Differential Expression of Circulating Damage-Associated Molecular Patterns in Patients with Coronary Artery Ectasia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Angiographic Evaluation
2.3. Laboratory Methods
2.4. Cell Culture
2.5. Luciferase Assay
2.6. RT-qPCR for mRNA and miR Expression
2.7. Statistical Analysis
3. Results
3.1. Baseline Clinical and Laboratory Characteristics
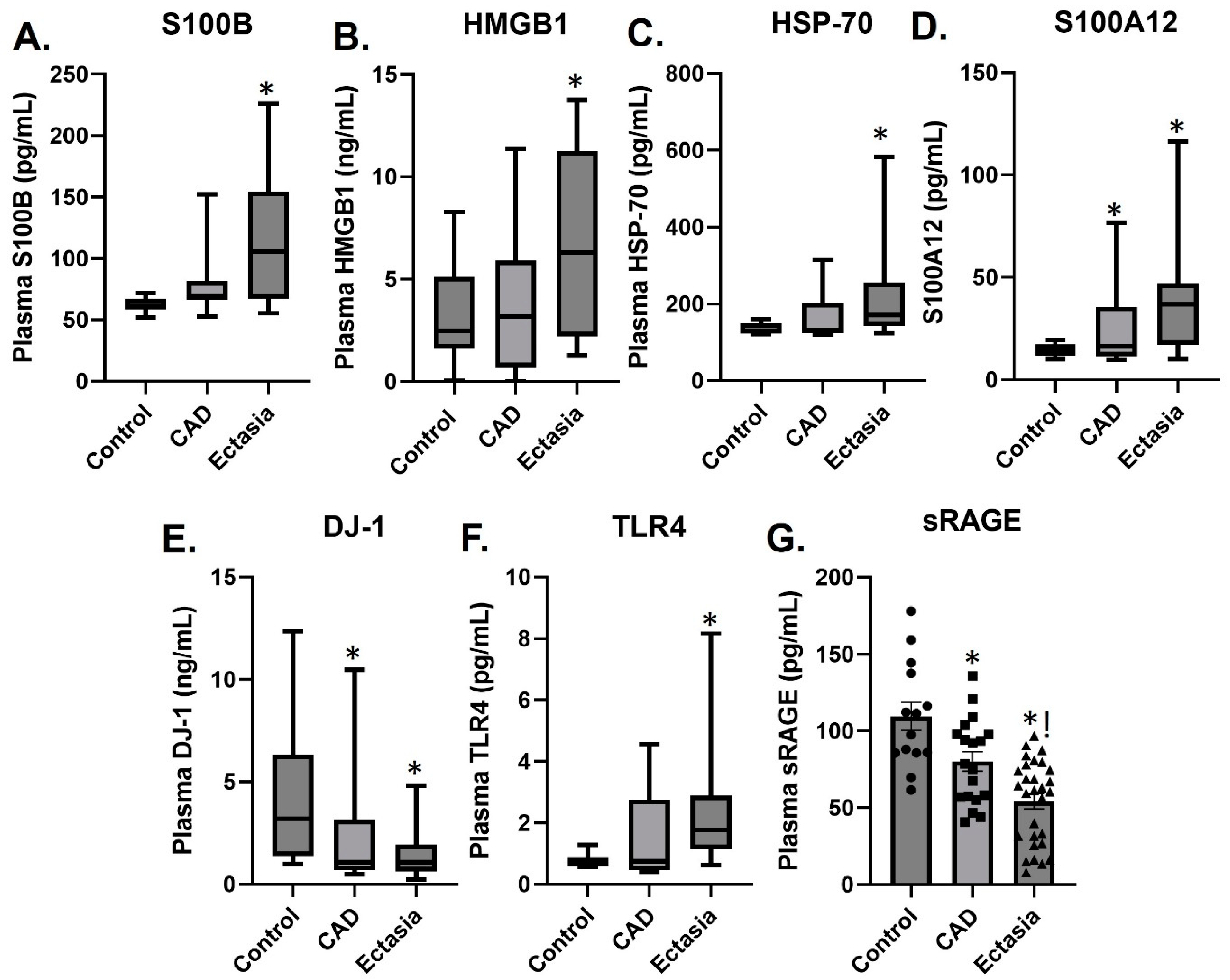
3.2. Plasma Levels of sRAGE, S100A12, S100B, HMGB1, TLR4, HSP70 and DJ-1
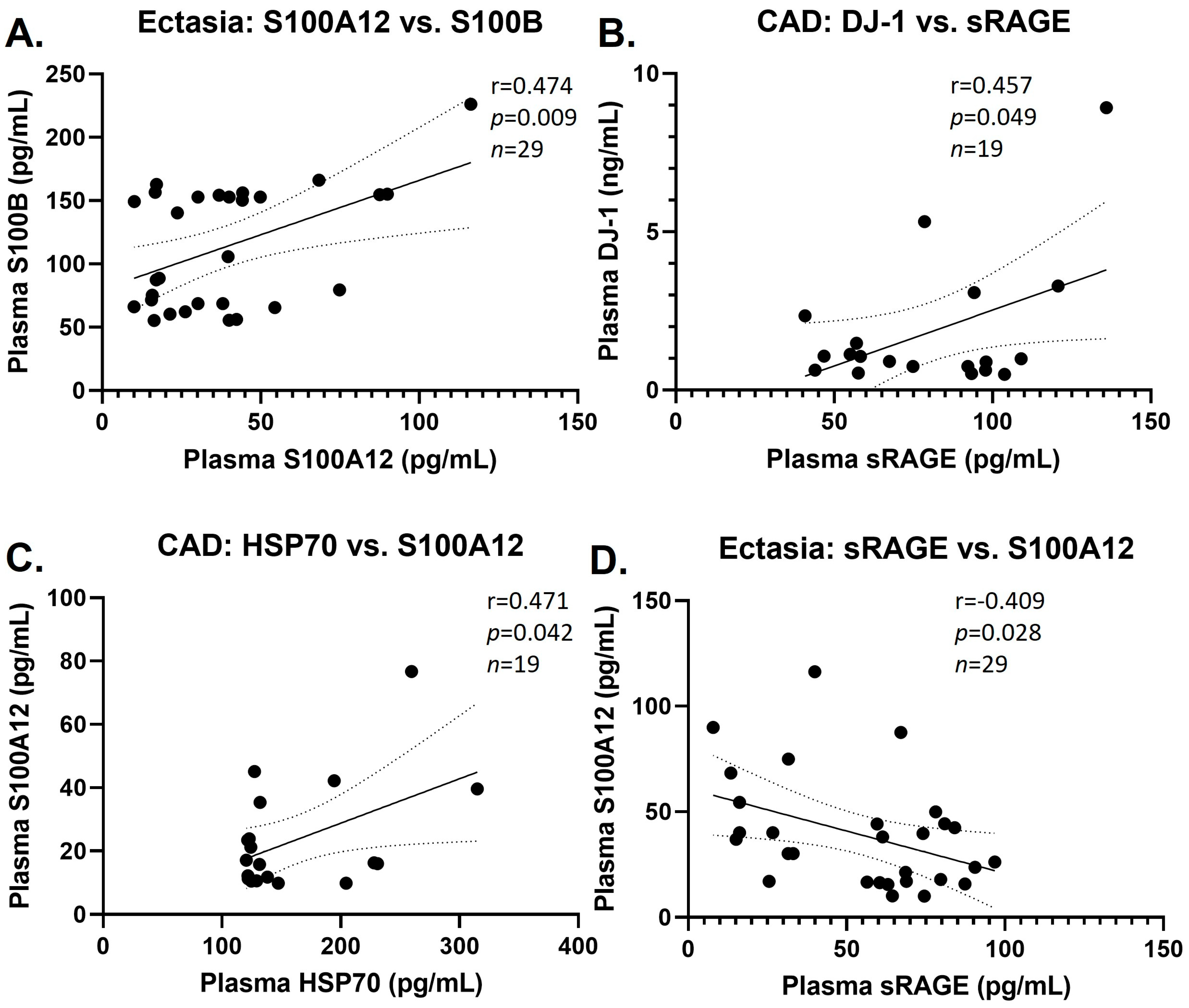
3.3. Group Correlations of DAMPs
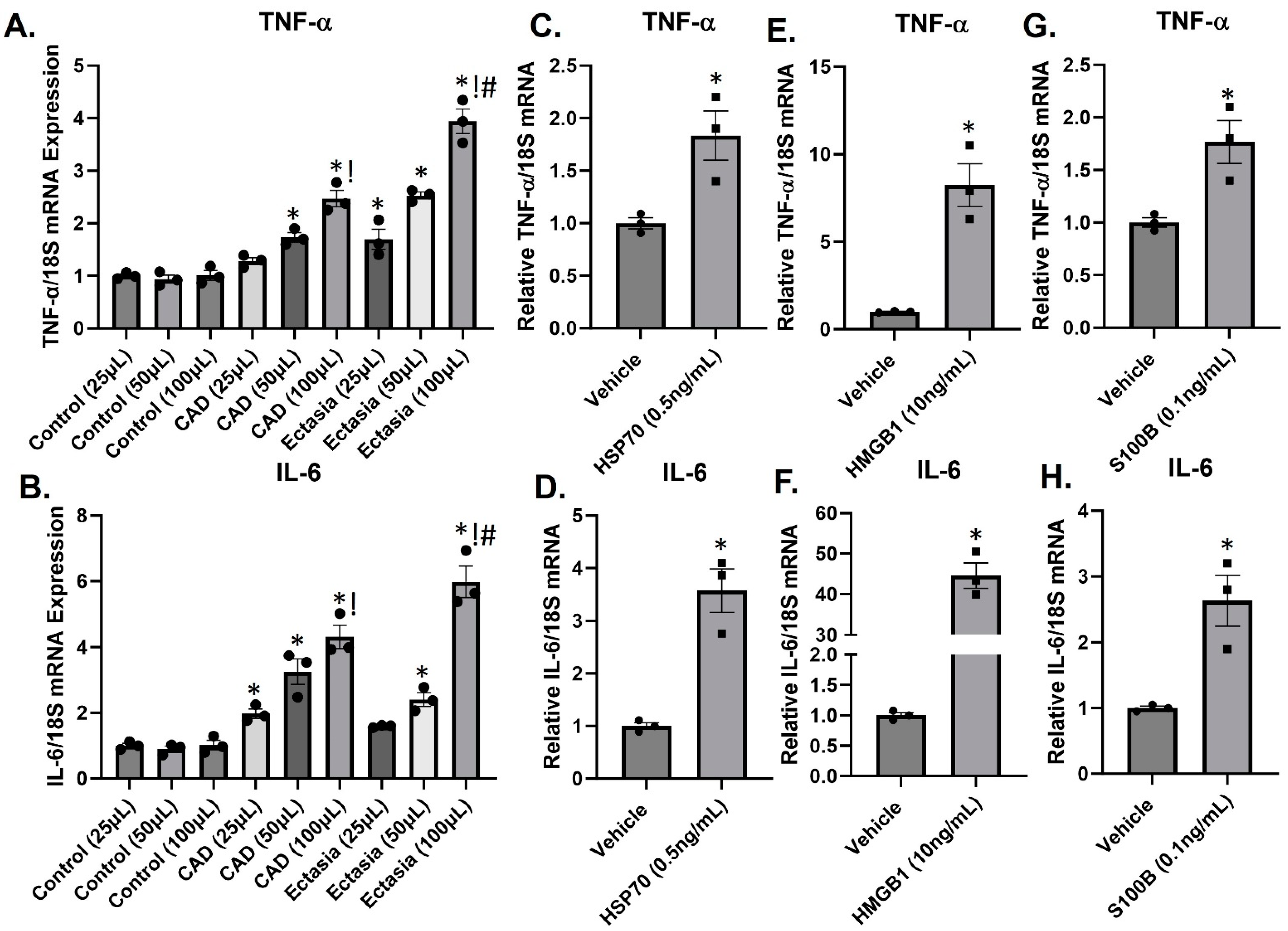
3.4. DAMPs Induce Inflammation
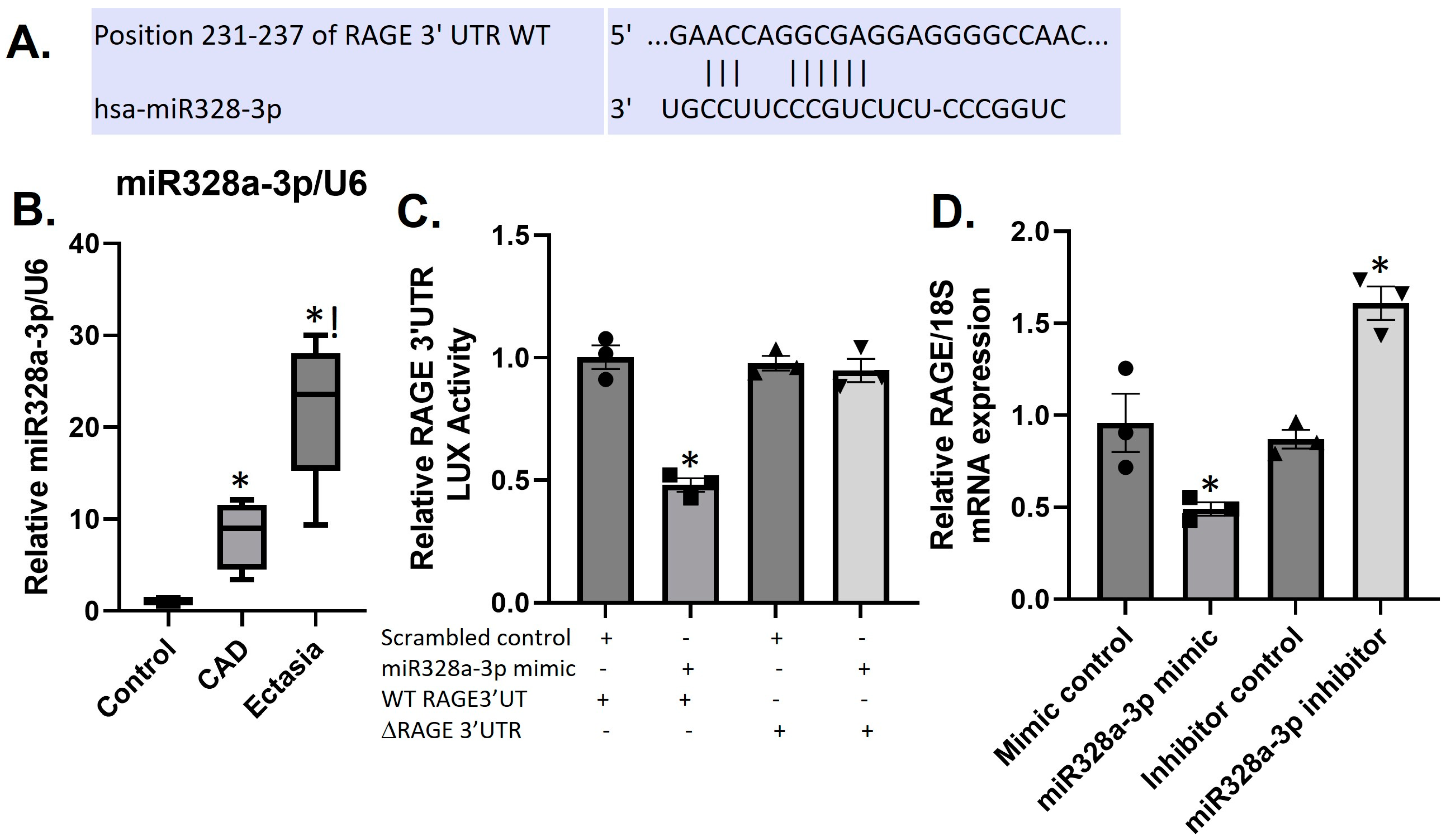
3.5. Differential Expression of sRAGE and miR328a-3p in CAE and CAD
3.6. MiR328a-3p Targets the RAGE-3′UTR in HUVECS
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Triantafyllis, A.S.; Kalogeropoulos, A.S.; Rigopoulos, A.G.; Sakadakis, E.A.; Toumpoulis, I.K.; Tsikrikas, S.; Kremastinos, D.T.; Rizos, I. Coronary artery ectasia and inflammatory cytokines: Link with a predominant Th-2 immune response? Cytokine 2013, 64, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Sorrell, V.L.; Davis, M.J.; Bove, A.A. Current knowledge and significance of coronary artery ectasia: A chronologic review of the literature, recommendations for treatment, possible etiologies, and future considerations. Clin. Cardiol. 1998, 21, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Swanton, R.H.; Thomas, M.L.; Coltart, D.J.; Jenkins, B.S.; Webb-Peploe, M.M.; Williams, B.T. Coronary artery ectasia–a variant of occlusive coronary arteriosclerosis. Br. Heart J. 1978, 40, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ramkumar, S.; Khav, N.; Dundon, B.K. Coronary artery ectasia presenting with ST-elevation myocardial infarction in a young indigenous man: A case report. Eur. Heart J. Case Rep. 2020, 4, 1–5. [Google Scholar] [CrossRef]
