Presenilin-1-Derived Circular RNAs: Neglected Epigenetic Regulators with Various Functions in Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
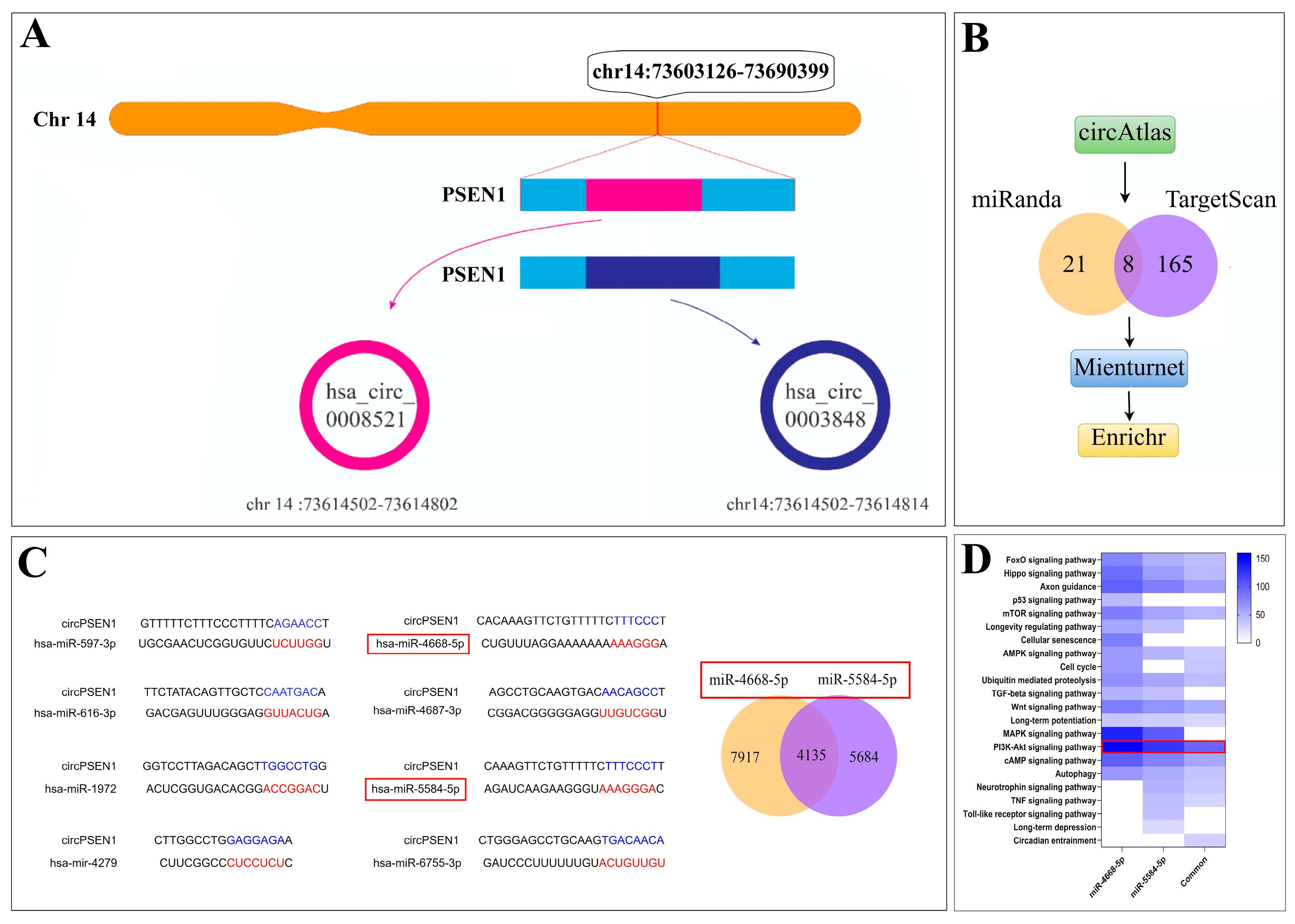
2.1. circPSEN1s–miRNAs and circRNA–Protein Interaction Prediction
2.2. miRNA Gene-Target Prediction and Pathway Enrichment Analysis
2.3. Protein–Protein Interaction (PPI)
2.4. circPSEN1s and Protein Molecular Docking
2.5. Microarray Data Analysis of Postmortem AD Patients
3. Results
3.1. circPSEN1s–miRNAs Interaction Prediction
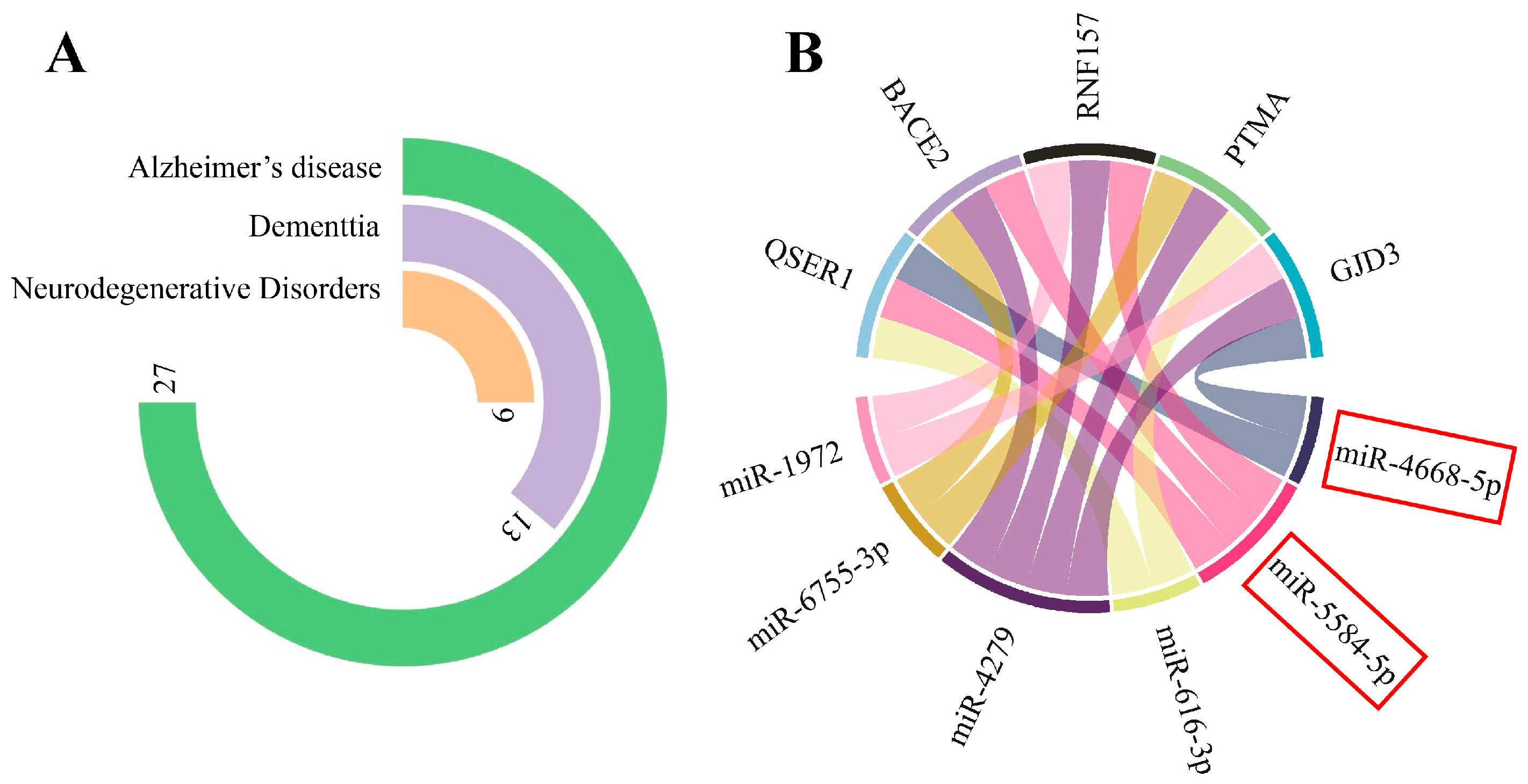
3.1.1. miRNA Gene-Target Prediction and Pathway Enrichment Analysis
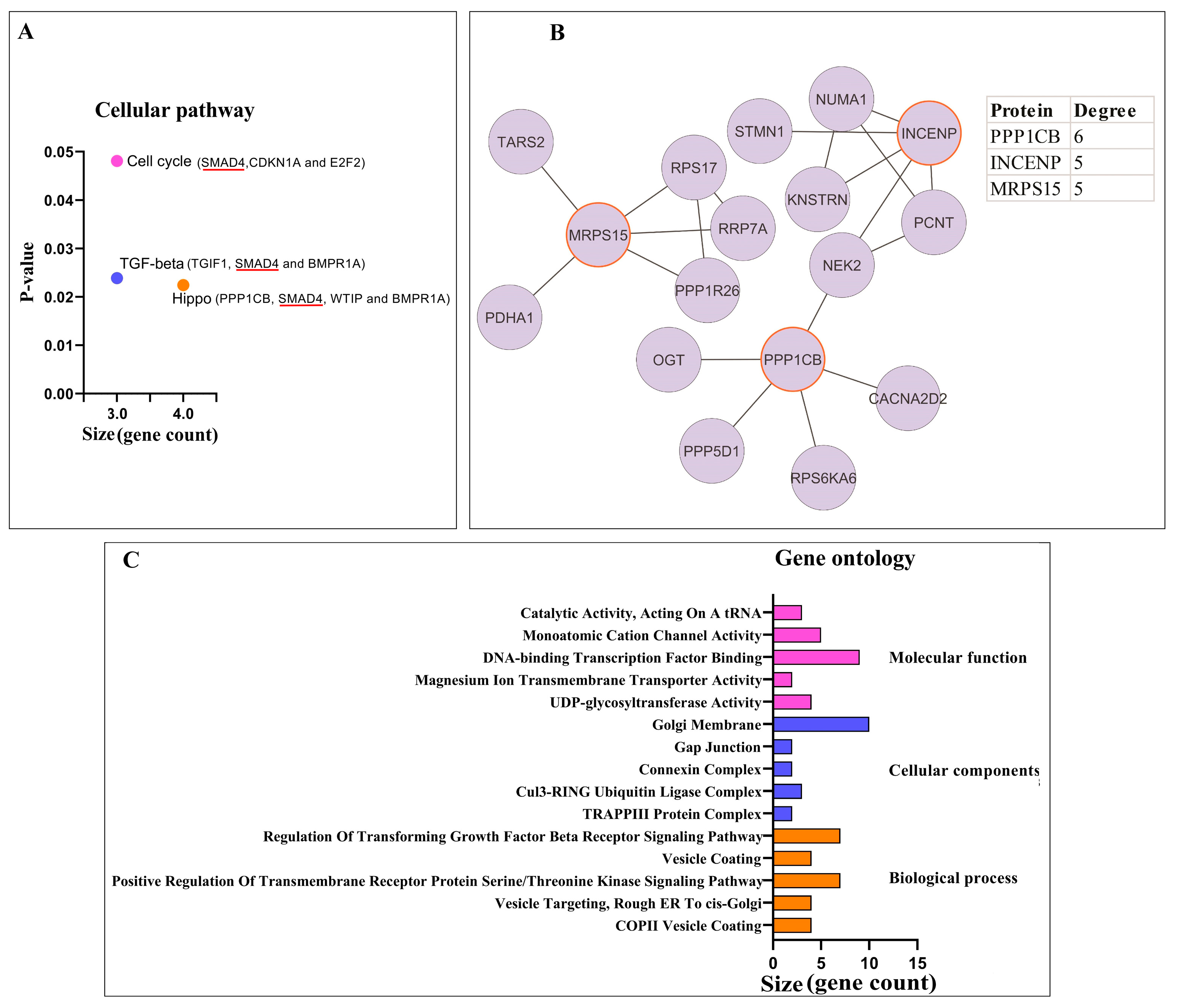
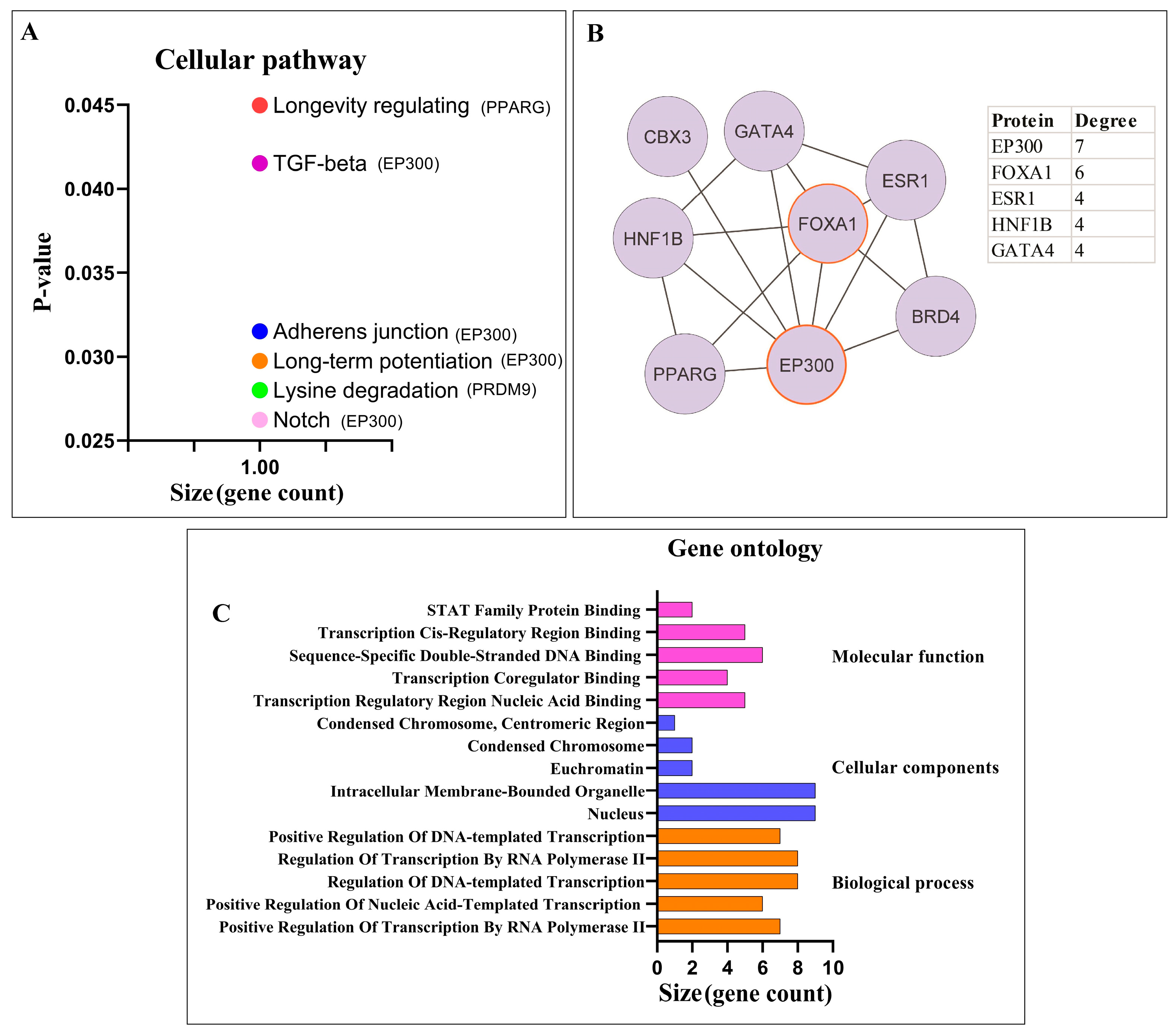
3.1.2. Pathway Enrichment Analysis and PPI Network Analysis of Proteins
3.2. Potential circPSEN1 Protein Targets
3.2.1. Pathway Enrichment Analysis and PPI Network Analysis of Proteins
3.2.2. Molecular Docking
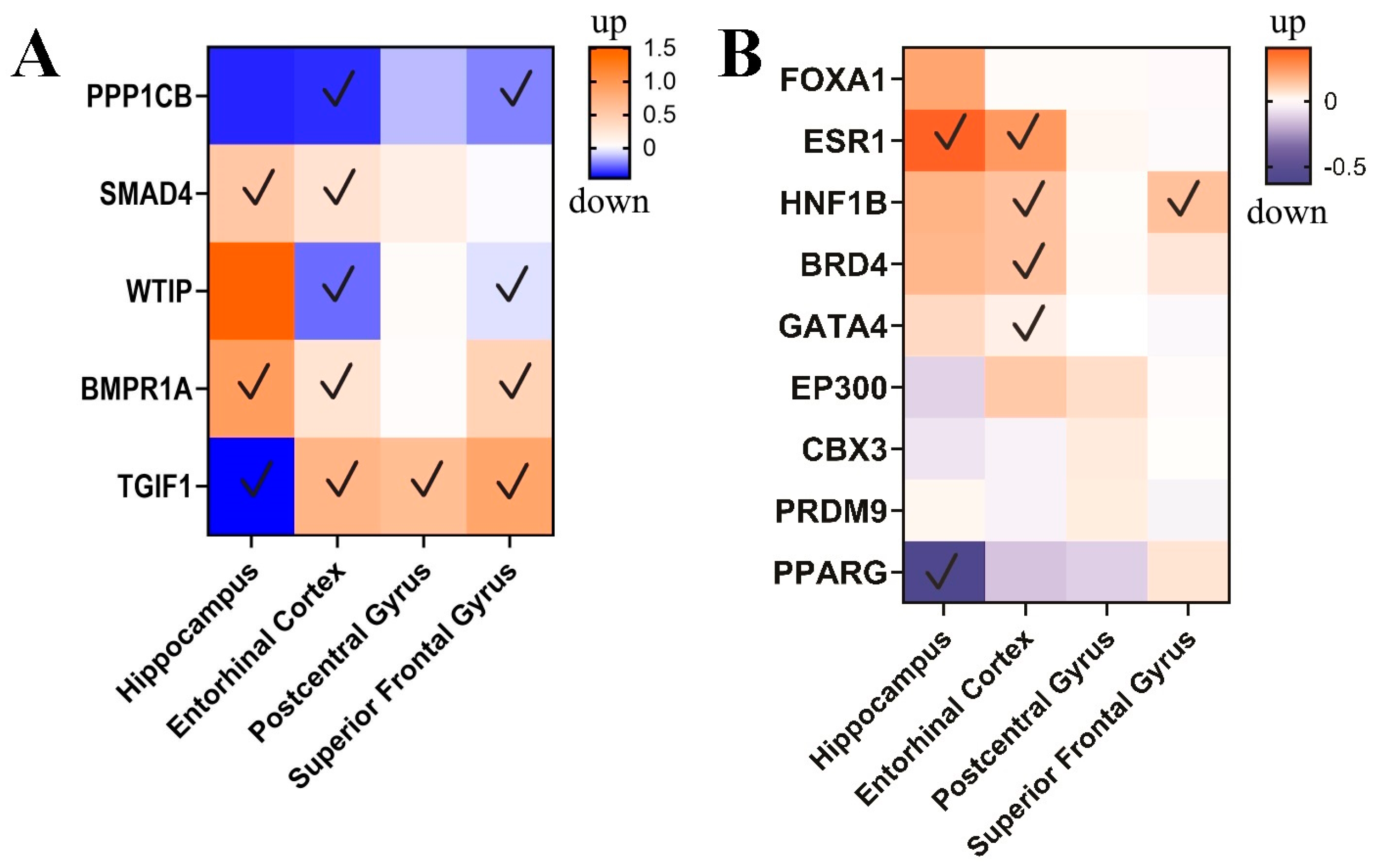
3.3. circPSEN1-Related Genes in the Brain of AD Patients
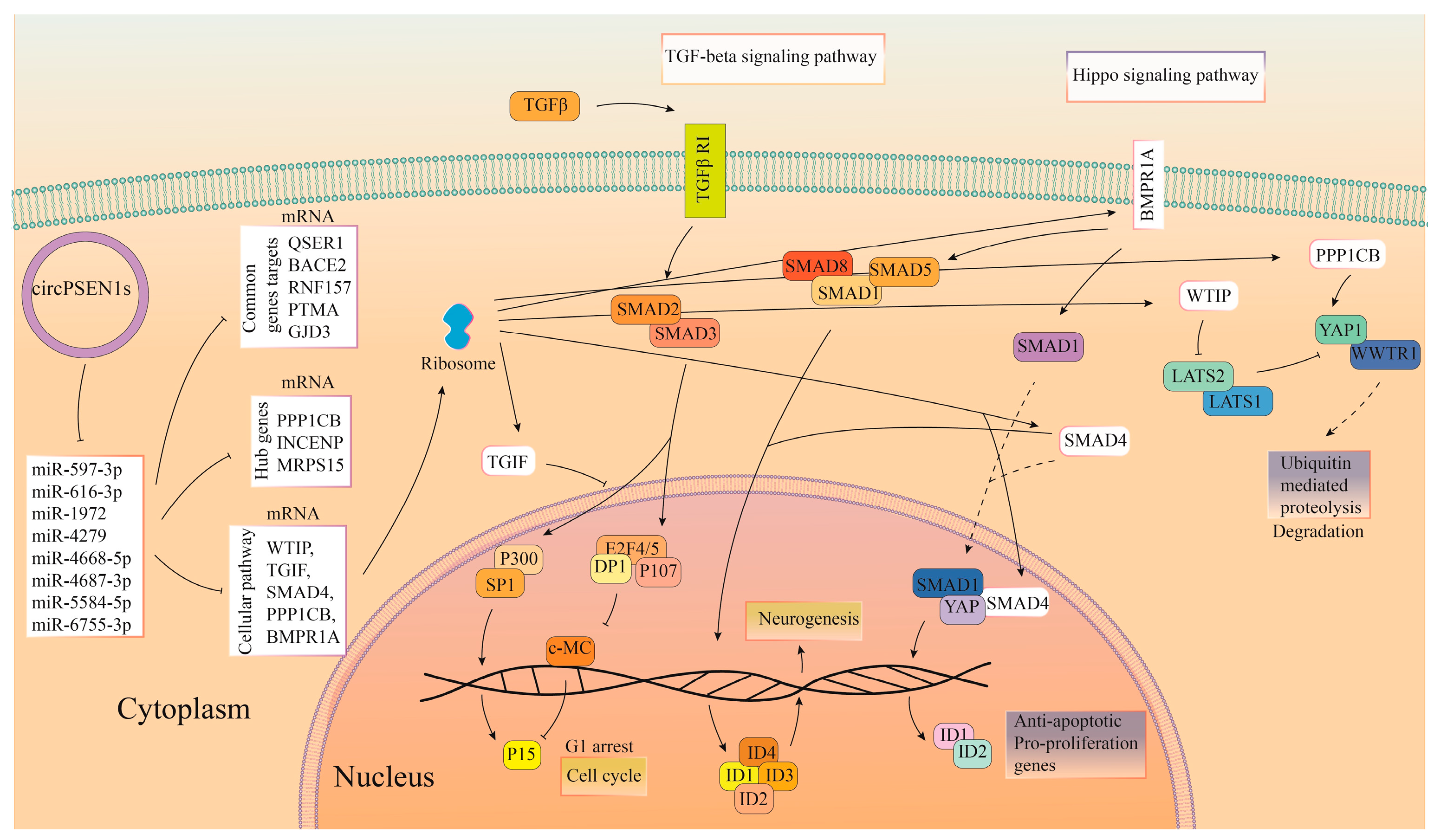
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 25, 5789. [Google Scholar] [CrossRef] [PubMed]
- Weller, J.; Budson, A. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000Research 2018, 7, 1161. [Google Scholar] [CrossRef] [PubMed]
