structure. These authors reported that, consistent with previously published data, the N-terminal annexin A2 hydrophobic residues (T3, V4, I7, L8, and L11) form contacts with multiple residues of S100A10 and are the top ten strongest interactions for both annexin A2 in the Ca2+-free and Ca2+-loaded models. These authors found that while the S100A10 residues (E6, M9, E10, M13, F14, F42, A82, and Y86) formed strong contacts with the N-terminal hydrophobic residues of annexin A2, five residues (F39, L79, C83, F87, and M91), thought to be important complex-forming residues, provided only weak interactions, suggesting they have a less significant contribution toward the total interaction between S100A10 and annexin A2.
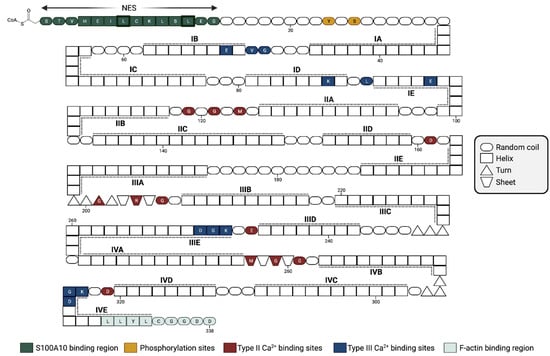
Figure 3.
Amino acid structure of annexin A2. Due to high sequence conservation, human annexins are structurally similar, with four annexin repeats (eight for annexin A6). The ~70-residue-long structural element (labeled domains I–IV) consists of five α-helices (labeled A–E) and is important for Ca2+-binding. Additionally, other important structural sites include the nuclear export sequence (NES) and multiple phosphorylation sites. Figure updated from [31]. Created with “Biorender.com (accessed on 8 September 2023)”.
It was particularly interesting to note that in the Ca2+-loaded model, the annexin A2 residue, R145, interacted with the S100A10 residue, D48, but that in the Ca2+-free model, the A2 residue, R145, interacted with the carboxyl-terminal S100A10 residue, K97 [30]. We had initially proposed that K97 was the residue that interacted with plasminogen [32]. This was substantiated by our report that several carboxypeptidases removed this residue, which resulted in the loss of the ability of S100A10 to bind and activate plasminogen [33,34]. However, more recently, we observed that the substitution of the carboxyl-terminal S100A10 residue with isoleucine did not affect plasminogen activation, and we subsequently proposed that the carboxyl-terminal S100A10 residue might actually regulate the exposure of an internal lysine residue [35]. We have also reported that Ca2+ did not stimulate S100A10-dependent plasminogen activation [36]. The report that the accessibility of carboxyl-terminal lysine of the S100A10 (K97) was affected by Ca2+ further suggested that the carboxyl-terminal lysine of S100A10 might not be critical for plasminogen activation.
Recently, we proposed that the formation of the AIIt complex was an example of a mutualistic symbiotic relationship [27]. In this regard, we suggested that the formation of the AIIt complex not only regulates the biological activity of each subunit but also confers new biological functions that were not possessed by either subunit. Furthermore, complex formation is also critical for the stability of S100A10 because, in the absence of annexin A2, S100A10 is rapidly degraded [37]. Similarly, annexin A2 requires S100A10 to increase its affinity for Ca2+, facilitating its participation in Ca2+-dependent processes such as membrane binding. These remarkable properties of the annexin A2/S100A10 complex are discussed in detail in our review [27].
The fundamental property that separates S100A10 from the other S100 proteins is that S100A10 cannot bind Ca2+ but is locked in the Ca2+-loaded conformation. The key question is what advantage the inability to respond to fluctuations in Ca2+ confers on the protein. The simplest explanation is that S100A10 will form a complex with annexin A2 in the presence or absence of Ca2+, whereas the formation of annexin complexes by other S100 proteins will require Ca2+. The ubiquitin-mediated degradation of newly formed S100A10 is Ca2+-independent, and therefore the complex formation will be finely tuned to the availability of annexin A2 and not to intracellular Ca2+ concentrations. However, any Ca2+-dependent interactions of the annexin A2/S100A10 complex with binding partners will be regulated by Ca2+-binding by the annexin A2 component of the complex. Since the binding of S100A10 to annexin A2 increases the affinity of annexin A2 for Ca2+, it could be argued that free annexin A2 and annexin A2 bound to S100A10 are conformationally distinct. In contrast, considering the micromolar Ca2+ binding affinity of most of the S100 proteins, their ability to form complexes with annexins would appear to be unlikely under resting submicromolar intracellular Ca2+ concentrations. The exception is S100A1, which, upon covalent modification, dramatically increases its Ca2+-binding affinity one-hundred-fold [38].
3. Binding Partners of S100A10
In addition to annexin A2, a number of additional proteins have been shown to interact with S100A10. For most of these protein–protein interactions, the physiological significance remains unknown. Additionally, it is unclear if S100A10 is the only protein in the S100 family that will interact with these proteins. A list of published S100A10 protein interactors was generated by summarizing public databases: NCBI (https://www.ncbi.nlm.nih.gov/), BioGRID (https://thebiogrid.org/) and ELIXIR (https://www.ebi.ac.uk/intact/home). Ontology analysis of the protein interactors was performed using DAVID 7.0 [39] (http://david.ncifcrf.gov/). Significant ontology findings with a p value < 0.05 and FDR < 0.1 are summarized in Table 1 and illustrated in Figure 4. Interactions that have been demonstrated to have physiological relevance are presented in Table 2. A full list of reported interacting proteins is summarized in Table 3.

Table 1.
Binding partners participating in different S100A10-related functions.
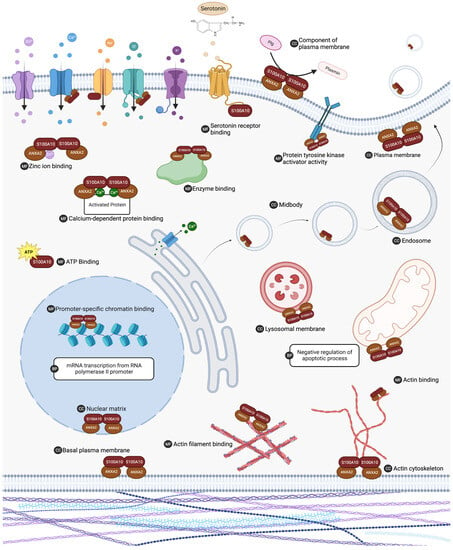
Figure 4.
The list of S100A10 interacting proteins was subjected to gene ontology analysis in DAVID 7.0 to characterize putative biological activity. Gene ontology analysis indicated that S100A10 interacting proteins are ubiquitous in the cell and are involved in many important processes. Biological processes (BP), molecular functions (MF), and cellular component (CC) ontology results with a p value < 0.05 and FDR < 0.1 are summarized. ANXA2 (annexin A2). Created with “Biorender.com (accessed on 6 August 2023)”.

Table 2.
Proteins with known functional interactions with p11.
To date, over 170 proteins have been proposed to interact with S100A10 and observed through various biochemical approaches. Mass spectrometry was the most commonly used technique to identify S100A10 interactors. This approach involved several techniques, including affinity capture (34% of identified interactors), proximity labeling (13%), and co-fractionation (13%). Two-hybrid yeast screening was the second most popular biochemical approach to identify interactors (13%). Other methods used to identify protein interactors included, but were not limited to, co-crystal structure, affinity capture-western blot, reconstituted complex, co-immunoprecipitation, and fluorescent imaging. It will be interesting to determine the physiological relevance of the predicted binding partners of S100A10 and, more specifically, to identify the binding regions or structural determinants in S100A10 that predict and demonstrate the binding affinity to the various proteins.
4. Regulation of S100A10
The expression and function of S100A10 are tightly regulated by various cytokines, growth factors, nitric oxide (reviewed in [69]), RAS oncogene [70], PML-RAR oncogene [71], and hypoxia-inducible factor (HIF) [72]. MicroRNA (miRNA) and long-non-coding RNA (LncRNA) are important non-protein-coding RNA machinery that make up a large percentage of the genome. They have been demonstrated to function in post-transcriptional and epigenetic regulation. LncRNA and miRNA play a crucial role in regulating development, stress-mediated processes, immune response and tumor progression, metastasis, and chemoresistance [73,74,75,76]. Most evidence of the regulation of S100A10 by miRNA and LncRNA has been obtained from studies in hepatocellular carcinoma (HCC) models. Shan et al. identified that mir-590-5p is decreased in some hepatocellular carcinoma cell lines with a concomitant upregulation of S100A10. They further showed that mir-590-5p regulated S100A10 expression by interacting with the 3′UTR of S100A10 mRNA [77]. Similarly, LINC00174 has been shown to be an oncogenic LncRNA in HCC, and it functions by sponging mir-320, which further upregulates the expression of S100A10, accelerating the proliferation and metastasis of HCC cells [78]. Another lncRNA, KCNMB2-AS1, functions by regulating S100A10 in bladder cancer through direct sponging of miR-374a-3p, which negatively regulates the expression of S100A10 [79].
5. Role of p11 in Fibrinolysis
S100A10 is present at the endothelial cell surface and bound to annexin A2, forming the AIIt complex. This complex has been shown to be one of the key receptor-binding plasminogens in endothelial cells. Plasminogen (Pg) is an inactive zymogen that is converted to the active serine protease plasmin (Pm) by plasminogen activators, tissue-plasminogen (tPA) activators, and urokinase plasminogen activator (uPA). This process is highly regulated; the principal inhibitor of Pm is α2-antiplasmin (AP), and the principal inhibitor of tPA and uPA is plasminogen activator inhibitor type 1 (PAI-1) [80,81,82,83]. Although tPA and uPA possess low activity, their activity dramatically increases when interacting with Pg receptors. Certain Pg receptors also protect Pg activators and Pm from inactivation. Thus, Pg receptors such as AIIt are the key regulators of Pm generation.
Plasmin is crucial for dissolving fibrin clots in a process known as fibrinolysis. Fibrin blood clot formation begins when a break in a blood vessel allows plasma to contact tissue factors in the extracellular matrix. Initially, factor VII binds to tissue factor and is activated, which participates in the activation of a clotting factor cascade and results in the cleavage of prothrombin into thrombin. This thrombin activates platelets, resulting in the generation of FXa/Va complexes and the generation of sufficient thrombin from prothrombin to convert fibrinogen to fibrin, resulting in the formation of a stable hemostatic plug [84,85]. The fibrinolytic surveillance system is critical in preventing the inappropriate formation of blood clots and in regulating the size of blood clots that form at the site of injury. Fibrinolysis is initiated by the secretion of plasminogen activators (tPA and uPA). The endothelium of blood vessels secretes tPA, which circulates in the blood as a single-chain protein (sct-PA). The kidney secretes uPA, which circulates in an inactive single-chain form (scu-PA), which Pm must convert into the active two-chain form (tcu-PA) [86,87].
Pm degrades the fibrin blood clot, producing D-dimer and other fibrin degradation products. Impaired breakdown of fibrin clots is associated with many diseases, including diabetes, insulin resistance, sepsis, stroke, and the metabolic syndrome [88,89,90,91]. Impaired fibrinolysis also contributes to deep venous thrombosis and pulmonary embolism [92]. Impaired fibrinolysis is a feature of ischemic stroke and is present in both the acute and convalescent phases of the disease [93,94,95].
Our laboratory has utilized the S100A10-null mouse to elucidate the function of S100A10 in fibrinolysis in vivo [96]. We began these studies with human endothelial cells (TIME), in which S100A10 had been depleted by shRNA. We observed that these cells bound 50% less plasminogen and generated 60% less plasmin. We also isolated lung endothelial cells from WT and S100A10-null mice and observed that the endothelial cells from the S100A10-null mice bound 40% less plasminogen and generated 40% less plasmin. These studies suggested an important role for S100A10 in the fibrinolytic surveillance system. To test this possibility, we injected 125I-fibrinogen into the tail vein of WT and S100A10-null mice, followed by the injection of batroxobin, to initiate clot formation. We then isolated tissues, and observed that the tissues of the S100A10-null mice had a significantly greater accumulation of 125I-label than the WT mice. This suggested that S100A10-null mice had an inability to clear fibrin clots. Consistent with this study, when we examined the tissues of WT and S100A10-null mice’s tissues, we observed the presence of fibrin clots in the S100A10-null mice but not in the WT mice. This study showcased the importance of S100A10 in the endothelial cell fibrinolytic surveillance system in vivo and has broad-reaching implications for stroke, cardiovascular diseases, and cancer progression.
6. Neurological Functions
Previous reviews of S100A10 have uncovered new insight into the protein’s role in regulating mood-related behavior [97]. S100A10 has been implicated in the pathophysiology of depression and is being examined as a critical modulator of neurological functions [97]. Several G protein-coupled receptors, channels, and transporters have been identified to interact with S100A10, including serotonin 5-HT receptors [40], metabotropic glutamate receptor mGluR5 [98], Na2+ ion channel NaV1.8 [62], acid-sensing channel ASIC-1 [58], K+ channel subfamily K member 3 (TASK-1) [65,66], and transient receptor potential cation Ca2+ channels subfamily TRPV5 and TRPV6 [67]. Global proteomic screens have additionally identified C-C chemokine receptor type 10 (CCR10) [99], flottilin-1 (FLOT1) [44], follicle-stimulating hormone receptor (FSHR) [100], olfactory receptor 4N2 (OR4N2), and 4N1 (OR14l1) [43] as S100A10 interactors.
S100A10 expression is widespread in the brain and spans multiple structures and cell types. Milosevic et al. suggest that the expression is distinct to specific regions when comparing neuronal and nonneuronal cell types [101]. Some identified areas include the hippocampus [45,102], amygdala [102], cerebral cortex [103], anterior cingulate cortex [40], and nucleus accumbens [104]. The expression of S100A10 in the brain has been extensively examined for its role in modulating mood-related behaviors, including major depressive disorder, Parkinson’s disease, and other neuropsychiatric disorders [105,106].
The role of S100A10 in depression and as a regulator of the antidepressant response has been examined in various mouse and human studies. Reduced S100A10 expression in multiple brain regions in depressed individuals implicated S100A10′s role in depression pathology [40,102,107]. Epigenetic studies of S100A10 propose it as a biomarker to predict treatment response and diagnose depressive-like behavior. Expression of S100A10 can be regulated by various factors, such as antidepressants, nitric oxide, growth factors, and dexamethasone [108,109]. While S100A10 is reduced in brain tissue from depressed patients, S100A10 expression increases in the peripheral blood. One study comparing S100A10 expression levels by quantitative PCR found that, compared to healthy patients, depressed and high-stressed individuals studied had higher expression of S100A10 in their blood cells [110].
The S100A10-knock-out mouse models have been instrumental in studying the function of S100A10 in depression, mood disorders, and anxiety. In one of the first hallmark studies, Svenningson et al. showed that S100A10-depleted mice demonstrate depression-like behaviors, and S100A10 expression is increased in rodent brains after anti-depressant therapy. S100A10 mediates its function in depression via interaction with the serotonin 1B receptor [5-hydroxytrptamin (5HT1B) receptor] [40]. More recently, Seo et al. observed that S100A10 is a key protein that modulates chronic stress-induced depression in rodent models, and anti-depressants reverse this phenotype by upregulating S100A10 in the prelimbic cortex (PrL) [109]. Furthermore, they showed that S100A10 in ependymal cells regulates depressive states in chronic stress by modulating the flow of cerebrospinal fluid (CSF) by maintaining the planar cell polarity in ependymal cells [109].