- Swaye, P.S.; Fisher, L.D.; Litwin, P.; Vignola, P.A.; Judkins, M.P.; Kemp, H.G.; Mudd, J.G.; Gosselin, A.J. Aneurysmal coronary artery disease. Circulation 1983, 67, 134–138. [Google Scholar] [CrossRef]
- Markis, J.E.; Joffe, C.; Cohn, P.F.; Feen, D.J.; Herman, M.V.; Gorlin, R. Clinical significance of coronary arterial ectasia. Am. J. Cardiol. 1976, 37, 217–222. [Google Scholar] [CrossRef]
- Boles, U.; Johansson, A.; Wiklund, U.; Sharif, Z.; David, S.; McGrory, S.; Henein, M.Y. Cytokine disturbances in coronary artery ectasia Do Not Support Atherosclerosis Pathogenesis. Int. J. Mol. Sci. 2018, 19, 260. [Google Scholar] [CrossRef]
- Ozturk, S.; Yetkin, E.; Waltenberger, J. Molecular and cellular insights into the pathogenesis of coronary artery ectasia. Cardiovasc. Pathol. 2018, 35, 37–47. [Google Scholar] [CrossRef]
- Tokgozoglu, L.; Ergene, O.; Kinay, O.; Nazli, C.; Hascelik, G.; Hoscan, Y. Plasma interleukin-6 levels are increased in coronary artery ectasia. Acta Cardiol. 2004, 59, 515–519. [Google Scholar] [CrossRef]
- Turhan, H.; Yetkin, E. Coronary artery ectasia: Is it a destructive inflammatory lesion of the vascular wall? Int. J. Cardiol. 2007, 118, 241. [Google Scholar] [CrossRef]
- Soto, M.E.; Reyes-Villatoro, M.A.; Márquez, R.; Cardoso, G.; Posadas-Sánchez, R.; Juárez-Orozco, L.E. Evaluation and analysis of plasma soluble adhesion molecules in patients with coronary ectasia and atherosclerotic coronary artery disease. Arch. Med. Res. 2014, 45, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Rigopoulos, A.G.; Kalogeropoulos, A.S.; Tsoporis, J.N.; Sakadakis, E.A.; Triantafyllis, A.S.; Noutsias, M.; Gupta, S.; Parker, T.G.; Rizos, I. Heat shock protein 70 is associated with cardioversion outcome and recurrence of symptomatic recent onset atrial fibrillation in hypertensive patients. Cardiovasc. Pharmacol. 2021, 77, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Roth, J.; Oppenheim, J.J.; Tracey, K.J.; Vogl, T.; Feldmann, M.; Horwood, N.; Nanchahal, J. Alarmins: Awaiting a clinical response. J. Clin. Investig. 2012, 122, 2711–2719. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.S.; Sohn, D.H. Damage-associated molecular patterns in inflammatory diseases. Immune Netw. 2018, 18, e27. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ali, S.A. Multifunctional role of S100 protein family in the immune system: An update. Cells 2022, 11, 2274. [Google Scholar] [CrossRef]
- Ismahil, M.A.; Hamid, T.; Bansal, S.S.; Patel, B.; Kingery, J.R.; Prabhu, S.D. Remodeling of the mononuclear phagocyte network underlies chronic inflammation and disease progression in heart failure: Critical importance of the cardiosplenic axis. Circ. Res. 2014, 114, 266–282. [Google Scholar] [CrossRef]
- Tsirebolos, G.; Tsoporis, J.N.; Drosatos, I.-A.; Izhar, S.; Gkavogiannakis, N.; Sakadakis, E.; Triantafyllis, A.S.; Parker, T.G.; Rallidis, L.S.; Rizos, I. Emerging markers of inflammation and oxidative stress as potential predictors of coronary artery disease. Int. J. Cardiol. 2023, 376, 127–133. [Google Scholar] [CrossRef]
- Yetkin, E.; Waltenberger, J. Novel insights into an old controversy: Is coronary artery ectasia a variant of coronary atherosclerosis? Clin. Res. Cardiol Off. J. Ger. Card. Soc. 2007, 96, 331–339. [Google Scholar] [CrossRef]