- Mantzavinos, V.; Alexiou, A. Biomarkers for Alzheimer’s Disease Diagnosis. Curr. Alzheimer Res. 2017, 14, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Qiu, C.; Kivipelto, M.; von Strauss, E. Epidemiology of Alzheimer’s disease: Occurrence, determinants, and strategies toward intervention. Dialogues Clin. Neurosci. 2009, 11, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. In search of pathogenic amyloid β-peptide in familial Alzheimer’s disease. Prog. Mol. Biol. Transl. Sci. 2019, 168, 71–78. [Google Scholar]
- Matuszyk, M.M.; Garwood, C.J.; Ferraiuolo, L.; Simpson, J.E.; Staniforth, R.A.; Wharton, S.B. Biological and methodological complexities of beta-amyloid peptide: Implications for Alzheimer’s disease research. J. Neurochem. 2022, 160, 434–453. [Google Scholar] [CrossRef]
- Karch, C.M.; Goate, A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 2015, 77, 43–51. [Google Scholar] [CrossRef]
- Ayodele, T.; Rogaeva, E.; Kurup, J.T.; Beecham, G.; Reitz, C. Early-Onset Alzheimer’s Disease: What Is Missing in Research? Curr. Neurol. Neurosci. Rep. 2021, 21, 4. [Google Scholar] [CrossRef]
- Bagaria, J.; Bagyinszky, E.; An, S.S.A. Genetics, Functions, and Clinical Impact of Presenilin-1 (PSEN1) Gene. Int. J. Mol. Sci. 2022, 23, 10970. [Google Scholar] [CrossRef]
- Qi, Y.; Han, W.; Chen, D.; Zhao, J.; Bai, L.; Huang, F.; Dai, Z.; Li, G.; Chen, C.; Zhang, W.; et al. Engineering circular RNA regulators to specifically promote circular RNA production. Theranostics 2021, 11, 7322–7336. [Google Scholar] [CrossRef]
- Yang, R.; Wang, R.C. Research Techniques Made Simple: Studying Circular RNA in Skin Diseases. J. Investig. Dermatol. 2021, 141, 2313–2319.e1. [Google Scholar] [CrossRef] [PubMed]