7. Inflammation and Wound Healing
The plasminogen activation system and S100A10 play an integral role in the inflammatory process. S100A10 functions on the plasma membrane and in the extracellular space, correlated with the immune response and regulation of immune activities [111]. The Ca2+ binding activity of S100A proteins plays an important role during the inflammatory process by regulating different molecules and inflammatory signaling pathways that lead to inflammation [112,113]. S100 isoforms can contribute to the immune response as pro-inflammatory stimulators, chemo-attractants, and antimicrobial peptides [111].
During inflammation and wound healing processes, extracellular matrix (ECM) degradation is a vital step, and the S100A10 dimer component of Allt is a crucial molecule in the process [112]. Pg binds to the S100A10 dimer of Allt, and plasminogen activators tPA and uPA mediate its activation into Pm, which can initiate downstream proteolytic cascades associated with the wound repair process [32,34,60,114,115,116]. This S100A10-mediated protease activation is utilized by highly motile cells, such as metastatic cancer cells and macrophages, to facilitate its migration through the ECM [114,116,117,118,119,120,121,122,123].
Macrophages play a critical role in the pathogenic inflammatory response through initiating, maintaining, and resolving inflammation processes [119,124]. Cell-surface generation of plasmin is required for macrophage recruitment [125]. This recruitment is partly mediated through the plasmin-dependent activation of matrix metallopeptidase 9 (MMP-9) [119,125]. Activation of plasmin occurs through four plasminogen receptors found on the cell surface, including S100A10 [32,34,35,126], α-enolase [127], Plg-RKT [128], and histone H2B [129].
We have previously demonstrated a direct involvement of S100A10 in response to inflammatory stimuli to recruit macrophages [119]. P11 was up-regulated in macrophages that were activated by inflammatory mediators. Thioglycollate-stimulated peritoneal macrophages had higher S100A10 and annexin A2 protein levels when compared to resident peritoneal macrophages. In S100A10-/- mice, macrophage migration into the peritoneal cavity across the peritoneal membrane was decreased. S100A10 and the other carboxyl-terminal Pg receptors contribute to Pm generation in macrophages, which allows macrophages to play a role in directly facilitating the proteolysis of the basement membrane, hydrolyzing ECM proteins, and activating MMP-9 during inflammatory responses [119].
S100A10 can also activate human and murine macrophages directly through the toll-like receptor 4 (TLR4) pathway [130,131]. Cell surface Pm generated by Allt can trigger the phosphorylation of PKC signaling molecules, which can activate mitogen-activated protein kinase (MAPK), TLR4, and NFκB signaling pathways [130]. Allt disassembly after Allt phosphorylation can activate the CD11b-dependent integrin-linked kinase (ILK) pathway [132], which, together with Pm, can induce NFκB nuclear translocation and promote pro-inflammatory factor production [26,130,133]. The release of these pro-inflammatory cytokines, including IL-1, IL-6, and TNFα, can facilitate immune-escape mechanisms by evading immunosurveillance and mitigating T cell cytotoxicity [26,130,133]. Allt-driven cytokine production is inhibited by TLR-4 knockdown [130], which suggests that TLR-4 is important for Allt-mediated inflammation.
More recently, S100A10 has been examined for its role in the inflammatory condition of COVID-19 patients. The S100 family of proteins has been suggested to be able to direct more monocytes and neutrophils to the target site of COVID-19 patients by controlling the cytokine release syndrome [113]. A study comparing NK and NK-T cell subsets between COVID-19 patients and healthy individuals identified S100A10 as a marker for COVID-19-derived NK-T cells [134]. Disease trajectory transcriptomic models of COVID-19 severity from peripheral mononuclear cells found that genes encoding Ca2+-binding proteins play important roles in regulating inflammatory pathways; however, p11 was not indicated as one of the S100 proteins involved [135]. Another study looking at peripheral blood samples from COVID-19 cases observed an upregulation of S100A10, S100A4, and S100A9 mRNA [113].
8. Exocytosis and Trafficking
The AIIt complex has long been implicated at the plasma membrane to play a role in early and late secretory events and membrane-cytoskeleton lineage [47,136,137]. At the cell membrane, the AIIt complex was found localized to the contact site of the plasma membrane/secretory granule, suggesting it played a role in the exocytotic process of the cell [138,139,140,141]. While the crosslinking activity of the AIIt complex is necessary for efficient, regulated exocytosis, it is not an obligatory component [137]. The exocytotic machinery of the Allt complex is Ca2+ sensitive [142] and is more efficient than monomeric annexin protein in regulating Ca2+-dependent exocytosis [138,143,144]. These features and functions of the AIIt complex have played an important role in the pathogenesis of many viruses, such as HBV, HPV, and HIV-1 [145,146,147,148,149]. The AIIt complex has been shown to facilitate the exocytosis of von Willebrand factor (vWF) in vascular endothelial cells [150,151], the release of the bluetongue virus (BTV) in BHK21 cells [152,153], and the human papilloma virus type 16 in human keratinocytes [154]. Finally, recent studies by Bai et al. have shown that the AIIt complex promotes hepatitis B virus (HBV) exocytosis in the trophoblasts by recruiting VAMP2 and SNAP25 for membrane fusion events [145].
9. Autophagy and Metabolism
Cancer cells require increased metabolism to maintain sustained proliferative growth and spread. S100A10 can respond to a variety of signals to maintain and accelerate cancer cell metabolism. Members of the S100 family of proteins are involved in a number of metabolic functions, including redox, energy, and sugar metabolism [15]. S100A10 has been shown to increase the malignant growth of cancer cells by activating the mTOR signaling pathway in osteosarcoma [155], gastric cancer [156], pancreatic ductal adenocarcinoma (PDAC) [157,158], and HCC [159,160]. Li et al. reported that S100A10, through its interaction with annexin A2, accelerated tumor glycolysis and lactate production and contributed to the switch from oxidative phosphorylation to aerobic glycolysis [156]. Glucose consumption was significantly increased in S100A10-overespressing cells and reduced in S100A10-knockout cells. This overexpression of S100A10 also reduced the amount of intracellular ATP production, indicating the important role of S100A10 in facilitating glycolysis. By modulating the Src/annexin A2/AKT/mTOR signaling pathway, S100A10 could promote pro-tumor aerobic glycolysis, suppress cell apoptosis, and maintain cell proliferation.
S100A10 knockdown was also shown to affect the malignant growth of osteosarcoma cells by modulating glycolysis [155]. Knockdown of S100A10 in the osteosarcoma cell line inhibited proliferation, migration, and invasion and induced apoptosis via the AKT/mTOR pathway by modulating glycolysis. S100A10 played a critical role in HCC progression through the epithelial-mesenchymal transition by upregulating the epidermal growth factor receptor and AKT/ERK signaling pathways. Furthermore, Lin et al. showed that S100A10 could activate LAMB3 through the JNK pathway in PANC-1 cells [157]. By activating the JNK/LAMB3-LAMC2 axis, S100A10 was able to promote PDAC cell proliferation, migration, and adhesion.
10. Regulation of CFTR Function
The cystic fibrosis conductance regulator protein (CFTR) is a cAMP/protein kinase A (PKA) and ATP-regulated Cl- channel. The mature, wild-type CFTR protein localizes to the apical membrane, where it controls fluid and ion transport. Mutations occurring in the Cl- channel can manifest themselves in a number of disorders, most notably cystic fibrosis (CF). CFTR is surrounded by an interconnected, dynamic network of components that impact its location, trafficking, and functioning within the cell. Mutations in CFTR have previously been shown to disrupt the intracellular trafficking of the protein [161,162]. Defective endocytosis, intracellular protein trafficking, and exocytosis have also been observed in CF [161,162], suggesting that CFTR may interact with and modulate proteins within the secretory pathway.
We’ve previously discussed how AIIt has been implicated in endocytic and secretory events occurring close to or at the plasma membrane. Annexins regulate vesicular traffic [163] and are involved in the regulation of inflammation. A well-characterized annexin involved in the inflammatory response is annexin A1 [164,165]. Annexin A1 expression was down-regulated in cftr-/- mice and CF patient epithelial samples, suggesting that in CF pathogenesis, the interaction of CFTR and annexin A1 may be a key process [166]. Annexins function at the plasma membrane and regulate vesicular traffic [163], and when compared to CFTR, they share significant sequence homology around the area of the most common CF mutation [167].
Annexins can interact with cytoskeletal proteins, such as ion channels, to modulate membrane events. CFTR is regulated by cAMP/PKA [168,169]. Borthwick et al. noted that cAMP/PKA could regulate Allt in cells, which led the group to speculate about the functional relationship among those different proteins [170]. They discovered that the calcineurin (CaN)-like phosphatase, mediated via cAMP/PKA, can dephosphorylate annexin A2, which results in it forming a complex with S100A10 [170]. Pre-treatment of the cells with a specific peptide corresponding to the annexin A2 binding site on S100A10 or with a CaN inhibitor before forskolin stimulation significantly reduced CFTR function in the cells. They concluded that the Allt/PKA/calcineurin/CFTR interacting complex is required for proper channel activity and tethering to the plasma membrane [170,171].
Compared to binding motifs on other ion channels, CFTR lacks a S100A10 binding motif [172]. The authors suggest that the cAMP and Ca2+-dependent pathways control the phosphorylation and dephosphorylation of annexin A2 within the Allt complex to regulate the interaction between Allt and CFTR. Another study suggests that the interaction between S100A10 and CFTR involves the NBD1 domain of CFTR [173]. S100A10 acts as an adapter, connecting the annexin A1/cytosolic phospholipase A2 (cPLA2) complex to CFTR. Annexin A1 failed to bind to the NBD1 region of CFTR, while S100A10 showed significant binding, indicating that in vitro, through NBD1/S100A10 binding, CFTR interacts with the cPLA2/annexin A1 complex to regulate inflammation. These studies provide evidence that S100A10 associates with CFTR, and this interaction plays a role in regulating ion homeostasis in epithelial cells. This research supports their hypothesis that AIIt affects CFTR function and provides additional evidence that its activation occurs through a cAMP/PKA-dependent process.
11. Mammalian Oviduct
S100 proteins that were associated with the mammalian oviduct were incidentally discovered by Nakamura et al., who reported S100 protein immunoreactivity in glandular cells of the cervix and placenta during early pregnancy [174]. S100 proteins were found to be linked to a differentiation phenomenon, and their expression was hypothesized to correlate with a defined phase of the cell cycle [175]. The presence of S100 in free-floating cells (differentiated) compared to epithelial cells (dedifferentiated cells) cultured from bovine oviducts further supported the hypothesis that S100 was associated with defined cell cycle phases [176]. When Nakamura et al. compared S100 protein immunoreactivity in non-pregnant women, the glandular cells of the cervix and endometrium showed no immunoreactivity to S100 protein staining. No immunoreactivity was observed in endometrial carcinomas or hyperplasia, suggesting a relationship between S100 proteins and early pregnancy. The number of S100-positive glands decreased by mid to late gestation, and in term placentas were almost absent, suggesting a relationship between the expression of S100 proteins and humoral factors related to pregnancy and the mammalian oviduct [174].
Reactivity for S100 proteins has been observed in all types of epithelial cells, segments of the fallopian tube [174,175,176], lymph vessels, blood capillaries, arterial vessels, and oocytes [177]. There was no change in reactivity observed during the oestrous cycle; however, S. Agarwal did observe a difference in reactivity between fallopian tubes that were removed for sterilization compared to ones removed for ectopic pregnancies, with the latter showing strong S100 reactivity [178].
The majority of studies examining S100A10 expression in the oviduct have been performed in non-human models. S100A10 is expressed at negligible levels in the mouse oviduct compared to S100A11, while both are found at substantial levels in the ovary [179]. PCR analysis of S100A10 in the mouse oviduct showed significantly lower expressions of S100A10 in the oviduct compared to the ovary, which suggests it is not the most prominent S100 protein in the oviduct. Similarly, Tingaud–Sequeira et al. reported significantly up-regulated S100A10 expression in the atretic ovaries and follicles of Senagalese sole fish [180]. The presence of S100A10 and annexin A2 has additionally been observed in the oviducts of rabbits, dogs, cats, cows, pigs, and humans [181].
12. Stemness and Cancer Pathogenesis
Tumors contain a subpopulation of cells, termed cancer stem cells (CSCs), that possess indefinite proliferative potential. CSCs possess embryonic characteristics and are drivers of tumor development [182,183]. Mechanistically, S100A10 has been examined to regulate expression levels of stem-cell-related genes. S100A10 was first suggested to confer stem-cell activity regulation by King et al., where they demonstrated that annexin A2 and S100A10 expression in Xenopus laevis larvae after limb amputation were up-regulated [184]. The expression of S100A10 was 4-fold higher in the regeneration-competent blastema compared to the time of amputation, in a pattern that was distinct from other immune-related genes [184]. This was the first evidence of S100A10 stemness-related activity.
CSCs have been examined for their role in tumor recurrence and metastasis. One of the reasons that chemotherapy resistance arises is the inability to eliminate the tumor stem cells completely. Alterations in mRNA and protein expression of the S100 protein family in various cancers participate in regulating drug resistance and conferring CSC properties [185]. S100A10 upregulation after chemotherapy treatment has been observed in colorectal cancer [186,187], leukemia [188], ovarian cancer [189,190], breast cancer [72,191], and neuroblastoma [192]. S100A10 can confer metastatic potential through mechanisms such as an upregulation of cellular invasion and CSC properties [191]. In addition to CSCs, S100A10 stimulation was also found to enhance bone marrow-derived stem cell osteogenesis [193].
S100A10 expression has been associated with the hallmarks of cancer in several cancers, such as the brain, breast, lung, colorectal, kidney, ovarian, acute lymphoblastic leukemia, and pancreatic [194]. Our laboratory has contributed to understanding the function of S100A10 in the pathogenesis of pro-myelocytic leukemia (PML), pancreatic cancer, and breast cancer [195,196]. Since S100A10 is a multi-functional protein, it plays both plasminogen-dependent and independent roles in the progression of cancer, depending on the cellular context. For example, S100A10 promotes hepatocellular carcinoma, gastric cancer, and osteosarcoma cell proliferation and suppresses apoptosis via enhanced aerobic glycolysis through mTOR signaling [155,156,159,197]. In acute promyelocytic leukemia (APL), we have shown that cell surface S100A10 expression in APL cells is responsible for the increased fibrinolytic events associated with APL patients [71]. Induction of PML-RAR-α oncoprotein resulted in increased S100A10 expression on the cell surface, with a subsequent increase in plasmin generation. In the MMTV-PyMT mouse mammary cancer model, loss of S100A10 resulted in dramatic impairment of tumor progression and pulmonary metastasis, but surprisingly, this was not mediated through loss of plasmin generation, as we did not observe a marked decrease in plasmin generation in the tumors lacking S100A10 [195]. Our previous studies using ectopic subcutaneous tumor models have suggested that plasmin generation and tumor cell infiltration by macrophages are substantially impaired in mice lacking S100A10, which substantially impedes tumor growth [119].
In hepatocellular carcinoma, S100A10 plays a pivotal role in tumor initiation, self-renewal capacity, chemoresistance, and metastasis, as validated in vivo and in experimental animal models. This study showed that S100A10 mediates these effects through two mechanisms. First, it enhanced metastasis by upregulating the expression of mesenchymal markers such as vimentin, fibronectin, and N-cadherin and promoting epithelial-mesenchymal transition. Second, the cancer cells also secreted S100A10 in the extracellular vesicles (EVs), which promoted motility, invasion, and metastasis. S100A10 potentially regulated the transfer of matrix metalloprotease 2 (MMP2), fibronectin, and epidermal growth factor (EGF) proteins into the EVs, thus indirectly regulating invasion and metastasis [159].
S100A10 is one of the 11 signature genes whose expression correlates with multidrug resistance in ovarian serous carcinoma [198]. Several other studies have also published similar observations in ovarian cancer, where increased S100A10 expression is associated with poor response to chemotherapy and overall survival [188,199]. The precise molecular mechanism by which S100A10 regulates drug resistance has yet to be discovered. One potential explanation is the role S100A10 plays in promoting cancer cell stemness, which has implications for promoting chemoresistance [72]. Nevertheless, elucidating the mechanism of S100A10 in therapy resistance is an interesting and necessary avenue for future research on S100A10.