- Li, J.J.; Nie, S.P.; Qian, X.W.; Zeng, H.S.; Zhang, C.Y. Chronic inflammatory status in patients with coronary artery ectasia. Cytokine 2009, 46, 61–64. [Google Scholar] [CrossRef]
- Davies, M.J. Aortic aneurysm formation: Lessons from human studies and experimental models. Circulation 1998, 98, 193–195. [Google Scholar] [CrossRef]
- Aydin, M.; Tekin, I.O.; Dogan, S.M.; Yildirim, N.; Arasli, M.; Sayin, M.R.; Aktop, Z. The levels of tumor necrosis factor-alpha and interleukin-6 in patients with isolated coronary artery ectasia. Mediat. Inflamm. 2009, 2009, 106145. [Google Scholar] [CrossRef] [PubMed]
- Prasad, K. AGE-RAGE stress plays a role in aortic aneurysm: A comprehensive review and novel potential therapeutic target. Rev. Cardiovasc. Med. 2019, 20, 201–208. [Google Scholar] [PubMed]
- Bangert, A.; Andrassy, M.; Müller, A.M. Critical role of RAGE and HMGB1 in inflammatory heart disease. Proc. Natl. Acad. Sci. USA 2016, 113, E155–E164. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Gong, F.; Zhang, Q.; Xie, C.; Wang, W.; Fu, S. Reverse regulation of soluble receptor for advanced glycation end products and proinflammatory factor resistin and S100A12 in Kawasaki disease. Arthritis Res. Ther. 2012, 14, R251. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Lv, P.; Zhao, X.; Wang, X.; Ma, X.; Meng, W.; Meng, X.; Dong, S. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol. Cell Biochem. 2014, 394, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, N.; Zhang, Y.; Ran, Y.; Pu, J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern. Med. 2011, 50, 1789–1795. [Google Scholar] [CrossRef]