- Gruhl, F.; Janich, P.; Kaessmann, H.; Gatfield, D. Circular RNA repertoires are associated with evolutionarily young transposable elements. eLife 2021, 10, e67991. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, B. Circular RNA in Diseased Heart. Cells 2020, 9, 1240. [Google Scholar] [CrossRef] [PubMed]
- Kameda, S.; Ohno, H.; Saito, H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 2023, 51, e24. [Google Scholar] [CrossRef]
- Amaya, L.; Grigoryan, L.; Li, Z.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T-cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef]
- Chen, H.H.; Eteleeb, A.; Wang, C.; Fernandez, M.V.; Budde, J.P.; Bergmann, K.; Norton, J.; Wang, F.; Ebl, C.; Morris, J.C.; et al. Circular RNA detection identifies circPSEN1 alterations in the brain specific to autosomal dominant Alzheimer’s disease. Acta Neuropathol. Commun. 2022, 10, 29. [Google Scholar] [CrossRef]
- Glažar, P.; Papavasileiou, P.; Rajewsky, N. circBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef]
- Wu, W.; Ji, P.; Zhao, F. CircAtlas: An integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020, 21, 101. [Google Scholar] [CrossRef]
- Huang, Z.; Shi, J.; Gao, Y.; Cui, C.; Zhang, S.; Li, J.; Zhou, Y.; Cui, Q. HMDD v3.0: A database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019, 47, D1013–D1017. [Google Scholar] [CrossRef]