Table 3.
Suggested interactors of p11.
Table 3.
Suggested interactors of p11.
| Interactor | Protein Name | Uniprot Accession | Cellular Location | Method | References |
|---|---|---|---|---|---|
| 5-HTR1B | 5-hydroxytryptamine receptor 1B | P28222 | Plasma membrane | Two-hybrid | [40,41] |
| 5-HTR1D | 5-hydroxytryptamine receptor 1D | P28221 | Plasma membrane | Two-hybrid | [41] |
| 5-HTR4 | 5-hydroxytryptamine receptor 4 | Q13639 | Plasma membrane | Two-hybrid | [41] |
| ABCE1 | ATP-binding cassette sub-family E member 1 | P61221 | Cytoplasm | AC-MS 1 | [200] |
| AHNAK | Neuroblast differentiation-associated protein AHNAK | Q09666 | Nucleus | AC-MS 1; AC-W 2; Co-crystal structure; Co-fractionation; PL-MS 3 | [42,43,44,45] |
| ANG | Angiogenin | P03950 | Extracellular; Nucleus | AC-W 2; Co-IP 4; Fluorescence imaging; Proximity ligation | [201] |
| ANLN | Anillin | Q9NQW6 | Nucleus; Cytoplasm | AC-MS 1 | [202] |
| ANTXR1 | Anthrax toxin receptor 1 | Q9H6X2 | Plasma membrane | AC-MS 1 | [203] |
| ANXA2 | Annexin A2 | P07355 | Nucleus; Cytoplasm; Plasma membrane; Extracellular | AC-MS 1; AC-W 2; Co-crystal structure; Co-fractionation; PL-MS 3; Reconstituted complex; Two-hybrid | [43,45,46,47,48,49,50,51,52,53,54,55,56,57] |
| ASIC-1 channels | Acid-sensing ion channel 1 | P78348 | Plasma membrane | Two hybrid | [58] |
| ATF2 | Cyclic AMP-dependent transcription factor ATF-2 | P15336 | Nucleus; Cytoplasm | AC-MS 1 | [204] |
| ATP6V1E1 | V-type proton ATPase subunit E | P36543 | Cytoplasm; Plasma membrane | Co-fractionation | [44] |
| BAD | BCL2-associated agonist of cell death | Q92934 | Cytoplasm | Reconstituted complex; Two-hybrid | [59] |
| BAP1 | Ubiquitin carboxyl-terminal hydrolase BAP1 | Q92560 | Nucleus | AC-MS 1 | [205] |
| BMI1 | Polycomb complex protein BMI-1 | P35226 | Nucleus; Cytoplasm | Co-IP 4 | [206] |
| Cathepsin B | Cathepsin B | P07858 | Cytoplasm; Plasma membrane; Extracellular | Two hybrid; Reconstituted complex | [60] |
| CCDC171 | Coiled-coil domain-containing protein 171 | Q6TFL3 | Nucleus | PL-MS 3 | [43] |
| CCNF | Cyclin-F | P41002 | Nucleus | AC-MS 1 | [207] |
| CCR10 | C-C chemokine receptor type 10 | P46092 | Plasma membrane | Proximity ligation; Pull-down | [99] |
| CD55 | Complement decay-accelerating factor | P08174 | Plasma membrane; Extracellular | Co-fractionation | [44] |
| CD81 | CD81 antigen | P60033 | Plasma membrane; Cytoplasm | Co-IP 4 | [208] |
| CDC25C | M-phase inducer phosphatase 3 | P30307 | Nucleus; Cytoplasm | AC-MS 1 | [209] |
| CDK16 | Cyclin-dependent kinase 16 | Q00536 | Cytoplasm | Two-hybrid | [210] |
| CDK9 | Cyclin-dependent kinase 9 | P50750 | Nucleus; Cytoplasm | AC-MS 1 | [211] |
| CEP192 | Centrosomal protein of 192 kDa | Q8TEP8 | Cytoplasm | AC-MS 1 | [212] |
| CFTR | Cystic fibrosis transmembrane conductance regulator | P13569 | Plasma membrane; Cytoplasm | Surface plasmon resonance; Pull-down | [170,173,213] |
| CHMP4B | Charged multivesicular body protein 4b | Q9H444 | Cytoplasm | AC-MS 1 | [202] |
| CHMP4C | Charged multivesicular body protein 4c | Q96CF2 | Cytoplasm | AC-MS 1 | [202] |
| CIT | Citron Rho-interacting kinase | O14578 | Cytoplasm | AC-MS 1 | [202] |
| COPS6 | COP9 signalosome complex subunit 6 | Q7L5N1 | Cytoplasm; Nucleus | Two-hybrid | [214] |
| Cytosolic Phospholipase A2 | Q9UP65 | Cytoplasm; Plasma membrane | Two-hybrid | [61] | |
| DLC1 | Rho GTPase-activating protein 7 | Q96QB1 | Cytoplasm | AC-W 2 | [215] |
| DNAAF10 | Dynein axonemal assembly factor 10 | Q96MX6 | Cytoplasm; Nucleus | Co-fractionation | [44] |
| DUSP19 | Dual specificity protein phosphatase 19 | Q8WTR2 | Cytoplasm | AC-MS 1 | [209] |
| DYNLT1 | Dynein light chain Tctex-type 1 | P63172 | Cytoplasm | PL-MS 3 | [216] |
| DYRK1A | Dual specificity tyrosine-phosphorylation-regulated kinase 1A | Q13627 | Nucleus | AC-MS 1 | [217] |
| ECT2 | Protein ECT2 | Q9H8V3 | Nucleus; Cytoplasm | AC-MS 1 | [202] |
| EFTUD2 | 116 kDa U5 small nuclear ribonucleoprotein component | Q15029 | Nucleus | AC-MS 1 | [218] |
| EGFR | Epidermal growth factor receptor | P00533 | Plasma membrane; Cytoplasm | AC-MS 1 | [219] |
| ELAVL1 | ELAV-like protein | Q15717 | Cytoplasm; Nucleus | Co-fractionation | [44] |
| ELMOD1 | ELMO domain-containing protein 1 | Q8N336 | Nucleus | PL-MS 3 | [43] |
| ERBB3 | Receptor tyrosine-protein kinase erbB-3 | P21860 | Plasma membrane; Extracellular | Two-hybrid; AC-MS 1 | [220,221] |
| ERBB4 | Receptor tyrosine-protein kinase erbB-4 | Q15303 | Plasma membrane; Cytoplasm | Two-hybrid | [220] |
| ESR1 | Estrogen receptor | P03372 | Nucleus; Cytoplasm; Plasma membrane | AC-MS1 | [222] |
| FAF2 | FAS-associated factor 2 | Q96CS3 | Cytoplasm | Co-fractionation | [44] |
| FBL | rRNA 2′-O-methyltransferase fibrillarin | P22087 | Nucleus | AC-MS 1 | [212] |
| FLNA | Filamin-A | P21333 | Cytoplasm | AC-MS 1 | [50] |
| FLOT1 | Flotillin-1 | O75955 | Plasma membrane | Co-fractionation | [44] |
| FSHR | Follicle-stimulating hormone receptor | P23945 | Plasma membrane | Two-hybrid | [100] |
| GBP3 | Guanylate-binding protein 3 | Q9H0R5 | Cytoplasm | PL-MS 3 | [43] |
| GDNF | Glial cell line-derived neurotrophic factor | P39905 | Extracellular | AC-MS 1 | [51] |
| GOLGA4 | Golgin subfamily A member 4 | Q13439 | Cytoplasm | PL-MS 3 | [43] |
| HDAC4 | Histone deacetylase 4 | P56524 | Cytoplasm; Nucleus | AC-MS 1 | [223] |
| HDAC6 | Histone deacetylase 6 | Q9UBN7 | Cytoplasm; Nucleus | AC-MS 1 | [224] |
| HDLBP | Vigilin | Q00341 | Cytoplasm; Nucleus | Co-fractionation; AC-MS 1 | [44,225] |
| Heparin | Heparin cofactor 2 | P05546 | Cytoplasm; Extracellular | Pull-down | [226] |
| HLTF | Helicase-like transcription factor | Q14527 | Nucleus; Cytoplasm | Co-fractionation; AC-MS 1; AC-W 2 | [44,45] |
| HMGN3 | High motility group nucleosome-binding domain-containing protein 3 | Q15651 | Nucleus | PL-MS 3 | [43] |
| HSPA8 | Heat shock cognate 71 kDa protein | P11142 | Cytoplasm; Nucleus; Plasma membrane | AC-MS 1 | [227] |
| KCNK3 | Potassium channel subfamily K member 3 | O14649 | Plasma membrane | Reconstituted complex; Two-hybrid | [65] |
| KIAA1429 | Protein virilizer homolog | Q69YN4 | Nucleus; Cytoplasm | AC-MS 1 | [228] |
| KIF14 | Kinesin-like protein KIF14 | Q15058 | Cytoplasm; Nucleus | AC-MS 1 | [202] |
| KIF20A | Kinesin-like protein KIF20A | O95235 | Cytoplasm; Nucleus | AC-MS 1 | [202] |
| LIMA1 | LIM domain and actin-binding protein 1 | Q9UHB6 | Cytoplasm | AC-MS 1 | [50] |
| LMX1B | LIM homeobox transcription factor 1β | O60663 | Nucleus | AC-MS 1 | [53] |
| LUC7L3 | Luc7-like protein 3 | O95232 | Nucleus | AC-MS 1 | [43] |
| MAEA | E3 ubiquitin-protein transferase MAEA | Q7L5Y9 | Cytoplasm; Nucleus | PL-MS 3 | [43] |
| MAPT | Microtubule-associated protein tau | P10636 | Cytoplasm | Two-hybrid | [229] |
| mGluR5 | Metabotropic glutamate receptor 5 | P41594 | Plasma membrane | AC-W 2 | [98] |
| MLKL | Mixed lineage kinase domain-like protein | Q8NB16 | Plasma membrane; Cytoplasm | Two-hybrid | [230] |
| MMGT1 | ER membrane protein complex subunit 5 | Q8N4V1 | Cytoplasm | AC-MS 1 | [231] |
| MYC | MYC proto-oncogene protein | P01106 | Nucleus | AC-MS 1 | [232] |
| MYH9 | Myosin-9 | P35579 | Cytoplasm | AC-MS 1 | [50] |
| Myo1c | Unconventional myosin-1c | O00159 | Cytoplasm; Nucleus | AC-MS 1 | [50] |
| NANOS2 | Nanos homolog 2 | P60321 | Cytoplasm | Two-hybrid | [230] |
| NARS1 | Asparagine—tRNA ligase, cytoplasmic | O43776 | Cytoplasm | PL-MS 3 | [43] |
| NaV1.8 | Sodium channel protein type 8 subunit α | Q9UQD0 | Plasma membrane | Two-hybrid | [62] |
| NBR1 | Next to BRCA1 gene protein | Q14596 | Cytoplasm | PL-MS 3 | [233] |
| NEB | Nebulin | P20929 | Cytoplasm | PL-MS 3 | [43] |
| NPM1 | Nucleophosmin | P06748 | Nucleus; Cytoplasm | AC-MS 1 | [234] |
| NUDCD1 | NudC domain-containing protein 1 | Q96RS6 | Nucleus; Cytoplasm | Co-IP 4 | [235] |
| OR14I1 | Olfactory receptor 14I1 | A6ND48 | Plasma membrane | PL-MS 3 | [43] |
| OR4N2 | Olfactory receptor 4N2 | Q8NGD1 | Plasma membrane | PL-MS 3 | [43] |
| PCTAIRE-1 | Cyclin-dependent kinase 16 | Q00536 | Cytoplasm | Two-hybrid | [63] |
| PDYN | Proenkephalin-B | P01213 | Extracellular | AC-MS 1 | [50] |
| PHF5A | PHD finger-like domain-containing protein 5A | Q7RTV0 | Nucleus | Co-fractionation | [44] |
| PIK3R6 | Phosphoinositide 3-kinase regulatory subunit 6 | Q5UE93 | Cytoplasm; Plasma membrane | PL-MS 3 | [53] |
| PLA2G4A | Cytosolic phospholipase A2 | P47712 | Cytoplasm; Nucleus | AC-W 2; Reconstituted complex | [46,55,236] |
| PLA2G4C | Cytosolic phospholipase A2γ | Q9UP65 | Cytoplasm | Reconstituted complex | [61] |
| PLA2R | Urokinase plasminogen activator surface receptor | Q03405 | Plasma membrane | AC-MS 1 | [237] |
| PLD1 | Phospholipase D1 | Q13393 | Cytoplasm | PL-MS 3 | [43] |
| Plg | Plasminogen | P00747 | Extracellular | AC-W 2; Surface plasmon resonance | [32,33] |
| PPFIA3 | Liprin-α-3 | O75145 | Plasma membrane; Cytoplasm | PL-MS 3 | [43] |
| PPIF | Peptidyl-prolyl cis-trans isomerase F | P30405 | Cytoplasm | Co-fractionation | [238] |
| PRC1 | Protein regulator of cytokinesis 1 | O43663 | Nucleus; Cytoplasm | AC-MS 1 | [202] |
| RAF1 | RAF proto-oncogene serine/threonine protein kinase | P04049 | Cytoplasm; Plasma membrane; Cytoplasm | AC-MS 1 | [225] |
| RPL10A | 60S ribosomal protein L10a | P62906 | Cytoplasm | Co-fractionation | [44] |
| RPL10L | 60S ribosomal protein L10-like | Q96L21 | Cytoplasm | Co-fractionation | [44] |
| RPL12 | 60S ribosomal protein L12 | P30050 | Nucleus; Cytoplasm | Co-fractionation | [44] |
| RYBP | RING1 and YY1-binring protein | Q8N488 | Nucleus; Cytoplasm | Co-IP 4 | [206] |
| S100A10 | Protein S100-A10 | P60903 | Plasma membrane; Cytoplasm; Nucleus; Extracellular | AC-MS 1; Co-crystal structure | [45,54] |
| S100A3 | Protein S100-A3 | P33764 | Cytoplasm | Two-hybrid | [230] |
| S100A7 | Protein S100-A7 | P31151 | Cytoplasm; Extracellular | AC-MS 1 | [64] |
| S100A8 | Protein S100-A8 | P05109 | Plasma membrane; Cytoplasm; Extracellular | AC-MS 1 | [64] |
| S100Z | Protein S100-Z | Q8WXG8 | Cytoplasm | Two-hybrid | [230] |
| SETDB1 | Histone-lysine N-methyltransferase SETDB1 | Q15047 | Nucleus | Two-hybrid | [214] |
| SIN3A | Paired amphipathic helix protein Sin3A | Q96ST3 | Nucleus; Cytoplasm | Co-fractionation | [44] |
| SLC25A46 | Mitochondrial outer membrane protein | Q96AG3 | Cytoplasm | PL-MS 3 | [43] |
| SLC8A1 | Sodium/calcium exchanger 1 | P32418 | Plasma membrane | Florescence polarization spectroscopy | [239] |
| SLFN14 | Protein SLFN14 | P0C7P3 | Nucleus | PL-MS 3 | [43] |
| Smarca3 | Helicase-like transcription factor | Q14527 | Nucleus; Cytoplasm | Co-crystal structure | [45] |
| SRGAP3 | SLIT-ROBO Rho GTPase-activating protein 3 | O43295 | Cytoplasm | PL-MS 3 | [43] |
| SRP9 | Signal recognition particle 9 kDa protein | P49458 | Cytoplasm | Co-fractionation | [44] |
| SRPRB | Signal recognition particle receptor subunit β | Q9Y5M8 | Cytoplasm | Co-fractionation | [44] |
| SUMO2 | Small ubiquitin-related modifier 2 | P61956 | Nucleus; Cytoplasm | Reconstituted complex | [240] |
| SUPT6H | Transcription elongation factor SPT6 | Q7KZ85 | Nucleus; Cytoplasm | AC-MS 1; AC-W 2 | [45] |
| SWI5 | DNA repair protein SWI homolog | Q1ZZU3 | Nucleus | PL-MS 3 | [43] |
| TAB1 | TGF-β-activated kinase 1 and MAP3K7-binding protein 1 | Q15750 | Cytoplasm | AC-MS 1 | [53] |
| TASK-1 | Potassium channel subfamily K member 3 | O14649 | Plasma membrane | Two-hybrid | [65,66] |
| TCEAL5 | Transcription elongation factor A protein-like 5 | Q5H9L2 | Nucleus | AC-MS 1 | [53] |
| TME65 | Transmembrane protein 65 | Q6PI78 | Plasma membrane | Co-fractionation | [44] |
| TNF | Tumor necrosis factor | P01375 | Plasma membrane; Extracellular | AC-MS 1 | [241] |
| TNFRSF10A | Tumor necrosis factor receptor superfamily member 10A | O00220 | Plasma membrane | AC-MS1; Co-IP 4 | [52] |
| TP53 | Cellular tumor antigen p53 | P04637 | Nucleus; Cytoplasm | Florescence polarization spectroscopy | [239] |
| Transglutaminase | Protein-glutamine-γ-glutamyltransferase K & 2 | P22735; P08587 | Plasma membrane; Cytoplasm | AC-W 2 | [68] |
| TRIM37 | E3 ubiquitin-protein ligase TRIM37 | O94972 | Cytoplasm | AC-MS 1 | [242] |
| TRIM67 | Tripartite motif-containing protein 67 | Q6ZTA4 | Cytoplasm | AC-MS 1 | [243] |
| TRPM4 | Transient receptor potential cation channel subfamily M member 4 | Q8TD43 | Plasma membrane | Florescence polarization spectroscopy | [239] |
| TRPV5 | Transient receptor potential cation channel subfamily V member 5 | Q9NQA5 | Plasma membrane | Reconstituted complex; Two-hybrid | [67] |
| TRPV6 | Transient receptor potential cation channel subfamily V member 6 | Q9H1D0 | Plasma membrane | Reconstituted complex | [67] |
| TTLL13 | Tubulin polyglutamylase TTLL13 | A6NNM8 | Cytoplasm | PL-MS 3 | [43] |
| UBAP2 | Ubiquitin-associated protein 2 | Q5T6F2 | Cytoplasm; Nucleus | Co-fractionation | [44] |
| ULK1 | Serine/threonine-protein kinase ULK1 | O75385 | Cytoplasm | Co-IP 4 | [244] |
| UQCRB | Cytochrome b-c1 complex subunit 7 | P14927 | Cytoplasm | AC-MS 1 | [52,53] |
| USP2 | Ubiquitin carboxyl-terminal hydrolase 2 | O75604 | Cytoplasm | Two-hybrid | [230] |
| VIRMA | Protein virilizer homolog | Q69YN4 | Nucleus; Cytoplasm | AC-MS 1 | [228] |
| WDR92 | Dynein axonemal assembly factor 10 | Q96MX6 | Nucleus | Co-fractionation | [44] |
| ZCCHC9 | Zinc finger CCHC domain-containing protein 9 | Q8N567 | Nucleus | AC-MS 1 | [53] |
| ZFR | Zinc finger RNA-binding protein | Q96KR1 | Nucleus | Co-fractionation | [44] |
| ZGPAT | Zinc finger CCCH-type with G patch domain-containing protein | Q8N5A5 | Nucleus | Two-hybrid | [230] |
| ZMPSTE24 | CAAX prenyl protease 1 homolog | O75844 | Cytoplasm | Co-fractionation | [44] |
| ZNF428 | Zinc finger protein 428 | Q96B54 | Nucleus | AC-MS 1 | [53] |
| ZRANB1 | Ubiquitin thioesterase ZRANB1 | Q9UGI0 | Cytoplasm; Nucleus | AC-MS 1 | [245] |
1—Affinity Capture—Mass Spectrometry, 2—Affinity Capture—Western Blot, 3—Proximity Label—Mass Spectrometry, 4—Anti bait Co-Immunoprecipitation.