- Iwańczyk, S.; Lehmann, T.; Cieślewicz, A.; Malesza, K.; Woźniak, P.; Hertel, A.; Krupka, G.; Jagodziński, P.P.; Grygier, M.; Lesiak, M.; et al. Circulating miRNA-451a and miRNA-328-3p as Potential Markers of Coronary Artery Aneurysmal Disease. Int. J. Mol. Sci. 2023, 24, 5817. [Google Scholar] [CrossRef]
- Kahle, P.J.; Oh, S.E.; Mouradian, M.M. Cytoprotective mechanisms of DJ-1 against oxidative stress through modulating ERK1/2 and ASK1 signal transduction. Redox Biol. 2018, 14, 211–217. [Google Scholar]
- Neves, M.; Grãos, M.; Anjo, S.I.; Manadas, B. Modulation of signaling pathways by DJ-1: An updated overview. Redox Biol. 2022, 51, 102283. [Google Scholar] [CrossRef] [PubMed]
- Tsoporis, J.N.; Drosatos, I.-A.; Gupta, S.; Amatullah, H.; Izhar, S.; dos Santos, C.C.; Salpeas, V.; Rigopoulos, A.G.; Toumpoulis, I.K.; Triantafyllis, A.S.; et al. Cytoprotective Mechanisms of DJ-1: Implications in Cardiac Pathophysiology. Molecules 2021, 26, 3795. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Sakai, S.; Tsuyama, J.; Nakamura, A.; Otani, K.; Kurabayashi, K.; Yogiashi, Y.; Masai, H.; Shichita, T. Extracellular DJ-1 induces sterile inflammation in the ischemic brain. PLoS Biol. 2021, 19, e3000939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, X.; Jiang, G.; Yu, Y.; Wu, J.; Su, Y.; Sun, A.; Zou, Y.; Jiang, H.; Ge, J. HMGB1 enhances mechanical stress-induced cardiomyocyte hypertrophy in vitro via the RAGE/ERK1/2 signaling pathway. Int. J. Mol. Med. 2019, 44, 885–892. [Google Scholar] [CrossRef]
- Qian, B.; Huang, H.; Cheng, M.; Qin, T.; Chen, T.; Zhao, J. Mechanism of HMGB1-RAGE in Kawasaki disease with coronary artery injury. Eur. J. Med. Res. 2020, 25, 8. [Google Scholar] [CrossRef]