- Licursi, V.; Conte, F.; Fiscon, G.; Paci, P. MIENTURNET: An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinform. 2019, 20, 545. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Xiao, Y. 3dRNA: 3D Structure Prediction from Linear to Circular RNAs. J. Mol. Biol. 2022, 434, 167452. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, C.V.; Dalenberg, K.; Duarte, J.M.; et al. RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 2022, 31, e4482. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Zarini-Gakiye, E.; Amini, J.; Sanadgol, N.; Vaezi, G.; Parivar, K. Recent Updates in the Alzheimer’s Disease Etiopathology and Possible Treatment Approaches: A Narrative Review of Current Clinical Trials. Curr. Mol. Pharmacol. 2020, 13, 273–294. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, M.; Sanadgol, N.; Abdollahi, M. A Systematic Review of Current Progresses in the Nucleic Acid-Based Therapies for Neurodegeneration with Implications for Alzheimer’s Disease. Mini. Rev. Med. Chem. 2020, 20, 1499–1517. [Google Scholar] [CrossRef] [PubMed]
- Lanoiselée, H.M.; Nicolas, G.; Wallon, D.; Rovelet-Lecrux, A.; Lacour, M.; Rousseau, S.; Richard, A.-C.; Pasquier, F.; Rollin-Sillaire, A.; Martinaud, O.; et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017, 14, e1002270. [Google Scholar] [CrossRef] [PubMed]
- Puri, S.; Hu, J.; Sun, Z.; Lin, M.; Stein, T.D.; Farrer, L.A.; Wolozin, B.; Zhang, X. Identification of circRNAs linked to Alzheimer’s disease and related dementias. Alzheimers Dement. 2023, 19, 3389–3405. [Google Scholar] [CrossRef]