Depletion of S100A10 in pancreatic cancer cell lines resulted in decreased cell surface plasminogen activation and an overall reduction in cell invasiveness and tumor growth [157,196]. Bydoun et al. established that S100A10 expression was driven by promoter methylation and oncogenic RAS in pancreatic cancer. Recent studies by Lin et al. have suggested that S100A10 positively modulates pancreatic cancer cell proliferation, migration, adhesion, and in vivo tumor growth by activating laminin subunit β3 (LAMB3) via the JNK pathway. Thus, two different mechanisms are activated by S100A10 in pancreatic cancer, with the overall goal of accelerating tumor progression.
13. Summary and Conclusions
This review highlights the recent advances in the structure, regulation, binding partners, interactions, and functions of S100A10. As a member of the S100 family of Ca2+-binding proteins, S100A10 is unique in its permanent active conformation. Our work has highlighted just a few of the many important cellular functions of S100A10. Other laboratories have extended our studies and also added to the repertoire of additional regulatory functions of S100A10. The most noteworthy of these studies is the role of S100A10 as a prognostic biomarker in cancer and depression. Especially interesting is the revelation that S100A10 mRNA levels may predict the severity of COVID-19 infection. On a final note, one of the most unexplored potential functions of S100A10 is its role in the brain fibrinolytic surveillance system. This has major implications in terms of a physiological role for S100A10 in mitigating the deleterious effects of stroke and as a potential therapeutic agent for augmenting tPA-dependent thrombolytic therapy.
Author Contributions
G.C.O., A.G.B. and D.M.W. wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research is supported by a grant from CIHR.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available at NCBI “https://www.ncbi.nlm.nih.gov/ (accessed on 3 July 2023)”, BioGRID “https://thebiogrid.org/ (accessed on 4 July 2023)”, and ELIXIR “https://www.ebi.ac.uk/intact/home (accessed on 5 July 2023)”.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ringer, S.; Buxton, D.W. Concerning the Action of Calcium, Potassium, and Sodium Salts upon the Eel’s Heart and upon the Skeletal Muscles of the Frog. J. Physiol. 1887, 8, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Heilbrunn, L.V.; Wiercinski, F.J. The Action of Various Cations on Muscle Protoplasm. J. Cell. Comp. Physiol. 1947, 29, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K. Myosin and Adenosinetriphosphatase. Biochem. J. 1942, 36, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Bozler, E. Relaxation in Extracted Muscle Fibers. J. Gen. Physiol. 1954, 38, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Ebashi, S. The Role of “Relaxing Factor” in Contraction-Relaxation Cycle of Muscle. Prog. Theor. Phys. Suppl. 1961, 17, 35–40. [Google Scholar] [CrossRef]
- Ebashi, S.; Kodama, A. Interaction of Troponin with F-Actin in the Presence of Tropomyosin. J. Biochem. 1966, 59, 425–426. [Google Scholar] [CrossRef]
- Kretsinger, R.H.; Nockolds, C.E. Carp Muscle Calcium-Binding Protein. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar] [CrossRef]
- Henikoff, S.; Greene, E.A.; Pietrokovski, S.; Bork, P.; Attwood, T.K.; Hood, L. Gene Families: The Taxonomy of Protein Paralogs and Chimeras. Science 1997, 278, 609–614. [Google Scholar] [CrossRef]
- Kawasaki, H.; Nakayama, S.; Kretsinger, R.H. Classification and Evolution of EF-Hand Proteins. BioMetals 1998, 11, 277–295. [Google Scholar] [CrossRef]
- Mäler, L.; Sastry, M.; Chazin, W.J. A Structural Basis for S100 Protein Specificity Derived from Comparative Analysis of Apo and Ca2+-Calcyclin. J. Mol. Biol. 2002, 317, 279–290. [Google Scholar] [CrossRef]
- Carafoli, E. Calcium Signaling: A Tale for All Seasons. Proc. Natl. Acad. Sci. USA 2002, 99, 1115–1122. [Google Scholar] [CrossRef]
- Yatime, L.; Betzer, C.; Jensen, R.K.; Mortensen, S.; Jensen, P.H.; Andersen, G.R. The Structure of the RAGE:S100A6 Complex Reveals a Unique Mode of Homodimerization for S100 Proteins. Structure 2016, 24, 2043–2052. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.W. A Soluble Protein Characteristic of the Nervous System. Biochem. Biophys. Res. Commun. 1965, 19, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Spratt, D.E.; Barber, K.R.; Marlatt, N.M.; Ngo, V.; Macklin, J.A.; Xiao, Y.; Konermann, L.; Duennwald, M.L.; Shaw, G.S. A Subset of Calcium-binding S100 Proteins Show Preferential Heterodimerization. FEBS J. 2019, 286, 1859–1876. [Google Scholar] [CrossRef] [PubMed]
- Donato, R.; Cannon, B.R.; Sorci, G.; Riuzzi, F.; Hsu, K.; Weber, D.J.; Geczy, C.L. Functions of S100 Proteins. Curr. Mol. Med. 2013, 13, 24–57. [Google Scholar] [CrossRef]
- Donato, R. S100: A Multigenic Family of Calcium-Modulated Proteins of the EF-Hand Type with Intracellular and Extracellular Functional Roles. Int. J. Biochem. Cell Biol. 2001, 33, 637–668. [Google Scholar] [CrossRef]
- Rintala-Dempsey, A.C.; Rezvanpour, A.; Shaw, G.S. S100-Annexin Complexes—Structural Insights: S100-Annexin Complexes—Structural Insights. FEBS J. 2008, 275, 4956–4966. [Google Scholar] [CrossRef]
- Leśniak, W.; Filipek, A. S100A6 Protein—Expression and Function in Norm and Pathology. Int. J. Mol. Sci. 2023, 24, 1341. [Google Scholar] [CrossRef]
- Heizmann, C.W. S100 Proteins Structure Functions and Pathology. Front. Biosci. 2002, 7, d1356–d1368. [Google Scholar] [CrossRef]
- Geisow, M.J. Common Domain Structure of Ca2+ and Lipid-Binding Proteins. FEBS Lett. 1986, 203, 99–103. [Google Scholar] [CrossRef]
- Rosengarth, A.; Luecke, H. Annexins: Calcium Binding Proteins with Unusual Binding Sites. In Handbook of Metalloproteins; Messerschmidt, A., Huber, R., Poulas, T., Wieghardt, K., Cygler, M., Bode, W., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; p. met052. ISBN 978-0-470-86981-9. [Google Scholar]
- Oliferenko, S.; Paiha, K.; Harder, T.; Gerke, V.; Schwärzler, C.; Schwarz, H.; Beug, H.; Günthert, U.; Huber, L.A. Analysis of CD44-Containing Lipid Rafts: Recruitment of Annexin II and Stabilization by the Actin Cytoskeleton. J. Cell Biol. 1999, 146, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Kassam, G.; Manro, A.; Braat, C.E.; Louie, P.; Fitzpatrick, S.L.; Waisman, D.M. Characterization of the Heparin Binding Properties of Annexin II Tetramer. J. Biol. Chem. 1997, 272, 15093–15100. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, S.L.; Kassam, G.; Manro, A.; Braat, C.E.; Louie, P.; Waisman, D.M. Fucoidan-Dependent Conformational Changes in Annexin II Tetramer. Biochemistry 2000, 39, 2140–2148. [Google Scholar] [CrossRef] [PubMed]
- Filipenko, N.R.; MacLeod, T.J.; Yoon, C.-S.; Waisman, D.M. Annexin A2 Is a Novel RNA-Binding Protein. J. Biol. Chem. 2004, 279, 8723–8731. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, A.; Bydoun, M.; Holloway, R.; Waisman, D. Annexin A2 Heterotetramer: Structure and Function. Int. J. Mol. Sci. 2013, 14, 6259–6305. [Google Scholar] [CrossRef]
- Bharadwaj, A.; Kempster, E.; Waisman, D.M. The Annexin A2/S100A10 Complex: The Mutualistic Symbiosis of Two Distinct Proteins. Biomolecules 2021, 11, 1849. [Google Scholar] [CrossRef]
- Gokhale, N.A.; Abraham, A.; Digman, M.A.; Gratton, E.; Cho, W. Phosphoinositide Specificity of and Mechanism of Lipid Domain Formation by Annexin A2-P11 Heterotetramer. J. Biol. Chem. 2005, 280, 42831–42840. [Google Scholar] [CrossRef]
- Filipenko, N.R.; Waisman, D.M. The C Terminus of Annexin II Mediates Binding to F-Actin. J. Biol. Chem. 2001, 276, 5310–5315. [Google Scholar] [CrossRef]
- Lindsay, S.; Bartolotti, L.; Li, Y. Ca2+ Ions Facilitate the Organization of the Annexin A2/S100A10 Heterotetramer. Proteins 2023, 91, 1042–1053. [Google Scholar] [CrossRef]
- Filipenko, N.R.; Waisman, D.M. Annexin II: Analysis of a Pleiotropic Protein. In Annexins; Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2003; pp. 127–156. ISBN 978-1-4613-4841-2. [Google Scholar]
- Kassam, G.; Le, B.-H.; Choi, K.-S.; Kang, H.-M.; Fitzpatrick, S.L.; Louie, P.; Waisman, D.M. The P11 Subunit of the Annexin II Tetramer Plays a Key Role in the Stimulation of T-PA-Dependent Plasminogen Activation. Biochemistry 1998, 37, 16958–16966. [Google Scholar] [CrossRef]
- Fogg, D.K.; Bridges, D.E.; Cheung, K.K.-T.; Kassam, G.; Filipenko, N.R.; Choi, K.-S.; Fitzpatrick, S.L.; Nesheim, M.; Waisman, D.M. The P11 Subunit of Annexin II Heterotetramer Is Regulated by Basic Carboxypeptidase. Biochemistry 2002, 41, 4953–4961. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, T.J.; Kwon, M.; Filipenko, N.R.; Waisman, D.M. Phospholipid-Associated Annexin A2-S100A10 Heterotetramer and Its Subunits: Characterization of The Interaction with Tissue Plasminogen Activator, Plasminogen, and Plasmin. J. Biol. Chem. 2003, 278, 25577–25584. [Google Scholar] [CrossRef]
- Miller, V.A.; Madureira, P.A.; Kamaludin, A.A.; Komar, J.; Sharma, V.; Sahni, G.; Thelwell, C.; Longstaff, C.; Waisman, D.M. Mechanism of Plasmin Generation by S100A10. Thromb. Haemost. 2017, 117, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, A.G.; Kempster, E.; Waisman, D.M. The ANXA2/S100A10 Complex—Regulation of the Oncogenic Plasminogen Receptor. Biomolecules 2021, 11, 1772. [Google Scholar] [CrossRef] [PubMed]
- Puisieux, A.; Ji, J.; Ozturk, M. Annexin II Up-Regulates Cellular Levels of P11 Protein by a Post-Translational Mechanism. Biochem. J. 1996, 313, 51–55. [Google Scholar] [CrossRef]
- Goch, G.; Vdovenko, S.; Kozłowska, H.; Bierzyñski, A. Affinity of S100A1 Protein for Calcium Increases Dramatically upon Glutathionylation: S100A1 Affinity for Calcium. FEBS J. 2005, 272, 2557–2565. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Svenningsson, P.; Chergui, K.; Rachleff, I.; Flajolet, M.; Zhang, X.; Yacoubi, M.E.; Vaugeois, J.-M.; Nomikos, G.G.; Greengard, P. Alterations in 5-HT1B Receptor Function by P11 in Depression-Like States. Science 2006, 311, 77–80. [Google Scholar] [CrossRef]
- Warner-Schmidt, J.L.; Flajolet, M.; Maller, A.; Chen, E.Y.; Qi, H.; Svenningsson, P.; Greengard, P. Role of P11 in Cellular and Behavioral Effects of 5-HT4 Receptor Stimulation. J. Neurosci. 2009, 29, 1937–1946. [Google Scholar] [CrossRef]
- Benaud, C.; Gentil, B.J.; Assard, N.; Court, M.; Garin, J.; Delphin, C.; Baudier, J. AHNAK Interaction with the Annexin 2/S100A10 Complex Regulates Cell Membrane Cytoarchitecture. J. Cell Biol. 2004, 164, 133–144. [Google Scholar] [CrossRef]
- Fasci, D.; van Ingen, H.; Scheltema, R.A.; Heck, A.J.R. Histone Interaction Landscapes Visualized by Crosslinking Mass Spectrometry in Intact Cell Nuclei. Mol. Cell Proteom. 2018, 17, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. A Census of Human Soluble Protein Complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.-S.; Gao, P.; Lee, K.-W.; Ceglia, I.; Seo, J.-S.; Zhang, X.; Ahn, J.-H.; Chait, B.T.; Patel, D.J.; Kim, Y.; et al. SMARCA3, a Chromatin-Remodeling Factor, Is Required for P11-Dependent Antidepressant Action. Cell 2013, 152, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Bailleux, A.; Wendum, D.; Audubert, F.; Jouniaux, A.-M.; Koumanov, K.; Trugnan, G.; Masliah, J. Cytosolic Phospholipase A2-P11 Interaction Controls Arachidonic Acid Release as a Function of Epithelial Cell Confluence. Biochem. J. 2004, 378, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Weber, K. Identity of p36K Phosphorylated upon Rous Sarcoma Virus Transformation with a Protein Purified from Brush Borders; Calcium-Dependent Binding to Non-Erythroid Spectrin and F-Actin. EMBO J. 1984, 3, 227–233. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Goel, R.K.; Phanse, S.; Youssef, A.; Padhorny, D.; Kotelnikov, S.; Kozakov, D.; Emili, A. Scalable Multiplex Co-Fractionation/Mass Spectrometry Platform for Accelerated Protein Interactome Discovery. Nat. Commun. 2022, 13, 4043. [Google Scholar] [CrossRef] [PubMed]