- de Souza, A.; Westra, J.; Limburg, P.; Bijl, M.; Kallenberg, C. HMGB1 in vascular diseases: Its role in vascular inflammation and atherosclerosis. Autoimmun. Rev. 2012, 11, 909–917. [Google Scholar] [CrossRef]
- Sharma, A.K.; Salmon, M.D.; Lu, G.; Su, G.; Pope, N.H.; Smith, J.R.; Weiss, M.L.; Upchurch, G.R., Jr. Mesenchymal Stem Cells Attenuate NADPH Oxidase-Dependent High Mobility Group Box 1 Production and Inhibit Abdominal Aortic Aneurysms. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 908–918. [Google Scholar] [CrossRef]
- Xia, C.; Braunstein, Z.; Toomey, A.C.; Zhong, J.; Rao, X. S100 Proteins as an Important Regulator of Macrophage Inflammation. Front. Immunol. 2017, 8, 1908. [Google Scholar] [CrossRef]
- Averill, M.M.; Kerkhoff, C.; Bornfeldt, K.E. S100A8 and S100A9 in cardiovascular biology and disease. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 223–229. [Google Scholar] [CrossRef]
- Goyette, J.; Yan, W.X.; Yamen, E.; Chung, Y.M.; Lim, S.Y.; Hsu, K.; Rahimi, F.; Di Girolamo, N.; Song, C.; Jessup, W.; et al. Pleiotropic roles of S100A12 in coronary atherosclerotic plaque formation and rupture. J. Immunol. 2009, 183, 593–603. [Google Scholar] [CrossRef]
- Tsoporis, J.N.; Marks, A.; Kahn, H.J.; Butany, J.W.; Liu, P.P.; O’Hanlon, D.; Parker, T.G. Inhibition of norepinephrine-induced cardiac hypertrophy in s100beta transgenic mice. J. Clin. Investig. 1998, 102, 1609–1616. [Google Scholar] [CrossRef] [PubMed]
- Tsoporis, J.N.; Izhar, S.; Leong-Poi, H.; Desjardins, J.F.; Huttunen, H.J.; Parker, T.G. S100B interaction with the receptor for advanced glycation end products (RAGE): A novel receptor-mediated mechanism for myocyte apoptosis postinfarction. Circ. Res. 2010, 106, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Zilaee, M.; Ferns, G.A.; Ghayour-Mobarhan, M. Heat shock proteins and cardiovascular disease. Adv. Clin. Chem. 2014, 64, 73–115. [Google Scholar] [PubMed]