- Razani, E.; Pourbagheri-Sigaroodi, A.; Safaroghli-Azar, A.; Zoghi, A.; Shanaki-Bavarsad, M.; Bashash, D. The PI3K/Akt signaling axis in Alzheimer’s disease: A valuable target to stimulate or suppress? Cell Stress Chaperones 2021, 26, 871–887. [Google Scholar] [CrossRef]
- Zhao, X.; Fang, K.; Liu, X.; Yao, R.; Wang, M.; Li, F.; Hao, S.; He, J.; Wang, Y.; Fan, M.; et al. QSER1 preserves the suppressive status of the pro-apoptotic genes to prevent apoptosis. Cell Death Differ. 2023, 30, 779–793. [Google Scholar] [CrossRef]
- Luo, J.; Zou, H.; Guo, Y.; Huang, K.; Ngan, E.S.; Li, P. BACE2 variant identified from HSCR patient causes AD-like phenotypes in hPSC-derived brain organoids. Cell Death Discov. 2022, 8, 47. [Google Scholar] [CrossRef]
- Matz, A.; Lee, S.J.; Schwedhelm-Domeyer, N.; Zanini, D.; Holubowska, A.; Kannan, M.; Farnworth, M.; Jahn, O.; Göpfert, M.C.; Stegmüller, J. Regulation of neuronal survival and morphology by the E3 ubiquitin ligase RNF157. Cell Death Differ. 2015, 22, 626–642. [Google Scholar] [CrossRef]
- Guan, H.; Mao, L.; Wang, J.; Wang, S.; Yang, S.; Wu, H.; Sun, W.; Chen, Z.; Chen, M. Exosomal RNF157 mRNA from prostate cancer cells contributes to M2 macrophage polarization through destabilizing HDAC1. Front. Oncol. 2022, 12, 1021270. [Google Scholar] [CrossRef]
- Chen, S.; He, R.; Lin, X.; Zhang, W.; Chen, H.; Xu, R.; Kang, M. PTMA binds to HMGB1 to regulate mitochondrial oxidative phosphorylation and thus affect the malignant progression of esophageal squamous cell carcinoma. J. Thorac. Dis. 2023, 15, 1302–1318. [Google Scholar] [CrossRef]