- He, K.-L.; Deora, A.B.; Xiong, H.; Ling, Q.; Weksler, B.B.; Niesvizky, R.; Hajjar, K.A. Endothelial Cell Annexin A2 Regulates Polyubiquitination and Degradation of Its Binding Partner S100A10/P11. J. Biol. Chem. 2008, 283, 19192–19200. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the Human Interactome Defines Protein Communities and Disease Networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Brukner, R.J.; Navarrete-Perea, J.; Cannon, J.R.; Baltier, K.; Gebreab, F.; Gygi, M.P.; Thornock, A.; Zarraga, G.; Tam, S.; et al. Dual Proteome-Scale Networks Reveal Cell-Specific Remodeling of the Human Interactome. Cell 2021, 184, 3022–3040.e28. [Google Scholar] [CrossRef] [PubMed]
- Réty, S.; Sopkova, J.; Renouard, M.; Osterloh, D.; Gerke, V.; Tabaries, S.; Russo-Marie, F.; Lewit-Bentley, A. The Crystal Structure of a Complex of P11 with the Annexin II N-Terminal Peptide. Nat. Struct. Biol. 1999, 6, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Wijewickrama, G.T.; Kim, J.H.; Das, S.; Tun, M.P.; Gokhale, N.; Jung, J.W.; Kim, K.P.; Cho, W. Mechanism of Regulation of Group IVA Phospholipase A2 Activity by Ser727 Phosphorylation. J. Biol. Chem. 2008, 283, 3960–3971. [Google Scholar] [CrossRef] [PubMed]
- Waisman, D.M. Annexin II Tetramer: Structure and Function. Mol. Cell Biochem. 1995, 149/150, 301–322. [Google Scholar] [CrossRef] [PubMed]
- Zobiack, N.; Rescher, U.; Ludwig, C.; Zeuschner, D.; Gerke, V. The Annexin 2/S100A10 Complex Controls the Distribution of Transferrin Receptor-Containing Recycling Endosomes. MBoC 2003, 14, 4896–4908. [Google Scholar] [CrossRef]
- Donier, E.; Rugiero, F.; Okuse, K.; Wood, J.N. Annexin II Light Chain P11 Promotes Functional Expression of Acid-Sensing Ion Channel ASIC1a. J. Biol. Chem. 2005, 280, 38666–38672. [Google Scholar] [CrossRef]
- Hsu, S.Y.; Kaipia, A.; McGee, E.; Lomeli, M.; Hsueh, A.J.W. Bok Is a Pro-Apoptotic Bcl-2 Protein with Restricted Expression in Reproductive Tissues and Heterodimerizes with Selective Anti-Apoptotic Bcl-2 Family Members. Proc. Natl. Acad. Sci. USA 1997, 94, 12401–12406. [Google Scholar] [CrossRef]
- Mai, J.; Waisman, D.M.; Sloane, B.F. Cell Surface Complex of Cathepsin B/Annexin II Tetramer in Malignant Progression. Biochim. Biophys. Acta 2000, 1477, 215–230. [Google Scholar] [CrossRef]
- Wu, T.; Angus, C.W.; Yao, X.-L.; Logun, C.; Shelhamer, J.H. P11, a Unique Member of the S100 Family of Calcium-Binding Proteins, Interacts with and Inhibits the Activity of the 85-kDa Cytosolic Phospholipase A2. J. Biol. Chem. 1997, 272, 17145–17153. [Google Scholar] [CrossRef]
- Okuse, K.; Malik-Hall, M.; Baker, M.D.; Poon, W.-Y.L.; Kong, H.; Chao, M.V.; Wood, J.N. Annexin II Light Chain Regulates Sensory Neuron-Specific Sodium Channel Expression. Nature 2002, 417, 653–656. [Google Scholar] [CrossRef]
- Le Bouffant, F.; Capdevielle, J.; Guillemot, J.-C.; Sladeczek, F. Characterization of Brain PCTAIRE-1 Kinase Immunoreactivity and Its Interactions with P11 and 14-3-3 Proteins. Eur. J. Biochem. 1998, 257, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, R.; Melle, C.; Escher, N.; Von Eggeling, F. Detection and Identification of Protein Interactions of S100 Proteins by ProteinChip Technology. J. Proteome Res. 2005, 4, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Girard, C.; Tinel, N.; Terrenoire, C.; Romey, G.; Lazdunski, M.; Borsotto, M. P11, an Annexin II Subunit, an Auxiliary Protein Associated with the Background K+ Channel, TASK-1. EMBO J. 2002, 21, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Renigunta, V.; Yuan, H.; Zuzarte, M.; Rinné, S.; Koch, A.; Wischmeyer, E.; Schlichthörl, G.; Gao, Y.; Karschin, A.; Jacob, R.; et al. The Retention Factor P11 Confers an Endoplasmic Reticulum-Localization Signal to the Potassium Channel TASK-1. Traffic 2006, 7, 168–181. [Google Scholar] [CrossRef]
- van de Graaf, S.F.J.; Hoenderop, J.G.J.; Gkika, D.; Lamers, D.; Prenen, J.; Rescher, U.; Gerke, V.; Staub, O.; Nilius, B.; Bindels, R.J.M. Functional Expression of the Epithelial Ca2+ Channels (TRPV5 and TRPV6) Requires Association of the S100A10-Annexin 2 Complex. EMBO J. 2003, 22, 1478–1487. [Google Scholar] [CrossRef]
- Ruse, M.; Lambert, A.; Robinson, N.; Ryan, D.; Shon, K.-J.; Eckert, R.L. S100A7, S100A10, and S100A11 Are Transglutaminase Substrates. Biochemistry 2001, 40, 3167–3173. [Google Scholar] [CrossRef]
- Głowacka, A.; Bieganowski, P.; Jurewicz, E.; Leśniak, W.; Wilanowski, T.; Filipek, A. Regulation of S100A10 Gene Expression. Biomolecules 2021, 11, 974. [Google Scholar] [CrossRef]
- Madureira, P.A.; Bharadwaj, A.G.; Bydoun, M.; Garant, K.; O’Connell, P.; Lee, P.; Waisman, D.M. Cell Surface Protease Activation during RAS Transformation: Critical Role of the Plasminogen Receptor, S100A10. Oncotarget 2016, 7, 47720–47737. [Google Scholar] [CrossRef]
- O’Connell, P.A.; Madureira, P.A.; Berman, J.N.; Liwski, R.S.; Waisman, D.M. Regulation of S100A10 by the PML-RAR-α Oncoprotein. Blood 2011, 117, 4095–4105. [Google Scholar] [CrossRef]
- Lu, H.; Xie, Y.; Tran, L.; Lan, J.; Yang, Y.; Murugan, N.L.; Wang, R.; Wang, Y.J.; Semenza, G.L. Chemotherapy-Induced S100A10 Recruits KDM6A to Facilitate OCT4-Mediated Breast Cancer Stemness. J. Clin. Investig. 2020, 130, 4607–4623. [Google Scholar] [CrossRef]
- Fernandes, J.; Acuña, S.; Aoki, J.; Floeter-Winter, L.; Muxel, S. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. ncRNA 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.-T.; Viereck, J.; Thum, T. Non-Coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Shan, X.; Miao, Y.; Fan, R.; Qian, H.; Chen, P.; Liu, H.; Yan, X.; Li, J.; Zhou, F. MiR-590-5P Inhibits Growth of HepG2 Cells via Decrease of S100A10 Expression and Inhibition of the Wnt Pathway. Int. J. Mol. Sci. 2013, 14, 8556–8569. [Google Scholar] [CrossRef]
- Zhao, J.; Chi, B.; Sun, Y.; Chi, N.; Zhang, X.; Sun, J.; Chen, Y.; Xia, Y. LINC00174 Is an Oncogenic lncRNA of Hepatocellular Carcinoma and Regulates miR-320/S100A10 Axis. Cell Biochem. Funct. 2020, 38, 859–869. [Google Scholar] [CrossRef]
- Zhu, J.; Huang, Y.; Zhang, Y.; Huang, R.; Huang, C. KCNMB2-AS1 Promotes Bladder Cancer Progression Through Sponging miR-374a-3p to Upregulate S100A10. Front. Genet. 2021, 12, 655569. [Google Scholar] [CrossRef]
- Medcalf, R.L. Fibrinolysis: From Blood to the Brain. J. Thromb. Haemost. 2017, 15, 2089–2098. [Google Scholar] [CrossRef]
- Medcalf, R.L.; Keragala, C.B. The Fibrinolytic System: Mysteries and Opportunities. HemaSphere 2021, 5, e570. [Google Scholar] [CrossRef]
- Ismail, A.A.; Shaker, B.T.; Bajou, K. The Plasminogen–Activator Plasmin System in Physiological and Pathophysiological Angiogenesis. Int. J. Mol. Sci. 2021, 23, 337. [Google Scholar] [CrossRef]
- Longstaff, C.; Kolev, K. Basic Mechanisms and Regulation of Fibrinolysis. J. Thromb. Haemost. 2015, 13, S98–S105. [Google Scholar] [CrossRef] [PubMed]
- Undas, A.; Ariëns, R.A.S. Fibrin Clot Structure and Function: A Role in the Pathophysiology of Arterial and Venous Thromboembolic Diseases. ATVB 2011, 31, e88–e99. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.A.; Travers, R.J.; Morrissey, J.H. How It All Starts: Initiation of the Clotting Cascade. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Hoylaerts, M.; Rijken, D.C.; Lijnen, H.R.; Collen, D. Kinetics of the Activation of Plasminogen by Human Tissue Plasminogen Activator. Role of Fibrin. J. Biol. Chem. 1982, 257, 2912–2919. [Google Scholar] [CrossRef] [PubMed]
- Stump, D.C.; Lijnen, H.R.; Collen, D. Purification and Characterization of Single-Chain Urokinase-Type Plasminogen Activator from Human Cell Cultures. J. Biol. Chem. 1986, 261, 1274–1278. [Google Scholar] [CrossRef]
- Anand, S.S.; Yi, Q.; Gerstein, H.; Lonn, E.; Jacobs, R.; Vuksan, V.; Teo, K.; Davis, B.; Montague, P.; Yusuf, S. Relationship of Metabolic Syndrome and Fibrinolytic Dysfunction to Cardiovascular Disease. Circulation 2003, 108, 420–425. [Google Scholar] [CrossRef]
- Fujii, S.; Goto, D.; Zaman, T.; Ishimori, N.; Watano, K.; Kaneko, T.; Okada, H.; Makiguchi, M.; Nakagawa, T.; Kitabatake, A. Diminished Fibrinolysis and Thrombosis: Clinical Implications for Accelerated Atherosclerosis. J. Atheroscler. Thromb. 1998, 5, 76–81. [Google Scholar] [CrossRef][Green Version]
- Mehra, A.; Ali, C.; Parcq, J.; Vivien, D.; Docagne, F. The Plasminogen Activation System in Neuroinflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2016, 1862, 395–402. [Google Scholar] [CrossRef]
- Schneider, D.J.; Nordt, T.K.; Sobel, B.E. Attenuated Fibrinolysis and Accelerated Atherogenesis in Type II Diabetic Patients. Diabetes 1993, 42, 1–7. [Google Scholar] [CrossRef]
- Eichinger, S. D-Dimer Levels and Risk of Recurrent Venous Thromboembolism. JAMA 2003, 290, 1071. [Google Scholar] [CrossRef]
- Rooth, E.; Wallen, N.H.; Blombäck, M.; He, S. Decreased Fibrin Network Permeability and Impaired Fibrinolysis in the Acute and Convalescent Phase of Ischemic Stroke. Thromb. Res. 2011, 127, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, W.M.; Bruck, D.C.; Jeter, M.A.; Corrigan, J.J. Fibrinolysis after Acute Ischemic Stroke. Thromb. Res. 1991, 64, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, S.; Hirose, G.; Hori, A.; Shirakawa, T.; Saigan, T. Activation of Thrombosis and Fibrinolysis Following Brain Infarction. J. Neurol. Sci. 2000, 181, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Surette, A.P.; Madureira, P.A.; Phipps, K.D.; Miller, V.A.; Svenningsson, P.; Waisman, D.M. Regulation of Fibrinolysis by S100A10 in Vivo. Blood 2011, 118, 3172–3181. [Google Scholar] [CrossRef]
- Svenningsson, P.; Kim, Y.; Warner-Schmidt, J.; Oh, Y.-S.; Greengard, P. P11 and Its Role in Depression and Therapeutic Responses to Antidepressants. Nat. Rev. Neurosci. 2013, 14, 673–680. [Google Scholar] [CrossRef]
- Lee, K.-W.; Westin, L.; Kim, J.; Chang, J.C.; Oh, Y.-S.; Amreen, B.; Gresack, J.; Flajolet, M.; Kim, D.; Aperia, A.; et al. Alteration by P11 of mGluR5 Localization Regulates Depression-like Behaviors. Mol. Psychiatry 2015, 20, 1546–1556. [Google Scholar] [CrossRef]
- Hessner, F.; Dlugos, C.P.; Chehab, T.; Schaefer, C.; Homey, B.; Gerke, V.; Weide, T.; Pavenstädt, H.; Rescher, U. CC Chemokine Receptor 10 Cell Surface Presentation in Melanocytes Is Regulated by the Novel Interaction Partner S100A10. Sci. Rep. 2016, 6, 22649. [Google Scholar] [CrossRef]
- Cohen, B.D.; Nechamen, C.A.; Dias, J.A. Human Follitropin Receptor (FSHR) Interacts with the Adapter Protein 14-3-3τ. Mol. Cell. Endocrinol. 2004, 220, 1–7. [Google Scholar] [CrossRef]
- Milosevic, A.; Liebmann, T.; Knudsen, M.; Schintu, N.; Svenningsson, P.; Greengard, P. Cell- and Region-Specific Expression of Depression-Related Protein P11 (S100a10) in the Brain. J. Comp. Neurol. 2017, 525, 955–975. [Google Scholar] [CrossRef]
- Anisman, H.; Du, L.; Palkovits, M.; Faludi, G.; Kovacs, G.G.; Szontagh-Kishazi, P.; Merali, Z.; Poulter, M.O. Serotonin Receptor Subtype and P11 mRNA Expression in Stress-Relevant Brain Regions of Suicide and Control Subjects. J. Psychiatry Neurosci. 2007, 33, 131–141. [Google Scholar]