- Zhu, J.; Quyyumi, A.A.; Wu, H.; Csako, G.; Rott, D.; Zalles-Ganley, A.; Ogunmakinwa, J.; Halcox, J.; Epstein, S.E. Increased serum levels of heat shock protein 70 are associated with low risk of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1055–1059. [Google Scholar] [CrossRef]
- Ren, B.; Zou, G.; Huang, Y.; Xu, G.; Xu, F.; He, J.; Zhu, H.; Yu, P. Serum levels of HSP70 and other DAMP proteins can aid in patient diagnosis after traumatic injury. Cell Stress. Chaperones 2016, 21, 677–686. [Google Scholar] [CrossRef]
| General Demographics * | Group A (n = 29) | Group B (n = 19) | Group C (n = 14) | p Value |
|---|---|---|---|---|
| Age ± SD (years) | 62.35 ± 10.34 | 66.42 ± 9.06 | 61.07 ± 10.19 | 0.25 |
| Gender (males/females) | 23/6 | 14/5 | 6/8 | 0.06 |
| BMI ± SD (kg/m2) | 29.14 ± 2.66 | 30.28 ± 3.84 | 28.01 ± 3.93 | 0.24 |
| Hypertension (%) | 23 (79.3) | 17 (89.5) | 8 (61.5) | 0.16 |
| Hyperlipidemia (%) | 22 (75.9) | 14 (73.68) | 7 (53.8) | 0.33 |
| Diabetes (%) | 8 (28.6) | 11 (57.9) | 2 (28.6) | 0.03 * |
| Smoking (%) | 18 (62.1) | 5 (26.3) | 5 (38.5) | 0.04 * |
| LVEF (%), median (IQR) | 65 (60.0–70.0) | 61.0 (60.0–65.0) | 65.0 (60.0–70.0) | 0.42 |
| SBP ± SD (mmHg) | 129.77 ± 15.69 | 129.14 ± 18.11 | 130.00 ± 13.69 | 0.99 |
| DBP ± SD (mmHg) | 70.91 ± 11.41 | 73.50 ± 12.09 | 77.22 ± 12.78 | 0.41 |
| Medications | ||||
| Aspirin (%) | 20 (71.4) | 10 (52.6) | 9 (75.0) | 0.32 |
| ACEIs (%) | 15 (46.4) | 8 (42.1) | 6 (50.0) | 0.91 |
| ARBs (%) | 7 (25.0) | 6 (31.6) | 3 (0.25) | 0.87 |
| Statins (%) | 18 (64.3) | 10 (52.6) | 3 (25.0) | 0.07 |