- Puthiyedth, N.; Riveros, C.; Berretta, R.; Moscato, P. Identification of Differentially Expressed Genes through Integrated Study of Alzheimer’s Disease Affected Brain Regions. PLoS ONE 2016, 11, e0152342. [Google Scholar] [CrossRef] [PubMed]
- Helkkula, P.; Hassan, S.; Saarentaus, E.; Vartiainen, E.; Ruotsalainen, S.; Leinonen, J.T.; FinnGen; Palotie, A.; Karjalainen, J.; Kurki, M.; et al. Genome-wide association study of varicose veins identifies a protective missense variant in GJD3 enriched in the Finnish population. Commun. Biol. 2023, 6, 71. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.L.; McPhie, D.L. The cell cycle as a therapeutic target for Alzheimer’s disease. Pharmacol. Ther. 2006, 111, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Hu, Y.; Lan, T.; Guan, K.-L.; Luo, T.; Luo, M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct. Target Ther. 2022, 7, 376. [Google Scholar] [CrossRef]
- Sahu, M.R.; Mondal, A.C. The emerging role of Hippo signaling in neurodegeneration. J. Neurosci. Res. 2020, 98, 796–814. [Google Scholar] [CrossRef]
- Bruno, L.; Karagil, S.; Mahmood, A.; Elbediwy, A.; Stolinski, M.; Mackenzie, F.E. Mechanosensing and the Hippo Pathway in Microglia: A Potential Link to Alzheimer’s Disease Pathogenesis? Cells 2021, 10, 3144. [Google Scholar] [CrossRef]