- Schmidt, E.F.; Warner-Schmidt, J.L.; Otopalik, B.G.; Pickett, S.B.; Greengard, P.; Heintz, N. Identification of the Cortical Neurons That Mediate Antidepressant Responses. Cell 2012, 149, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Warner-Schmidt, J.L.; Schmidt, E.F.; Marshall, J.J.; Rubin, A.J.; Arango-Lievano, M.; Kaplitt, M.G.; Ibañez-Tallon, I.; Heintz, N.; Greengard, P. Cholinergic Interneurons in the Nucleus Accumbens Regulate Depression-like Behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 11360–11365. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.X.; Oh, Y.-S.; Kim, Y. S100A10 and Its Binding Partners in Depression and Antidepressant Actions. Front. Mol. Neurosci. 2022, 15, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hedhli, N.; Falcone, D.J.; Huang, B.; Cesarman-Maus, G.; Kraemer, R.; Zhai, H.; Tsirka, S.E.; Santambrogio, L.; Hajjar, K.A. The Annexin A2/S100A10 System in Health and Disease: Emerging Paradigms. J. Biomed. Biotechnol. 2012, 2012, 406273. [Google Scholar] [CrossRef] [PubMed]
- Alexander, B.; Warner-Schmidt, J.; Eriksson, T.M.; Tamminga, C.; Arango-Lievano, M.; Ghose, S.; Vernov, M.; Stavarache, M.; Musatov, S.; Flajolet, M.; et al. Reversal of Depressed Behaviors in Mice by P11 Gene Therapy in the Nucleus Accumbens. Sci. Transl. Med. 2010, 2, 54ra76. [Google Scholar] [CrossRef]
- Svenningsson, P.; Greengard, P. P11 (S100A10)—An Inducible Adaptor Protein That Modulates Neuronal Functions. Curr. Opin. Pharmacol. 2007, 7, 27–32. [Google Scholar] [CrossRef]
- Seo, J.-S.; Zhong, P.; Liu, A.; Yan, Z.; Greengard, P. Elevation of P11 in Lateral Habenula Mediates Depression-like Behavior. Mol. Psychiatry 2018, 23, 1113–1119. [Google Scholar] [CrossRef]
- Becerril-Villanueva, E.; Olvera-Alvarez, M.I.; Alvarez-Herrera, S.; Maldonado-García, J.L.; López-Torres, A.; Ramírez-Marroquín, O.A.; González-Ruiz, O.; Nogueira-Fernández, J.M.; Mendoza-Contreras, J.M.; Sánchez-García, H.O.; et al. Screening of SERT and P11 mRNA Levels in Airline Pilots: A Translational Approach. Front. Psychiatry 2022, 13, 859768. [Google Scholar] [CrossRef]
- Singh, P.; Ali, S.A. Multifunctional Role of S100 Protein Family in the Immune System: An Update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef]
- Tong, L.; Lan, W.; Lim, R.R.; Chaurasia, S.S. S100A Proteins as Molecular Targets in the Ocular Surface Inflammatory Diseases. Ocul. Surf. 2014, 12, 23–31. [Google Scholar] [CrossRef]
- Bagheri-Hosseinabadi, Z.; Abbasi, M.; Kahnooji, M.; Ghorbani, Z.; Abbasifard, M. The Prognostic Value of S100A Calcium Binding Protein Family Members in Predicting Severe Forms of COVID-19. Inflamm. Res. 2022, 71, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Brownstein, C.; Deora, A.B.; Jacovina, A.T.; Weintraub, R.; Gertler, M.; Khan, K.M.F.; Falcone, D.J.; Hajjar, K.A. Annexin II Mediates Plasminogen-Dependent Matrix Invasion by Human Monocytes: Enhanced Expression by Macrophages. Blood 2004, 103, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Dassah, M.; Deora, A.B.; He, K.; Hajjar, K.A. The Endothelial Cell Annexin A2 System and Vascular Fibrinolysis. Gen. Physiol. Biophys. 2009, 28, F20–F28. [Google Scholar]
- Hitchcock, J.K.; Katz, A.A.; Schäfer, G. Dynamic Reciprocity: The Role of Annexin A2 in Tissue Integrity. J. Cell Commun. Signal. 2014, 8, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Fogg, D.K.; Yoon, C.; Waisman, D.M. P11 Regulates Extracellular Plasmin Production and Invasiveness of HT1080 Fibrosarcoma Cells. FASEB J. 2003, 17, 235–246. [Google Scholar] [CrossRef]
- Lokman, N.A.; Ween, M.P.; Oehler, M.K.; Ricciardelli, C. The Role of Annexin A2 in Tumorigenesis and Cancer Progression. Cancer Microenviron. 2011, 4, 199–208. [Google Scholar] [CrossRef]
- O’Connell, P.A.; Surette, A.P.; Liwski, R.S.; Svenningsson, P.; Waisman, D.M. S100A10 Regulates Plasminogen-Dependent Macrophage Invasion. Blood 2010, 116, 1136–1146. [Google Scholar] [CrossRef]
- Phipps, K.D.; Surette, A.P.; O’Connell, P.A.; Waisman, D.M. Plasminogen Receptor S100A10 Is Essential for the Migration of Tumor-Promoting Macrophages into Tumor Sites. Cancer Res. 2011, 71, 6676–6683. [Google Scholar] [CrossRef]
- Shiozawa, Y.; Havens, A.M.; Jung, Y.; Ziegler, A.M.; Pedersen, E.A.; Wang, J.; Wang, J.; Lu, G.; Roodman, G.D.; Loberg, R.D.; et al. Annexin II/Annexin II Receptor Axis Regulates Adhesion, Migration, Homing, and Growth of Prostate Cancer. J. Cell. Biochem. 2008, 105, 370–380. [Google Scholar] [CrossRef]
- Zhang, L.; Fogg, D.K.; Waisman, D.M. RNA Interference-Mediated Silencing of the S100A10 Gene Attenuates Plasmin Generation and Invasiveness of Colo 222 Colorectal Cancer Cells. J. Biol. Chem. 2004, 279, 2053–2062. [Google Scholar] [CrossRef]
- Zhong, L.; Wei, K.; Yang, X.; Zhang, L.; Zhou, X.; Pan, H.; Li, J.; Chen, W.; Zhang, Z. Increased Expression of Annexin A2 in Oral Squamous Cell Carcinoma. Arch. Oral. Biol. 2009, 54, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, N.; Kobayashi, K. Macrophages in Inflammation. CDTIA 2005, 4, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Crossley, J.; Xing, Z. Macrophage Engulfment of Apoptotic Neutrophils Contributes to the Resolution of Acute Pulmonary Inflammation in Vivo. Am. J. Respir. Cell Mol. Biol. 1995, 12, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Kassam, G.; Choi, K.-S.; Ghuman, J.; Kang, H.-M.; Fitzpatrick, S.L.; Zackson, T.; Zackson, S.; Toba, M.; Shinomiya, A.; Waisman, D.M. The Role of Annexin II Tetramer in the Activation of Plasminogen. J. Biol. Chem. 1998, 273, 4790–4799. [Google Scholar] [CrossRef]
- Miles, L.A.; Dahlberg, C.M.; Plescia, J.; Felez, J.; Kato, K.; Plow, E.F. Role of Cell-Surface Lysines in Plasminogen Binding to Cells: Identification of α-Enolase as a Candidate Plasminogen Receptor. Biochemistry 1991, 30, 1682–1691. [Google Scholar] [CrossRef]
- Miles, L.A.; Andronicos, N.M.; Chen, E.I.; Baik, N.; Bai, H.; Parmer, C.M.; Lighvani, S.; Nangia, S.; Kiosses, W.B.; Kamps, M.P.; et al. Identification of the Novel Plasminogen Receptor, Plg-RKT. In Proteomics—Human Diseases and Protein Functions; IntechOpen: London, UK, 2012; ISBN 978-953-307-832-8. [Google Scholar]
- Das, R.; Burke, T.; Plow, E.F. Histone H2B as a Functionally Important Plasminogen Receptor on Macrophages. Blood 2007, 110, 3763–3772. [Google Scholar] [CrossRef]
- Swisher, J.F.A.; Burton, N.; Bacot, S.M.; Vogel, S.N.; Feldman, G.M. Annexin A2 Tetramer Activates Human and Murine Macrophages through TLR4. Blood 2010, 115, 549–558. [Google Scholar] [CrossRef]
- Hajjar, K.A. The Biology of Annexin A2: From Vascular Fibrinolysis to Innate Immunity. Trans. Am. Clin. Climatol. Assoc. 2015, 126, 144–155. [Google Scholar]
- Lin, L.; Wu, C.; Hu, K. Tissue Plasminogen Activator Activates NF-κB through a Pathway Involving Annexin A2/CD11b and Integrin-Linked Kinase. J. Am. Soc. Nephrol. 2012, 23, 1329–1338. [Google Scholar] [CrossRef]
- Hoesel, B.; Schmid, J.A. The Complexity of NF-κB Signaling in Inflammation and Cancer. Mol. Cancer 2013, 12, 86. [Google Scholar] [CrossRef]
- Sansico, F.; Miroballo, M.; Bianco, D.S.; Tamiro, F.; Colucci, M.; Santis, E.D.; Rossi, G.; Rosati, J.; Di Mauro, L.; Miscio, G.; et al. COVID-19 Specific Immune Markers Revealed by Single Cell Phenotypic Profiling. Biomedicines 2021, 9, 1794. [Google Scholar] [CrossRef] [PubMed]
- Gisby, J.S.; Buang, N.B.; Papadaki, A.; Clarke, C.L.; Malik, T.H.; Medjeral-Thomas, N.; Pinheiro, D.; Mortimer, P.M.; Lewis, S.; Sandhu, E.; et al. Multi-Omics Identify Falling LRRC15 as a COVID-19 Severity Marker and Persistent pro-Thrombotic Signals in Convalescence. Nat. Commun. 2022, 13, 7775. [Google Scholar] [CrossRef] [PubMed]
- Gerke, V.; Moss, S.E. Annexins and Membrane Dynamics. Biochim. Biophys. Acta Mol. Cell Res. 1997, 1357, 129–154. [Google Scholar] [CrossRef] [PubMed]
- König, J.; Prenen, J.; Nilius, B.; Gerke, V. The Annexin II-P11 Complex Is Involved in Regulated Exocytosis in Bovine Pulmonary Artery Endothelial Cells. J. Biol. Chem. 1998, 273, 19679–19684. [Google Scholar] [CrossRef]
- Umbrecht-Jenck, E.; Demais, V.; Calco, V.; Bailly, Y.; Bader, M.-F.; Chasserot-Golaz, S. S100A10-Mediated Translocation of Annexin-A2 to SNARE Proteins in Adrenergic Chromaffin Cells Undergoing Exocytosis. Traffic 2010, 11, 958–971. [Google Scholar] [CrossRef]
- Nakata, T.; Sobue, K.; Hirokawa, N. Conformational Change and Localization of Calpactin I Complex Involved in Exocytosis as Revealed by Quick-Freeze, Deep-Etch Electron Microscopy and Immunocytochemistry. J. Cell Biol. 1990, 110, 13–25. [Google Scholar] [CrossRef]
- Senda, T.; Okabe, T.; Matsuda, M.; Fujita, H. Quick-Freeze, Deep-Etch Visualization of Exocytosis in Anterior Pituitary Secretory Cells: Localization and Possible Roles of Actin and Annexin II. Cell Tissue Res. 1994, 277, 51–60. [Google Scholar] [CrossRef]
- Liu, L.; Tao, J.-Q.; Zimmerman, U.-J.P. Annexin II Binds to the Membrane of A549 Cells in a Calcium-Dependent and Calcium-Independent Manner. Cell. Signal. 1997, 9, 299–304. [Google Scholar] [CrossRef]
- Neher, E.; Zucker, R.S. Multiple Calcium-Dependent Processes Related to Secretion in Bovine Chromaffin Cells. Neuron 1993, 10, 21–30. [Google Scholar] [CrossRef]
- Chasserot-Golaz, S.; Vitale, N.; Sagot, I.; Delouche, B.; Dirrig, S.; Pradel, L.A.; Henry, J.P.; Aunis, D.; Bader, M.F. Annexin II in Exocytosis: Catecholamine Secretion Requires the Translocation of P36 to the Subplasmalemmal Region in Chromaffin Cells. J. Cell Biol. 1996, 133, 1217–1236. [Google Scholar] [CrossRef]
- Sarafian, T.; Pradel, L.-A.; Henry, J.-P.; Aunis, D.; Bader, M.-F. The Participation of Annexin II (Calpactin I) in Calcium-Evoked Exocytosis Requires Protein Kinase C. J. Cell Biol. 1991, 114, 1135–1147. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ran, J.; Zhao, X.; Liang, Y.; Yang, X.; Xi, Y. The S100A10–AnxA2 Complex Is Associated with the Exocytosis of Hepatitis B Virus in Intrauterine Infection. Lab. Investig. 2022, 102, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.R.; Skeate, J.G.; Kast, W.M. Annexin A2 in Virus Infection. Front. Microbiol. 2018, 9, 2954. [Google Scholar] [CrossRef] [PubMed]
- Dziduszko, A.; Ozbun, M.A. Annexin A2 and S100A10 Regulate Human Papillomavirus Type 16 Entry and Intracellular Trafficking in Human Keratinocytes. J. Virol. 2013, 87, 7502–7515. [Google Scholar] [CrossRef]
- Woodham, A.W.; Da Silva, D.M.; Skeate, J.G.; Raff, A.B.; Ambroso, M.R.; Brand, H.E.; Isas, J.M.; Langen, R.; Kast, W.M. The S100A10 Subunit of the Annexin A2 Heterotetramer Facilitates L2-Mediated Human Papillomavirus Infection. PLoS ONE 2012, 7, e43519. [Google Scholar] [CrossRef]
- Woodham, A.W.; Sanna, A.M.; Taylor, J.R.; Skeate, J.G.; Da Silva, D.M.; Dekker, L.V.; Kast, W.M. Annexin A2 Antibodies but Not Inhibitors of the Annexin A2 Heterotetramer Impair Productive HIV-1 Infection of Macrophages in Vitro. Virol. J. 2016, 13, 187. [Google Scholar] [CrossRef]
- Chehab, T.; Santos, N.C.; Holthenrich, A.; Koerdt, S.N.; Disse, J.; Schuberth, C.; Nazmi, A.R.; Neeft, M.; Koch, H.; Man, K.N.M.; et al. A Novel Munc13-4/S100A10/Annexin A2 Complex Promotes Weibel–Palade Body Exocytosis in Endothelial Cells. MBoC 2017, 28, 1688–1700. [Google Scholar] [CrossRef]
- Holthenrich, A.; Gerke, V. Regulation of Von-Willebrand Factor Secretion from Endothelial Cells by the Annexin A2-S100A10 Complex. Int. J. Mol. Sci. 2018, 19, 1752. [Google Scholar] [CrossRef]
- Celma, C.C.P.; Roy, P. Interaction of Calpactin Light Chain (S100A10/P11) and a Viral NS Protein Is Essential for Intracellular Trafficking of Nonenveloped Bluetongue Virus. J. Virol. 2011, 85, 4783–4791. [Google Scholar] [CrossRef]