| CCBs (%) | 5 (17.9) | 7 (36.8) | 5 (41.7) | 0.20 |
| Beta blockers (%) | 15 (53.6) | 11 (57.9) | 4 (33.3) | 0.28 |
| Nitrates (%) | 8 (28.6) | 3 (15.8) | 4 (33.3) | 0.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsoporis, J.N.; Triantafyllis, A.S.; Kalogeropoulos, A.S.; Izhar, S.; Rigopoulos, A.G.; Rallidis, L.S.; Sakadakis, E.; Toumpoulis, I.K.; Salpeas, V.; Leong-Poi, H.; et al. Differential Expression of Circulating Damage-Associated Molecular Patterns in Patients with Coronary Artery Ectasia. Biomolecules 2024, 14, 10. https://doi.org/10.3390/biom14010010
Tsoporis JN, Triantafyllis AS, Kalogeropoulos AS, Izhar S, Rigopoulos AG, Rallidis LS, Sakadakis E, Toumpoulis IK, Salpeas V, Leong-Poi H, et al. Differential Expression of Circulating Damage-Associated Molecular Patterns in Patients with Coronary Artery Ectasia. Biomolecules. 2024; 14(1):10. https://doi.org/10.3390/biom14010010
Chicago/Turabian StyleTsoporis, James N., Andreas S. Triantafyllis, Andreas S. Kalogeropoulos, Shehla Izhar, Angelos G. Rigopoulos, Loukianos S. Rallidis, Eleftherios Sakadakis, Ioannis K. Toumpoulis, Vasileios Salpeas, Howard Leong-Poi, and et al. 2024. "Differential Expression of Circulating Damage-Associated Molecular Patterns in Patients with Coronary Artery Ectasia" Biomolecules 14, no. 1: 10. https://doi.org/10.3390/biom14010010
APA StyleTsoporis, J. N., Triantafyllis, A. S., Kalogeropoulos, A. S., Izhar, S., Rigopoulos, A. G., Rallidis, L. S., Sakadakis, E., Toumpoulis, I. K., Salpeas, V., Leong-Poi, H., Parker, T. G., & Rizos, I. (2024). Differential Expression of Circulating Damage-Associated Molecular Patterns in Patients with Coronary Artery Ectasia. Biomolecules, 14(1), 10. https://doi.org/10.3390/biom14010010