- Estrada, L.D.; Oliveira-Cruz, L.; Cabrera, D. Transforming Growth Factor Beta Type I Role in Neurodegeneration: Implications for Alzheimer´s Disease. Curr. Protein Pept. Sci. 2018, 19, 1180–1188. [Google Scholar] [CrossRef]
- Zheng, C.; Zhou, X.W.; Wang, J.Z. The dual roles of cytokines in Alzheimer’s disease: Update on interleukins, TNF-α, TGF-β, and IFN-γ. Transl. Neurodegener. 2016, 5, 7. [Google Scholar] [CrossRef]
- Kapoor, A.; Nation, D.A. Role of Notch signaling in neurovascular aging and Alzheimer’s disease. Semin. Cell Dev. Biol. 2021, 116, 90–97. [Google Scholar] [CrossRef]
- Ono, H.; Basson, M.D.; Ito, H. P300 inhibition enhances gemcitabine-induced apoptosis of pancreatic cancer. Oncotarget 2016, 7, 51301–513010. [Google Scholar] [CrossRef]
- Hedges, D.; Janis, R.; Mickelson, S.; Keith, C.; Bennett, D.; Brown, B.L. P300 Amplitude in Alzheimer’s Disease: A Meta-Analysis and Meta-Regression. Clin. EEG Neurosci. 2016, 47, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Parra, M.A.; Ascencio, L.L.; Urquina, H.F.; Manes, F.; Ibáñez, A.M. P300 and neuropsychological assessment in mild cognitive impairment and Alzheimer dementia. Front. Neurol. 2012, 3, 172. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.Y.; Ding, L.; Zhou, T.R.; Yan, T.; Li, J.; Liang, C. FOXA1 in prostate cancer. Asian J. Androl. 2023, 25, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Cong, Y.; Feng, N.; Liang, W.; Wu, Y. Up-regulated microRNA-132 reduces the cognition-damaging effect of sevoflurane on Alzheimer’s disease rats by inhibiting FOXA1. Genomics 2021, 113, 3452–3644. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, Y.; Wang, Y.; Chen, L.; Wang, N.; Su, Y.; Diwu, Y.; Zhang, Q. LncRNA NKILA Exacerbates Alzheimer’s Disease Progression by Regulating the FOXA1-Mediated Transcription of TNFAIP1. Neurochem. Res. 2023, 48, 2895–2910. [Google Scholar] [CrossRef]
- Kim, I.B.; Park, S.C. The Entorhinal Cortex and Adult Neurogenesis in Major Depression. Int. J. Mol. Sci. 2021, 22, 11725. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanadgol, N.; Amini, J.; Beyer, C.; Zendedel, A. Presenilin-1-Derived Circular RNAs: Neglected Epigenetic Regulators with Various Functions in Alzheimer’s Disease. Biomolecules 2023, 13, 1401. https://doi.org/10.3390/biom13091401
Sanadgol N, Amini J, Beyer C, Zendedel A. Presenilin-1-Derived Circular RNAs: Neglected Epigenetic Regulators with Various Functions in Alzheimer’s Disease. Biomolecules. 2023; 13(9):1401. https://doi.org/10.3390/biom13091401
Chicago/Turabian StyleSanadgol, Nima, Javad Amini, Cordian Beyer, and Adib Zendedel. 2023. "Presenilin-1-Derived Circular RNAs: Neglected Epigenetic Regulators with Various Functions in Alzheimer’s Disease" Biomolecules 13, no. 9: 1401. https://doi.org/10.3390/biom13091401
APA StyleSanadgol, N., Amini, J., Beyer, C., & Zendedel, A. (2023). Presenilin-1-Derived Circular RNAs: Neglected Epigenetic Regulators with Various Functions in Alzheimer’s Disease. Biomolecules, 13(9), 1401. https://doi.org/10.3390/biom13091401