- Beaton, A.R.; Rodriguez, J.; Reddy, Y.K.; Roy, P. The Membrane Trafficking Protein Calpactin Forms a Complex with Bluetongue Virus Protein NS3 and Mediates Virus Release. Proc. Natl. Acad. Sci. USA 2002, 99, 13154–13159. [Google Scholar] [CrossRef]
- Woodham, A.W.; Taylor, J.R.; Jimenez, A.I.; Skeate, J.G.; Schmidt, T.; Brand, H.E.; Da Silva, D.M.; Kast, W.M. Small Molecule Inhibitors of the Annexin A2 Heterotetramer Prevent Human Papillomavirus Type 16 Infection. J. Antimicrob. Chemother. 2015, 70, 1686–1690. [Google Scholar] [CrossRef]
- Ling, F.; Lu, Q. S100 Calcium-Binding Protein A10 Contributes to Malignant Traits in Osteosarcoma Cells by Regulating Glycolytic Metabolism via the AKT/mTOR Pathway. Bioengineered 2022, 13, 12298–12308. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.-Y.; Li, L.-X.; Zhou, R.-C.; Sikong, Y.; Gu, X.; Jin, B.-Y.; Li, B.; Li, Y.-Q.; Zuo, X.-L. S100A10 Accelerates Aerobic Glycolysis and Malignant Growth by Activating mTOR-Signaling Pathway in Gastric Cancer. Front. Cell Dev. Biol. 2020, 8, 559486. [Google Scholar] [CrossRef]
- Lin, H.; Yang, P.; Li, B.; Chang, Y.; Chen, Y.; Li, Y.; Liu, K.; Liang, X.; Chen, T.; Dai, Y.; et al. S100A10 Promotes Pancreatic Ductal Adenocarcinoma Cells Proliferation, Migration and Adhesion through JNK/LAMB3-LAMC2 Axis. Cancers 2022, 15, 202. [Google Scholar] [CrossRef] [PubMed]
- Kirtonia, A.; Pandey, A.K.; Ramachandran, B.; Mishra, D.P.; Dawson, D.W.; Sethi, G.; Ganesan, T.S.; Koeffler, H.P.; Garg, M. Overexpression of Laminin-5 Gamma-2 Promotes Tumorigenesis of Pancreatic Ductal Adenocarcinoma through EGFR/ERK1/2/AKT/mTOR Cascade. Cell. Mol. Life Sci. 2022, 79, 362. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, H.; Sze, K.M.-F.; Wang, J.; Tian, L.; Lu, J.; Tsui, Y.-M.; Ma, H.T.; Lee, E.; Chen, A.; et al. S100A10 Promotes HCC Development and Progression via Transfer in Extracellular Vesicles and Regulating Their Protein Cargos. Gut 2023, 72, 1370–1384. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Feng, O.Q.; Hui, Y.Z.; Huang, X.; Ping, L.S. Prognostic Value of the S100 Calcium-Binding Protein Family Members in Hepatocellular Carcinoma. Biosci. Rep. 2023, 43, BSR20222523. [Google Scholar] [CrossRef] [PubMed]
- Weixel, K.M.; Bradbury, N.A. The Carboxyl Terminus of the Cystic Fibrosis Transmembrane Conductance Regulator Binds to AP-2 Clathrin Adaptors. J. Biol. Chem. 2000, 275, 3655–3660. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, N.A. Intracellular CFTR: Localization and Function. Physiol. Rev. 1999, 79, S175–S191. [Google Scholar] [CrossRef]
- Creutz, C.E. The Annexins and Exocytosis. Science 1992, 258, 924–931. [Google Scholar] [CrossRef]
- D’Acquisto, F.; Perretti, M.; Flower, R.J. Annexin-A1: A Pivotal Regulator of the Innate and Adaptive Immune Systems: Anx-A1: A Pivotal Regulator of the Innate and Adaptive Immune Systems. Br. J. Pharmacol. 2008, 155, 152–169. [Google Scholar] [CrossRef] [PubMed]
- Perretti, M.; D’Acquisto, F. Annexin A1 and Glucocorticoids as Effectors of the Resolution of Inflammation. Nat. Rev. Immunol. 2009, 9, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Bensalem, N.; Ventura, A.P.; Vallée, B.; Lipecka, J.; Tondelier, D.; Davezac, N.; Santos, A.D.; Perretti, M.; Fajac, A.; Sermet-Gaudelus, I.; et al. Down-Regulation of the Anti-Inflammatory Protein Annexin A1 in Cystic Fibrosis Knock-out Mice and Patients. MCP 2005, 4, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Chap, H.; Fauvel, J.; Gassama-Diagne, A.; Ragb-Thomas, J.; Simon, M. Une Homologie Frappante Entre Le CFTR et Les Annexines. Med Sci. (Paris) 1991, 7, 8. [Google Scholar] [CrossRef]
- Dahan, D.; Evagelidis, A.; Hanrahan, J.; Jia, Y.; Hinkson, D.; Luo, J.; Zhu, T. Regulation of the CFTR Channel by Phosphorylation. Pflügers Archiv—Eur. J. Physiol. 2001, 443, S92–S96. [Google Scholar] [CrossRef]
- Naren, A.P.; Cormet-Boyaka, E.; Fu, J.; Villain, M.; Blalock, J.E.; Quick, M.W.; Kirk, K.L. CFTR Chloride Channel Regulation by an Interdomain Interaction. Science 1999, 286, 544–548. [Google Scholar] [CrossRef]
- Borthwick, L.A.; Mcgaw, J.; Conner, G.; Taylor, C.J.; Gerke, V.; Mehta, A.; Robson, L.; Muimo, R. The Formation of the cAMP/Protein Kinase A-Dependent Annexin 2–S100A10 Complex with Cystic Fibrosis Conductance Regulator Protein (CFTR) Regulates CFTR Channel Function. Mol. Biol. Cell. 2007, 18, 3388–3397. [Google Scholar] [CrossRef]
- Borthwick, L.A.; Riemen, C.; Goddard, C.; Colledge, W.H.; Mehta, A.; Gerke, V.; Muimo, R. Defective Formation of PKA/CnA-Dependent Annexin 2–S100A10/CFTR Complex in ΔF508 Cystic Fibrosis Cells. Cell. Signal. 2008, 20, 1073–1083. [Google Scholar] [CrossRef]
- Muimo, R. Regulation of CFTR Function by Annexin A2-S100A10 Complex in Health and Disease. Gen. Physiol. Biophys. 2009, 28, F14–F19. [Google Scholar]
- Borot, F.; Vieu, D.-L.; Faure, G.; Fritsch, J.; Colas, J.; Moriceau, S.; Baudouin-Legros, M.; Brouillard, F.; Ayala-Sanmartin, J.; Touqui, L.; et al. Eicosanoid Release Is Increased by Membrane Destabilization and CFTR Inhibition in Calu-3 Cells. PLoS ONE 2009, 4, e7116. [Google Scholar] [CrossRef]
- Nakamura, Y.; Moritsuka, Y.; Ohta, Y.; Itoh, S.; Haratake, A.; Kage, M. S-100 Protein in Glands within Decidua and Cervical Glands during Early Pregnancy. Hum. Pathol. 1989, 20, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Kligman, D.; Hilt, D.C. The S100 Protein Family. Trends Biochem. Sci. 1988, 13, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Walter, I.; Miller, I. S-100 Protein Subunits in Bovine Oviduct Epithelium:In Situ Distribution and Changes during Primary Cell Culture. Histochem. J. 1996, 28, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, S.; Tsukushi, M.; Yamano, S.; Daigo, M. S-100 Protein-Immunoreactive Cells in the Bovine Ovary. Anat. Rec. 1989, 223, 384–386. [Google Scholar] [CrossRef] [PubMed]
- Agarwa, S. Cutaneous Ciliated Cyst. Histopathology 1991, 19, 99–101. [Google Scholar] [CrossRef]
- Hanaue, M.; Miwa, N.; Uebi, T.; Fukuda, Y.; Katagiri, Y.; Takamatsu, K. Characterization of S100A11, a Suppressive Factor of Fertilization, in the Mouse Female Reproductive Tract. Mol. Reprod. Dev. 2011, 78, 91–103. [Google Scholar] [CrossRef]
- Tingaud-Sequeira, A.; Chauvigné, F.; Lozano, J.; Agulleiro, M.J.; Asensio, E.; Cerdà, J. New Insights into Molecular Pathways Associated with Flatfish Ovarian Development and Atresia Revealed by Transcriptional Analysis. BMC Genom. 2009, 10, 434. [Google Scholar] [CrossRef]
- Teijeiro, J.M.; Roldán, M.L.; Marini, P.E. Annexin A2 and S100A10 in the Mammalian Oviduct. Cell Tissue Res. 2016, 363, 567–577. [Google Scholar] [CrossRef]
- Capp, J.-P. Cancer Stem Cells: From Historical Roots to a New Perspective. J. Oncol. 2019, 2019, 5189232. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem Cells, Cancer, and Cancer Stem Cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef]
- King, M.W.; Neff, A.W.; Mescher, A.L. Proteomics Analysis of Regenerating Amphibian Limbs: Changes during the Onset of Regeneration. Int. J. Dev. Biol. 2009, 53, 955–969. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Zhang, H.; Jia, J.; Chen, S.; Sun, Y.; Zhu, X. Roles of S100 Family Members in Drug Resistance in Tumors: Status and Prospects. Biomed. Pharmacother. 2020, 127, 110156. [Google Scholar] [CrossRef]
- Fenouille, N.; Grosso, S.; Yunchao, S.; Mary, D.; Pontier-Bres, R.; Imbert, V.; Czerucka, D.; Caroli-Bosc, F.-X.; Peyron, J.-F.; Lagadec, P. Calpain 2-Dependent IκBα Degradation Mediates CPT-11 Secondary Resistance in Colorectal Cancer Xenografts. J. Pathol. 2012, 227, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Tanigawara, Y. Forced Expression of S100A10 Reduces Sensitivity to Oxaliplatin in Colorectal Cancer Cells. Proteome Sci. 2014, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Spijkers-Hagelstein, J.A.P.; Schneider, P.; Hulleman, E.; de Boer, J.; Williams, O.; Pieters, R.; Stam, R.W. Elevated S100A8/S100A9 Expression Causes Glucocorticoid Resistance in MLL-Rearranged Infant Acute Lymphoblastic Leukemia. Leukemia 2012, 26, 1255–1265. [Google Scholar] [CrossRef]
- Nymoen, D.A.; Hetland Falkenthal, T.E.; Holth, A.; Ow, G.S.; Ivshina, A.V.; Tropé, C.G.; Kuznetsov, V.A.; Staff, A.C.; Davidson, B. Expression and Clinical Role of Chemoresponse-Associated Genes in Ovarian Serous Carcinoma. Gynecol. Oncol. 2015, 139, 30–39. [Google Scholar] [CrossRef]
- Wang, L.; Yan, W.; Li, X.; Liu, Z.; Tian, T.; Chen, T.; Zou, L.; Cui, Z. S100A10 Silencing Suppresses Proliferation, Migration and Invasion of Ovarian Cancer Cells and Enhances Sensitivity to Carboplatin. J. Ovarian Res. 2019, 12, 113. [Google Scholar] [CrossRef]
- Yanagi, H.; Watanabe, T.; Nishimura, T.; Hayashi, T.; Kono, S.; Tsuchida, H.; Hirata, M.; Kijima, Y.; Takao, S.; Okada, S.; et al. Upregulation of S100A10 in Metastasized Breast Cancer Stem Cells. Cancer Sci. 2020, 111, 4359–4370. [Google Scholar] [CrossRef]
- Zheng, X.; Naiditch, J.; Czurylo, M.; Jie, C.; Lautz, T.; Clark, S.; Jafari, N.; Qiu, Y.; Chu, F.; Madonna, M.B. Differential Effect of Long-Term Drug Selection with Doxorubicin and Vorinostat on Neuroblastoma Cells with Cancer Stem Cell Characteristics. Cell Death Dis. 2013, 4, e740. [Google Scholar] [CrossRef]
- Díaz-Payno, P.J.; Browe, D.C.; Cunniffe, G.M.; Kelly, D.J. The Identification of Articular Cartilage and Growth Plate Extracellular Matrix-Specific Proteins Supportive of Either Osteogenesis or Stable Chondrogenesis of Stem Cells. Biochem. Biophys. Res. Commun. 2020, 528, 285–291. [Google Scholar] [CrossRef]
- Noye, T.; Lokman, N.; Oehler, M.; Ricciardelli, C. S100A10 and Cancer Hallmarks: Structure, Functions, and Its Emerging Role in Ovarian Cancer. Int. J. Mol. Sci. 2018, 19, 4122. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, A.G.; Dahn, M.L.; Liu, R.-Z.; Colp, P.; Thomas, L.N.; Holloway, R.W.; Marignani, P.A.; Too, C.K.L.; Barnes, P.J.; Godbout, R.; et al. S100A10 Has a Critical Regulatory Function in Mammary Tumor Growth and Metastasis: Insights Using MMTV-PyMT Oncomice and Clinical Patient Sample Analysis. Cancers 2020, 12, 3673. [Google Scholar] [CrossRef] [PubMed]
- Bydoun, M.; Sterea, A.; Liptay, H.; Uzans, A.; Huang, W.; Rodrigues, G.J.; Weaver, I.C.G.; Gu, H.; Waisman, D.M. S100A10, a Novel Biomarker in Pancreatic Ductal Adenocarcinoma. Mol. Oncol. 2018, 12, 1895–1916. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, Y.; Li, J.; Cai, M.; Zhang, B.; Yu, Z.; Li, Y.; Huang, J.; Chen, X.; Song, Y.; et al. S100A Family Is a Group of Immune Markers Associated with Poor Prognosis and Immune Cell Infiltration in Hepatocellular Carcinoma. BMC Cancer 2023, 23, 637. [Google Scholar] [CrossRef]
- Gillet, J.-P.; Calcagno, A.M.; Varma, S.; Davidson, B.; Bunkholt Elstrand, M.; Ganapathi, R.; Kamat, A.A.; Sood, A.K.; Ambudkar, S.V.; Seiden, M.V.; et al. Multidrug Resistance–Linked Gene Signature Predicts Overall Survival of Patients with Primary Ovarian Serous Carcinoma. Clin. Cancer Res. 2012, 18, 3197–3206. [Google Scholar] [CrossRef] [PubMed]
- Lokman, N.A.; Pyragius, C.E.; Ruszkiewicz, A.; Oehler, M.K.; Ricciardelli, C. Annexin A2 and S100A10 Are Independent Predictors of Serous Ovarian Cancer Outcome. Transl. Res. 2016, 171, 83–95.e2. [Google Scholar] [CrossRef] [PubMed]
- Kärblane, K.; Gerassimenko, J.; Nigul, L.; Piirsoo, A.; Smialowska, A.; Vinkel, K.; Kylsten, P.; Ekwall, K.; Swoboda, P.; Truve, E.; et al. ABCE1 Is a Highly Conserved RNA Silencing Suppressor. PLoS ONE 2015, 10, e0116702. [Google Scholar] [CrossRef]
- Dutta, S.; Bandyopadhyay, C.; Bottero, V.; Veettil, M.V.; Wilson, L.; Pins, M.R.; Johnson, K.E.; Warshall, C.; Chandran, B. Angiogenin Interacts with the Plasminogen Activation System at the Cell Surface of Breast Cancer Cells to Regulate Plasmin Formation and Cell Migration. Mol. Oncol. 2014, 8, 483–507. [Google Scholar] [CrossRef]
- Capalbo, L.; Bassi, Z.I.; Geymonat, M.; Todesca, S.; Copoiu, L.; Enright, A.J.; Callaini, G.; Riparbelli, M.G.; Yu, L.; Choudhary, J.S.; et al. The Midbody Interactome Reveals Unexpected Roles for PP1 Phosphatases in Cytokinesis. Nat. Commun. 2019, 10, 4513. [Google Scholar] [CrossRef]
- Xu, J.; Yang, X.; Deng, Q.; Yang, C.; Wang, D.; Jiang, G.; Yao, X.; He, X.; Ding, J.; Qiang, J.; et al. TEM8 Marks Neovasculogenic Tumor-Initiating Cells in Triple-Negative Breast Cancer. Nat. Commun. 2021, 12, 4413. [Google Scholar] [CrossRef]
- Lau, E.; Ronai, Z.A. ATF2—At the Crossroad of Nuclear and Cytosolic Functions. J. Cell Sci. 2012, 125, 2815–2824. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-J.; Li, T.-N.; Huang, R.-S.; Pan, M.Y.-C.; Lin, S.-Y.; Lin, S.; Wu, K.-P.; Wang, L.H.-C.; Hsu, S.-T.D. Tumor Suppressor BAP1 Nuclear Import Is Governed by Transportin-1. J. Cell Biol. 2022, 221, e202201094. [Google Scholar] [CrossRef] [PubMed]
- Freire-Benéitez, V.; Pomella, N.; Millner, T.O.; Dumas, A.A.; Niklison-Chirou, M.V.; Maniati, E.; Wang, J.; Rajeeve, V.; Cutillas, P.; Marino, S. Elucidation of the BMI1 Interactome Identifies Novel Regulatory Roles in Glioblastoma. NAR Cancer 2021, 3, zcab009. [Google Scholar] [CrossRef] [PubMed]
- Siebert, A.; Gattringer, V.; Weishaupt, J.H.; Behrends, C. ALS-Linked Loss of Cyclin-F Function Affects HSP90. Life Sci. Alliance 2022, 5, e202101359. [Google Scholar] [CrossRef]
- Gerold, G.; Meissner, F.; Bruening, J.; Welsch, K.; Perin, P.M.; Baumert, T.F.; Vondran, F.W.; Kaderali, L.; Marcotrigiano, J.; Khan, A.G.; et al. Quantitative Proteomics Identifies Serum Response Factor Binding Protein 1 as a Host Factor for Hepatitis C Virus Entry. Cell Rep. 2015, 12, 864–878. [Google Scholar] [CrossRef]
- Kumar, P.; Munnangi, P.; Chowdary, K.R.; Shah, V.J.; Shinde, S.R.; Kolli, N.R.; Halehalli, R.R.; Nagarajaram, H.A.; Maddika, S. A Human Tyrosine Phosphatase Interactome Mapped by Proteomic Profiling. J. Proteome Res. 2017, 16, 2789–2801. [Google Scholar] [CrossRef]
- Sladeczek, F.; Camonis, J.H.; Burnol, A.-F.; Bouffant, F.L. The Cdk-like Protein PCTAIRE-1 from Mouse Brain Associates with P11 and 14-3-3 Proteins. Mol. Gen. Genet. 1997, 254, 571–577. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, Y.; Kalita, M.; Li, X.; Jamaluddin, M.; Tian, B.; Edeh, C.B.; Wiktorowicz, J.E.; Kudlicki, A.; Brasier, A.R. Systematic Determination of Human Cyclin Dependent Kinase (CDK)-9 Interactome Identifies Novel Functions in RNA Splicing Mediated by the DEAD Box (DDX)-5/17 RNA Helicases*. Mol. Cell Proteom. 2015, 14, 2701–2721. [Google Scholar] [CrossRef]
- Cho, N.H.; Cheveralls, K.C.; Brunner, A.-D.; Kim, K.; Michaelis, A.C.; Raghavan, P.; Kobayashi, H.; Savy, L.; Li, J.Y.; Canaj, H.; et al. OpenCell: Endogenous Tagging for the Cartography of Human Cellular Organization. Science 2022, 375, eabi6983. [Google Scholar] [CrossRef]
- Hutt, D.M.; Loguercio, S.; Campos, A.R.; Balch, W.E. A Proteomic Variant Approach (ProVarA) for Personalized Medicine of Inherited and Somatic Disease. J. Mol. Biol. 2018, 430, 2951–2973. [Google Scholar] [CrossRef]
- Stelzl, U.; Worm, U.; Lalowski, M.; Haenig, C.; Brembeck, F.H.; Goehler, H.; Stroedicke, M.; Zenkner, M.; Schoenherr, A.; Koeppen, S.; et al. A Human Protein-Protein Interaction Network: A Resource for Annotating the Proteome. Cell 2005, 122, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Popescu, N.C.; Zimonjic, D.B. DLC1 Interaction with S100A10 Mediates Inhibition of In Vitro Cell Invasion and Tumorigenicity of Lung Cancer Cells through a RhoGAP-Independent Mechanism. Cancer Res. 2011, 71, 2916–2925. [Google Scholar] [CrossRef] [PubMed]
- Redwine, W.B.; DeSantis, M.E.; Hollyer, I.; Htet, Z.M.; Tran, P.T.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Reck-Peterson, S.L. The Human Cytoplasmic Dynein Interactome Reveals Novel Activators of Motility. eLife 2017, 6, e28257. [Google Scholar] [CrossRef] [PubMed]
- Roewenstrunk, J.; Di Vona, C.; Chen, J.; Borras, E.; Dong, C.; Arató, K.; Sabidó, E.; Huen, M.S.Y.; De La Luna, S. A Comprehensive Proteomics-Based Interaction Screen That Links DYRK1A to RNF169 and to the DNA Damage Response. Sci. Rep. 2019, 9, 6014. [Google Scholar] [CrossRef] [PubMed]
- Malinová, A.; Cvačková, Z.; Matějů, D.; Hořejší, Z.; Abéza, C.; Vandermoere, F.; Bertrand, E.; Staněk, D.; Verheggen, C. Assembly of the U5 snRNP Component PRPF8 Is Controlled by the HSP90/R2TP Chaperones. J. Cell Biol. 2017, 216, 1579–1596. [Google Scholar] [CrossRef]
- Yao, Z.; Yaeger, R.; Rodrik-Outmezguine, V.S.; Tao, A.; Torres, N.M.; Chang, M.T.; Drosten, M.; Zhao, H.; Cecchi, F.; Hembrough, T.; et al. Tumours with Class 3 BRAF Mutants Are Sensitive to the Inhibition of Activated RAS. Nature 2017, 548, 234–238. [Google Scholar] [CrossRef]
- Kennedy, S.A.; Jarboui, M.-A.; Srihari, S.; Raso, C.; Bryan, K.; Dernayka, L.; Charitou, T.; Bernal-Llinares, M.; Herrera-Montavez, C.; Krstic, A.; et al. Extensive Rewiring of the EGFR Network in Colorectal Cancer Cells Expressing Transforming Levels of KRASG13D. Nat. Commun. 2020, 11, 499. [Google Scholar] [CrossRef]
- Li, J.; Bennett, K.; Stukalov, A.; Fang, B.; Zhang, G.; Yoshida, T.; Okamoto, I.; Kim, J.; Song, L.; Bai, Y.; et al. Perturbation of the Mutated EGFR Interactome Identifies Vulnerabilities and Resistance Mechanisms. Mol. Syst. Biol. 2013, 9, 705. [Google Scholar] [CrossRef]
- Nassa, G.; Giurato, G.; Salvati, A.; Gigantino, V.; Pecoraro, G.; Lamberti, J.; Rizzo, F.; Nyman, T.A.; Tarallo, R.; Weisz, A. The RNA-Mediated Estrogen Receptor α Interactome of Hormone-Dependent Human Breast Cancer Cell Nuclei. Sci. Data 2019, 6, 173. [Google Scholar] [CrossRef]
- Rosenbluh, J.; Mercer, J.; Shrestha, Y.; Oliver, R.; Tamayo, P.; Doench, J.G.; Tirosh, I.; Piccioni, F.; Hartenian, E.; Horn, H.; et al. Genetic and Proteomic Interrogation of Lower Confidence Candidate Genes Reveals Signaling Networks in β-Catenin-Active Cancers. Cell Syst. 2016, 3, 302–316.e4. [Google Scholar] [CrossRef]
- Xu, X.; Ding, P.; Shi, L.; Wu, G.; Ma, X. LukS-PV Inhibits Hepatocellular Carcinoma Cells Migration by Downregulating HDAC6 Expression. BMC Cancer 2022, 22, 630. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Lv, T.; Yang, J.; Wu, Z.; Yan, L.; Yang, J.; Shi, Y.; Jiang, L. HDLBP Promotes Hepatocellular Carcinoma Proliferation and Sorafenib Resistance by Suppressing Trim71-Dependent RAF1 Degradation. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Gesslbauer, B.; Derler, R.; Handwerker, C.; Seles, E.; Kungl, A.J. Exploring the Glycosaminoglycan-Protein Interaction Network by Glycan-Mediated Pull-down Proteomics. Electrophoresis 2016, 37, 1437–1447. [Google Scholar] [CrossRef]
- Ryu, S.W.; Stewart, R.; Pectol, D.C.; Ender, N.A.; Wimalarathne, O.; Lee, J.-H.; Zanini, C.P.; Harvey, A.; Huibregtse, J.M.; Mueller, P.; et al. Proteome-Wide Identification of HSP70/HSC70 Chaperone Clients in Human Cells. PLoS Biol. 2020, 18, e3000606. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Sügis, E.; Dauvillier, J.; Leontjeva, A.; Adler, P.; Hindie, V.; Moncion, T.; Collura, V.; Daudin, R.; Loe-Mie, Y.; Herault, Y.; et al. HENA, Heterogeneous Network-Based Data Set for Alzheimer’s Disease. Sci. Data 2019, 6, 151. [Google Scholar] [CrossRef] [PubMed]
- Luck, K.; Kim, D.-K.; Lambourne, L.; Spirohn, K.; Begg, B.E.; Bian, W.; Brignall, R.; Cafarelli, T.; Campos-Laborie, F.J.; Charloteaux, B.; et al. A Reference Map of the Human Binary Protein Interactome. Nature 2020, 580, 402–408. [Google Scholar] [CrossRef]
- Savidis, G.; McDougall, W.M.; Meraner, P.; Perreira, J.M.; Portmann, J.M.; Trincucci, G.; John, S.P.; Aker, A.M.; Renzette, N.; Robbins, D.R.; et al. Identification of Zika Virus and Dengue Virus Dependency Factors Using Functional Genomics. Cell Rep. 2016, 16, 232–246. [Google Scholar] [CrossRef]
- Wang, L.; Chen, C.; Song, Z.; Wang, H.; Ye, M.; Wang, D.; Kang, W.; Liu, H.; Qing, G. EZH2 Depletion Potentiates MYC Degradation Inhibiting Neuroblastoma and Small Cell Carcinoma Tumor Formation. Nat. Commun. 2022, 13, 12. [Google Scholar] [CrossRef]
- Zellner, S.; Schifferer, M.; Behrends, C. Systematically Defining Selective Autophagy Receptor-Specific Cargo Using Autophagosome Content Profiling. Mol. Cell 2021, 81, 1337–1354.e8. [Google Scholar] [CrossRef]
- Chen, R.; Wang, Y.; Liu, Y.; Zhang, Q.; Zhang, X.; Zhang, F.; Shieh, C.-H.P.; Yang, D.; Zhang, N. Quantitative Study of the Interactome of PKCζ Involved in the EGF-Induced Tumor Cell Chemotaxis. J. Proteome Res. 2013, 12, 1478–1486. [Google Scholar] [CrossRef] [PubMed]
- Asselin-Mullen, P.; Chauvin, A.; Dubois, M.-L.; Drissi, R.; Lévesque, D.; Boisvert, F.-M. Protein Interaction Network of Alternatively Spliced NudCD1 Isoforms. Sci. Rep. 2017, 7, 12987. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Pawliczak, R.; Cowan, M.J.; Gladwin, M.T.; Madara, P.; Logun, C.; Shelhamer, J.H. Epidermal Growth Factor Induces P11 Gene and Protein Expression and Down-Regulates Calcium Ionophore-Induced Arachidonic Acid Release in Human Epithelial Cells. J. Biol. Chem. 2002, 277, 38431–38440. [Google Scholar] [CrossRef] [PubMed]
- Fresquet, M.; Jowitt, T.A.; McKenzie, E.A.; Ball, M.D.; Randles, M.J.; Lennon, R.; Brenchley, P.E. PLA2R Binds to the Annexin A2-S100A10 Complex in Human Podocytes. Sci. Rep. 2017, 7, 6876. [Google Scholar] [CrossRef] [PubMed]
- Wan, C.; Borgeson, B.; Phanse, S.; Tu, F.; Drew, K.; Clark, G.; Xiong, X.; Kagan, O.; Kwan, J.; Bezginov, A.; et al. Panorama of Ancient Metazoan Macromolecular Complexes. Nature 2015, 525, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.A.; Ecsédi, P.; Kovács, G.M.; Póti, Á.L.; Reményi, A.; Kardos, J.; Gógl, G.; Nyitray, L. High-throughput Competitive Fluorescence Polarization Assay Reveals Functional Redundancy in the S100 Protein Family. FEBS J. 2020, 287, 2834–2846. [Google Scholar] [CrossRef]
- Brüninghoff, K.; Aust, A.; Taupitz, K.F.; Wulff, S.; Dörner, W.; Mootz, H.D. Identification of SUMO Binding Proteins Enriched after Covalent Photo-Cross-Linking. ACS Chem. Biol. 2020, 15, 2406–2414. [Google Scholar] [CrossRef]
- Kupka, S.; Reichert, M.; Draber, P.; Walczak, H. Formation and Removal of Poly-Ubiquitin Chains in the Regulation of Tumor Necrosis Factor-Induced Gene Activation and Cell Death. FEBS J. 2016, 283, 2626–2639. [Google Scholar] [CrossRef]
- Chen, S.; He, Z.; Zhu, C.; Liu, Y.; Li, L.; Deng, L.; Wang, J.; Yu, C.; Sun, C. TRIM37 Mediates Chemoresistance and Maintenance of Stemness in Pancreatic Cancer Cells via Ubiquitination of PTEN and Activation of the AKT–GSK-3β–β-Catenin Signaling Pathway. Front. Oncol. 2020, 10, 554787. [Google Scholar] [CrossRef]
- Demirdizen, E.; Al-Ali, R.; Narayanan, A.; Sun, X.; Varga, J.P.; Steffl, B.; Brom, M.; Krunic, D.; Schmidt, C.; Schmidt, G.; et al. TRIM67 Drives Tumorigenesis in Oligodendrogliomas through Rho GTPase-Dependent Membrane Blebbing. Neuro-Oncology 2022, 25, 1031–1043. [Google Scholar] [CrossRef]
- Chen, Y.-D.; Fang, Y.-T.; Chang, C.-P.; Lin, C.-F.; Hsu, L.-J.; Wu, S.-R.; Chiu, Y.-C.; Anderson, R.; Lin, Y.-S. S100A10 Regulates ULK1 Localization to ER–Mitochondria Contact Sites in IFN-γ-Triggered Autophagy. J. Mol. Biol. 2017, 429, 142–157. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, X. TRABID Targets DDB2 for Deubiquitination to Promote Proliferation of Hepatocellular Carcinoma Cells. Biochem. Biophys. Res. Commun. 2022, 625, 23–30. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).